Increasing evidence has shown that heterotrophic microeukaryotes are an important component in global marine ecosystems, while their diversity and ecological functions remain largely unknown. Without appropriately incorporating these organisms into the food web models, our current understanding of marine microbial community ecology is incomplete, which may further hamper broader studies of biogeochemistry and climate change. This study focuses on a major group of unicellular fungus-like protists (Labyrinthulomycetes) and reveals their distinct annual community patterns relative to fungi and bacteria. Results of our observations provide new information on the community structure and ecology of this protistan group and shed light on the intricate ecological roles of unicellular heterotrophic eukaryotes in the coastal oceans.
KEYWORDS: fungus-like protist, coastal ocean, time series, community structure, niche partitioning, microbial interaction
ABSTRACT
Heterotrophic microbes play a key role in remineralizing organic material in the coastal ocean. While there is a significant body of literature examining heterotrophic bacterioplankton and phytoplankton communities, much less is known about the diversity, dynamics, and ecology of eukaryotic heterotrophs. Here, we focus on the Labyrinthulomycetes, a fungus-like protistan group whose biomass can exceed that of the bacterioplankton in coastal waters. We examined their diversity and community structure in a weekly temperate coastal ocean time series. Their seasonal community patterns were related to temperature, insolation, dissolved inorganic carbon, fungal abundance, ammonia, chlorophyll a, pH, and other environmental variables. Similar to the bacterioplankton, annual community patterns of the Labyrinthulomycetes were dominated by a few persistent taxa with summer or winter preferences. However, like the patterns of fungi at this site, the majority of the Labyrinthulomycetes phylotypes occurred mostly as short, reoccurring, season-specific blooms. Furthermore, some specific phylotypes of Labyrinthulomycetes displayed time-lagged correlations or cooccurrences with bacterial, algal, or fungal phylotypes, suggesting their potentially multifaceted involvement in the marine food webs. Overall, this study reports niche partitioning between closely related Labyrinthulomycetes and identifies distinct ecotypes and temporal patterns compared to bacterioplankton and fungi.
IMPORTANCE Increasing evidence has shown that heterotrophic microeukaryotes are an important component in global marine ecosystems, while their diversity and ecological functions remain largely unknown. Without appropriately incorporating these organisms into the food web models, our current understanding of marine microbial community ecology is incomplete, which may further hamper broader studies of biogeochemistry and climate change. This study focuses on a major group of unicellular fungus-like protists (Labyrinthulomycetes) and reveals their distinct annual community patterns relative to fungi and bacteria. Results of our observations provide new information on the community structure and ecology of this protistan group and shed light on the intricate ecological roles of unicellular heterotrophic eukaryotes in the coastal oceans.
INTRODUCTION
Planktonic heterotrophic microbes dominate secondary production and biogeochemical processes in the world’s oceans (1, 2). Until recently, most efforts to understand microbial ecology and biogeochemical cycling of organic material have focused on bacterioplankton, archaea, and algae (3–7). Although a high biomass of heterotrophic microbial eukaryotes (e.g., heterotrophic protists and fungi) has long been known to exist in the oceans, our understanding of their ecological and biogeochemical functions remains limited in coastal ecosystems, especially in comparison with eukaryotic photoautotrophs (8–14).
One important group of heterotrophic eukaryotes, the fungus-like protists, also known as Labyrinthulomycetes, are ubiquitous unicellular protists found in the global ocean (9, 15), where their biomass has been reported to approach and occasionally surpass that of bacterioplankton (16–20). Beyond their significant contribution to secondary production, they play important ecological roles in the degradation of organic matter and in provisioning higher trophic levels with ω-3 polyunsaturated fatty acids (21, 22). Although some members have been observed to live as symbionts, parasites, bacterivores, or diatom predators, most of the Labyrinthulomycetes are presumed to be saprophytes, feeding on both terrigenous and autochthonous dissolved and particulate organic matter (23–27). This protistan group has been proposed to serve as a “leftover scavenger,” metabolizing the organic material remaining following bacterial metabolism (16, 28, 29). Nevertheless, limited previous investigations mean that the diversity and ecological role of the Labyrinthulomycetes protists remain poorly understood (15).
However, recent studies in the coastal waters of Japan and China have demonstrated that the Labyrinthulomycetes communities can change dramatically across seasons and habitats (16, 17, 21, 30). Due to the limitations of these studies’ culture-based methods or the observational scales (e.g., limited sampling period or temporal resolution), we have not fully captured their diversity or identified their ecological drivers. Nevertheless, previous studies have revealed that patterns of Labyrinthulomycetes protists are associated with a number of physical, chemical, and biological variables in the coastal ocean, including temperature, salinity, pH, dissolved and particulate organic matter, inorganic nutrients, phytoplankton, bacterioplankton, and viruses (16–18, 20, 21, 30–38). Inconsistencies in reported key drivers for this clade suggest that, like other microbial groups, closely related taxa within this heterotrophic protistan group may exhibit ecological partitioning (14, 39–41).
High-resolution time series can provide insight into microbial responses to potential environmental drivers (11, 12, 14, 39, 42, 43). Moreover, comparisons among taxa can identify differences in their ecologies as well as potential associations between taxa. Previous studies have revealed strong summer and winter associations in the bacterioplankton (14), while some eukaryotes, including fungi, occur as episodic, ephemeral blooms (42, 43). Labyrinthulomycetes and fungi have been considered ecologically similar and often cooccur in particulate habitats containing ample organic resources (e.g., marine snow, mangrove detritus), but few community-level observations have been made for either group in coastal waters, leaving open a number of questions about their ecological similarities and differences (9, 44). Further, Labyrinthulomycetes’ relationship with heterotrophic bacterioplankton and phytoplankton remains elusive. Given the foregone diversity within marine microbiomes, we hypothesize that these groups have complex associations and interactions.
To better characterize Labyrinthulomycetes diversity and dynamics and understand their ecology, we employed time-series observations to identify potential environmental drivers and compare their distribution patterns with those of other microbes. In this study, we examined 3 years of weekly observations at the temperate, mesotrophic Piver’s Island Coastal Observatory (PICO) near the Beaufort Inlet of North Carolina, USA, to identify patterns in the diversity and dynamics of Labyrinthulomycetes.
RESULTS AND DISCUSSION
Temporal dynamics of Labyrinthulomycetes abundance and α-diversity measures.
The total abundance of the Labyrinthulomycetes 18S rRNA gene for representative winter (mid-January and mid-February, the coldest weeks of the year) and summer (mid-July and mid-August, the warmest weeks of the year) was estimated to be 3.74 × 105 to 1.39 × 106 copies/liter (see Fig. S1 in the supplemental material), which is comparable to the abundance in the Pearl River Delta in southern China (30), where their biomass has been repeatedly reported to constitute a significant fraction of the heterotrophic microbial community (17, 45). Unlike for bacterioplankton, their gene abundances in winter and summer were not significantly different (analysis of variance [ANOVA], P > 0.05, n = 12).
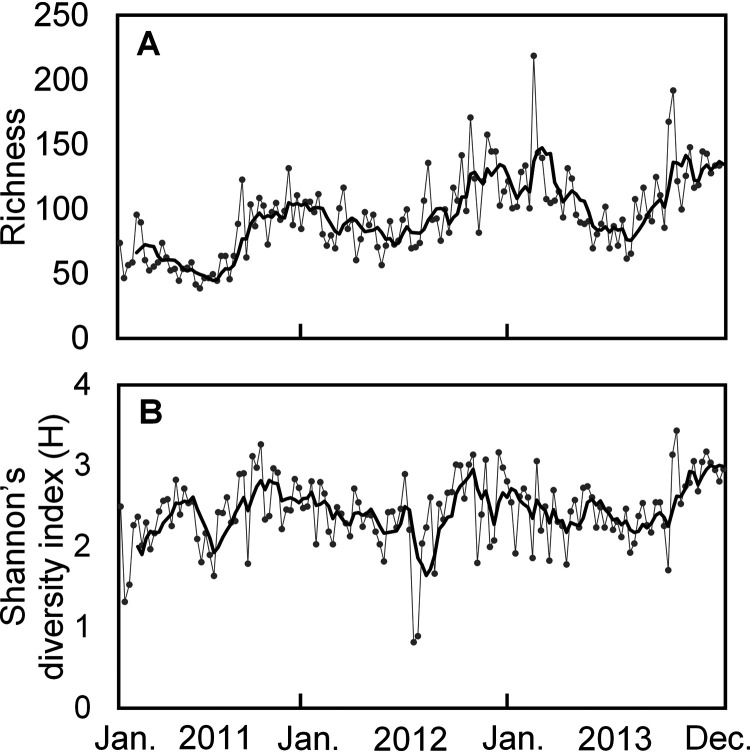
Across the time series, the 18S rRNA gene amplicon sequence variant (ASV) richness of the Labyrinthulomycetes protists was lower than the operational taxonomic unit (OTU) richness of either the bacterioplankton or the fungi and exhibited a reoccurring peak in early winter (Fig. 1, Fig. S2). The community diversity of Labyrinthulomycetes is correlated with a number of seasonally associated environmental factors (e.g., insolation and temperature; Table S1). While the fungi also exhibited a winter peak in diversity (richness, evenness, and Shannon’s diversity) (12), the Labyrinthulomycetes showed weaker seasonality and a peak preceding that of the fungi (Fig. S2). These patterns also sharply contrast with the bacterioplankton diversity, which peaks in spring and fall (14). Compared to the bacterioplankton and fungi, the Labyrinthulomycetes showed significantly lower (ANOVA, P < 0.001) and less-seasonal evenness (Fig. S2). These results suggest that the fungus-like Labyrinthulomycetes exhibit distinct responses to environmental factors compared to either “true fungi” or bacteria.
FIG 1.
(A and B) The richness (A) and Shannon’s diversity index (B) of the Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) across the weekly time series at the Piver’s Island Coastal Observatory (PICO) from 2011 to 2013, with 5-week moving average curves indicated by bold lines.
Seasonal community-level patterns of Labyrinthulomycetes.
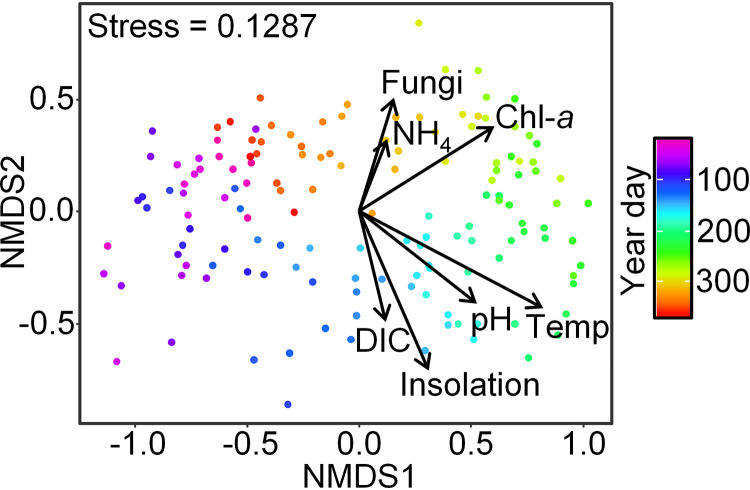
Nonmetric multidimensional scaling (NMDS) analyses revealed a seasonal pattern for the Labyrinthulomycetes that is related to a number of environmental factors, including temperature, insolation, dissolved inorganic carbon, fungal abundance, ammonia, chlorophyll a, and pH (Fig. 2) (permutation tests, P < 0.01). As with the coastal waters of northern China and Japan (16, 21), temperature was identified here as a major factor associated with community structure (Fig. 2), which also aligns with the importance of temperature for both bacterioplankton and fungal communities at this site (12, 14). While the ∼20°C annual temperature cycle in this temperate coastal ocean environment is important across diverse microbial lineages (46–50), it is difficult to disentangle highly correlated seasonal factors (e.g., light and temperature) or indirect effects of temperature mediated by interactions with other temperature-sensitive organisms. Therefore, we can interpret statistically significant relationships between their community composition and fungal abundance, chlorophyll a, insolation, dissolved inorganic carbon, and ammonia (Fig. 2) as either potential drivers or as proxies for unmeasured factors. Previously, Labyrinthulomycetes were observed to increase with high phytoplankton biomass (16, 18, 35, 37), potentially due to the ability to either degrade phytoplankton detritus (51–54) or associate with phytoplankton as parasites or symbionts (23, 55). However, coastal and estuarine Labyrinthulomycetes abundance has also been linked to terrigenous particulate organic matter (21, 33), suggesting the potential for diverse substrate utilization within this class. Additionally, the significant relationship between Labyrinthulomycetes composition and fungal abundance suggests that some Labyrinthulomycetes cooccur with fungi, as previously observed in particulate-associated habitats (9, 44). In order to better understand different ecological roles within closely related members of Labyrinthulomycetes, we examined the temporal patterns of individual ASVs.
FIG 2.
The nonmetric multidimensional scaling (NMDS) biplot of Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) across the weekly time series at the Piver’s Island Coastal Observatory (PICO) from 2011 to 2013. Each circle represents the Labyrinthulomycetes composition at a specific time point, colored by year-day. Vectors indicate environmental factors that were related (permutation tests, P < 0.01) to the community composition. DIC, dissolved inorganic carbon; Fungi, fungal ITS abundance; Chl-a, chlorophyll a; Temp, water temperature.
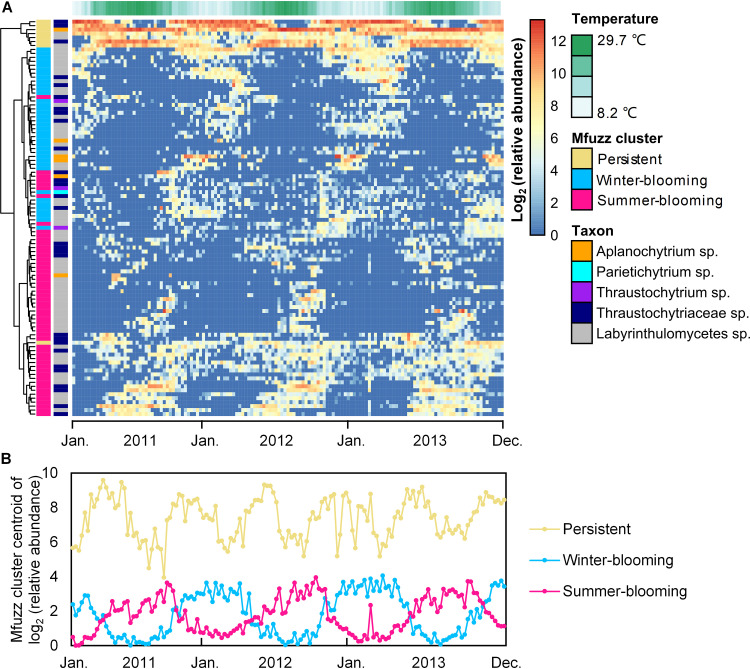
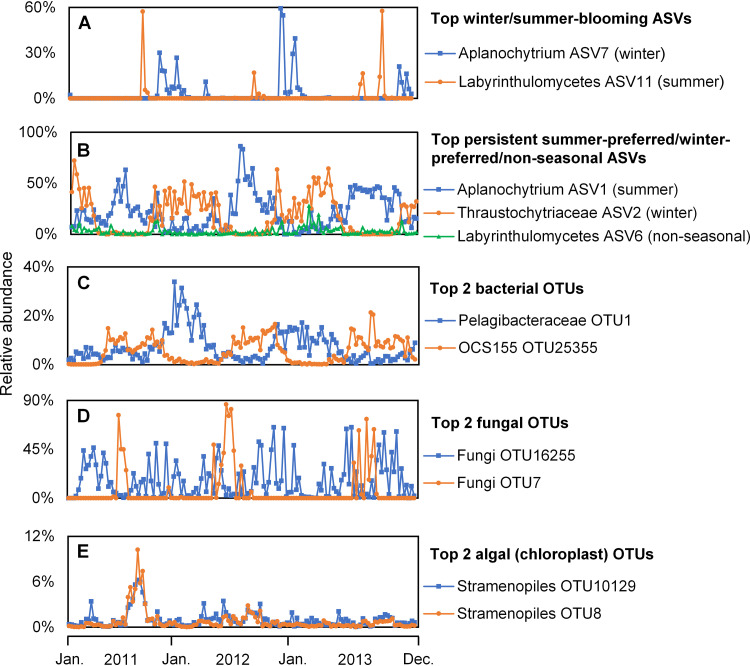
Of the 1,629 ASVs, the 100 most abundant ASVs accounted for 93.11% of the total sequences (Fig. S3); thus, the ASV patterns in Fig. 3 are representative of the community. Soft clustering of 3 years of weekly samples revealed that the ASVs group into summer-blooming, winter-blooming, and persistent (which also exhibit seasonality) patterns, which consist of 54, 38, and 8 ASVs, respectively (Fig. 3). The summer- and winter-blooming ASVs reoccurred annually as transient blooms within a preferred season (Fig. S4). For example, ASV11 (unclassified Labyrinthulomycetes) and ASV7 (aplanochytrid) were generally not abundant (below 0.01% of the library during most weeks) but exhibited blooms where they comprised up to 60% of the Labyrinthulomycetes community for a single week (Fig. 4A). Although many of these seasonal-blooming ASVs have short-duration blooms, they generally reoccurred annually over the 3-year time series (Fig. 3), suggesting a response to seasonal environmental conditions rather than episodic disturbances (e.g., storms). In contrast, the persistent cluster contains the 6 most abundant as well as the 14th and 15th most abundant ASVs and together amounts to an average of 65.02% of the Labyrinthulomycetes community. In contrast to taxa that exhibited short-term blooms, these 8 ASVs persisted over several months but also exhibited seasonal preferences and interannual differences (Fig. S5), demonstrating that the Labyrinthulomycetes are not transient members of the coastal microbial community. Further, the two most abundant ASVs, representing 21.74% and 17.52% of the total sequences, respectively, dominated the seasonal shift between aplanochytrids and thraustochytrids (Fig. 4B), which are the two most abundant groups of Labyrinthulomycetes at this site (Fig. S6) as well as in the coastal waters of southern and northern China (16, 17). This observation is generally consistent with the seasonal patterns of culturable thraustochytrids and aplanochytrids observed previously (21). However, these data also reveal distinct dynamics within the same genus of Labyrinthulomycetes protists (e.g., ASV1 and ASV7) (Fig. 4A and B).
FIG 3.
Relative abundances of the 100 most abundant Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) across the weekly time series at Piver’s Island Coastal Observatory (PICO) from 2011 to 2013. (A) The heatmap for the log2-transformed relative abundances of the 100 most abundant ASVs (y axis), which were clustered by Ward’s minimum variance method (dendrogram). Columns to the left of the heatmap annotate each ASV with its Mfuzz soft-cluster and taxonomic grouping (at the genus or most specific classified level). The temperature heatmap across the top indicates the water temperature. (B) The centroid of the log2-transformed abundance for each Mfuzz soft cluster across the time series.
FIG 4.
(A to E) Relative abundance patterns of the most abundant winter/summer-blooming (A) and persistent summer-preferred/winter-preferred/nonseasonal (B) Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) across the weekly time series at Piver’s Island Coastal Observatory (PICO) from 2011 to 2013 in comparison with those of the two most abundant bacterial (C), fungal (D), and algal (E) OTUs. “Winter/summer-blooming” and “persistent” refer to the Mfuzz clusters (see Fig. 3). ASV1 and ASV2 are the two most abundant Labyrinthulomycetes ASVs. Each ASV/OTU is labeled with the most specific available taxonomic assignment and its identification number.
In comparing this clade with other microbial groups, we observed similar seasonal preferences in both the bacterioplankton and the fungi (Fig. 4A to D). While some bacterial OTUs were abundant across all time points, most bacterial OTUs were present across either the summer or winter as observed for the persistent Labyrinthulomycetes cluster (Fig. 4B and C, Fig. S7 and S8). In contrast, the short, seasonally associated bloom pattern observed for most Labyrinthulomycetes ASVs is more similar to that of fungal phylotypes (Fig. 4D, Fig. S7). Some eukaryotic chloroplast sequences also exhibited large weekly changes in their relative abundance, but with a weaker seasonal reoccurrence compared to the Labyrinthulomycetes (Fig. 4E, Fig. S7). This strong seasonal signal across microbial groups could be explained through direct control by environmental factors (e.g., temperature) or by interactions with seasonally responsive organisms. For example, ASV1, the most abundant Labyrinthulomycetes phylotype, belongs to a genus (Aplanochytrium) commonly observed with phytoplankton (21, 32, 55–57), suggesting that this ASV may utilize phytoplankton-derived organic carbon, which should be more available during the summer. The second most abundant ASV2 (Thraustochytriaceae) is more abundant in winter and may serve as a complementary decomposer to bacteria, potentially degrading recalcitrant organic material when more labile phytoplankton-derived organic material is scarce. However, the majority of the 100 most abundant phylotypes exhibit season-specific ephemeral blooms more similar to patterns observed in the fungi (Fig. 3A, Fig. S8), including the most abundant fungal phylotypes (Fig. 4D). Unlike for the bacterioplankton, patterns in these eukaryotic heterotrophs are possibly driven by distinct resources or specific habitat/host conditions. Our observations provide new hypotheses regarding the ecologies of Labyrinthulomycetes, which can guide new research into drivers and functions using metagenomics, targeted culture work, or other approaches.
Phylogeny and niche partitioning of Labyrinthulomycetes.
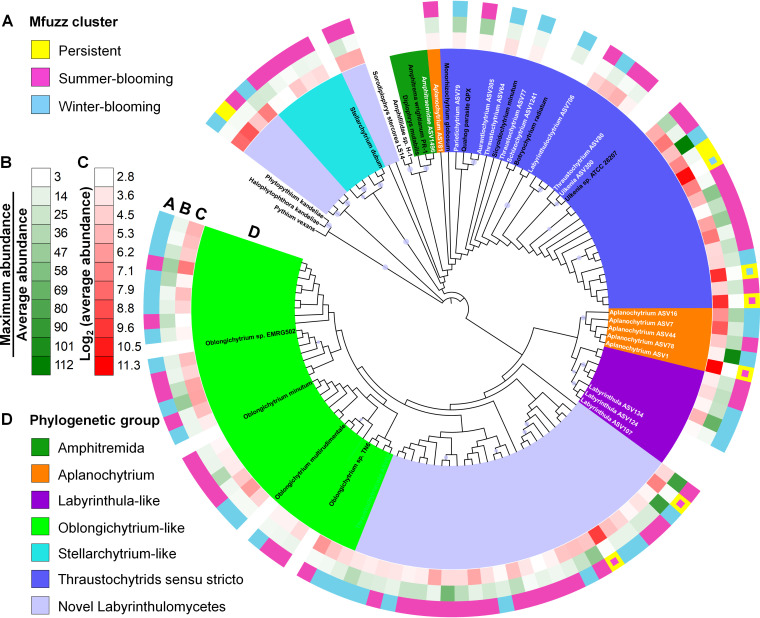
In order to further understand ecological partitioning within Labyrinthulomycetes, we constructed a phylogenetic tree containing the 100 most abundant ASVs, reference sequences, and 8 rare ASVs that were identified at the genus level (or at the family level for the poorly resolved Amphitraemidae). The resulting phylogenetic tree contains 6 major clades of known taxa, including Amphitremida, Aplanochytrium, Labyrinthula-like, Oblongichytrium-like, Stellarchytrium-like, and thraustochytrids sensu stricto (comprising closely related genera within the family Thraustochytriaceae following the latest taxonomic classification) (21, 58, 59), as well as a number of potentially novel lineages (Fig. 5). Notably, the Aplanochytrium clade is the dominant genus (26.10% of total sequences) across the time series, consistent with previous reports of this clade’s prevalence in both coastal and pelagic waters (16–18, 21, 30, 60, 61). The thraustochytrids sensu stricto clade, another major subgroup of the Labyrinthulomycetes protists, however, contains mostly unclassified thraustochytrids and a very low abundance of identified genera, Aurantiochytrium, Labyrinthulochytrium, Parietichytrium, Schizochytrium, Thraustochytrium, and Ulkenia, which account for <1% of the total sequences. These thraustochytrid genera, as well as Botryochytrium and Sicyoidochytrium, which were not found here, are commonly observed in the coastal waters of China and Japan (16, 17, 21, 30). Overall, high phylogenetic diversity of Labyrinthulomycetes has been identified at our sampling site, but the majority of the phylotypes do not belong to described genera, and many of these phylotypes, including some dominant ones (e.g., ASV5 and ASV6), are not closely related to previously described clades and thus likely represent new lineages.
FIG 5.
Maximum likelihood tree constructed using the representative sequences of the 100 most abundant Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) and 8 rare but classified ASVs identified from the weekly time series at Piver’s Island Coastal Observatory (PICO) from 2011 to 2013. Reference sequences from NCBI GenBank are included to identify the major clades. The labels of the classified ASVs and reference sequences are indicated by white and black text, respectively. Bootstrap values higher than 80% are indicated by circles. (A to D) The 100 most abundant ASVs are annotated with a number of features, including (A) soft-clustering assignment consistent with Fig. 3, but the persistent ASVs with obvious summer or winter preferences marked with small pink and blue squares, respectively, (B) bloom potential as defined by its maximum relative abundance divided by the average relative abundance across the time series, (C) average relative abundance, and (D) phylogenetic affiliation.
Additionally, in the phylogenetic tree, closely related ASVs display distinct seasonal partitioning (Fig. 5). For example, within the thraustochytrid sensu stricto clade, 14 ASVs (accounting for 13.95% of total sequences) are summer associated and 9 ASVs (accounting for 26.32% of total sequences) are winter associated, while ASV15 (accounting for 0.96% of total sequences) is generally more abundant in spring and autumn. In contrast, the Stellarchytrium-like clade only contains summer-associated ASVs. Overall, this study emphasizes that closely related strains exhibit distinct environmental preferences, highlighting the importance of evaluating Labyrinthulomycetes phylotypes at fine phylogenetic scales.
Correlations between Labyrinthulomycetes phylotypes and other organisms.
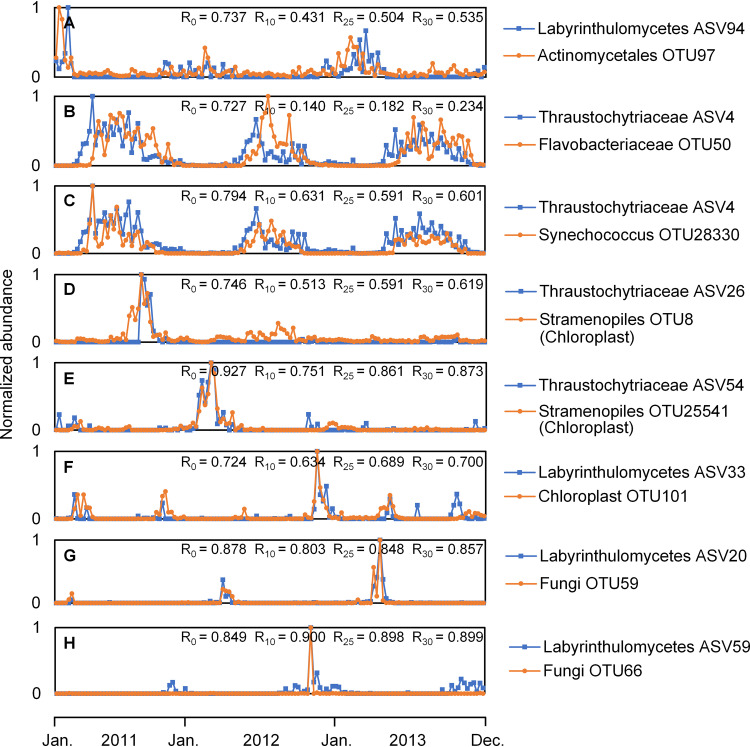
Recent evidence from cooccurrence patterns of coastal microbial communities including bacteria, algae, fungi, and protists have identified correlations between taxon pairs across microbial groups (12, 43, 62). Here, we identified a number of correlations between Labyrinthulomycetes ASVs and other microbial taxa (Fig. 6, Tables S2 to S4). Correlations between Labyrinthulomycetes and heterotrophic bacteria were largely driven by seasonal preferences, with the strongest correlations mostly involving the two most abundant ASVs (Table S1). Some ASVs exhibit time-lagged correlations with heterotrophic bacterial OTUs (R > 0.7, P < 5 × 10−23), which were still significant (P < 0.01) even when seasonal trends were removed (Fig. 6A and B). Based on observational data, it is not possible to determine whether these taxa respond to similar environmental conditions or potentially interact; however, these results might generate new hypotheses about the functional partitioning of Labyrinthulomycetes protists based on their potential relationships with heterotrophic bacteria and inferred nutritional modes. The ASVs that increased following the heterotrophic bacterial OTUs are potentially amoebic bacterivores or subsist upon resources not consumed by these bacteria (16, 28). In contrast, the ASVs which peak prior to heterotrophic bacteria might be initial decomposers, consistent with previous reports that Labyrinthulomycetes can secrete hydrolytic enzymes to metabolize protein, lipid, cellulose, starch, xylan, gelatin, urea, organophosphate, chitin, and glucoside (53, 54, 57, 61, 63–66). In addition to correlations with heterotrophic bacteria, our analysis identified cooccurrences of Labyrinthulomycetes with phytoplankton and fungi (Fig. 6C to H, Tables S3 and S4). For example, a summer-associated Synechococcus OTU (Cyanobacteria) showed a significant correlation (R = 0.794, P = 5 × 10−32) and detrended correlation (R = 0.631 at 10% span, R = 0.591 at 25% span, and R = 0.601 at 30% span; P < 1 × 10−14) with a thraustochytrid ASV (Fig. 6C). Similarly, one Labyrinthulomycetes ASV cooccurred with a eukaryotic chloroplast (Fig. 6D to F) and a fungus (Fig. 6G and H) as episodic, transient spikes, with all the illustrated pairs showing strong correlations (R = 0.746, R = 0.927, R = 0.724, R= 0.878, and R = 0.849; P < 3 × 10−24) and significant detrended correlations (R > 0.5, P < 7 × 10−11). Prior culture-dependent reports of species-specific associations between Labyrinthulomycetes and phototrophs (23), such as the endophyte Aplanochytrium minuta and the brown alga Sargassum cinereum (55), and between the wasting disease pathogen Labyrinthula zosterae and the eelgrass Zostera marina (67) also suggest that Labyrinthulomycetes protists may be closely tied to other organisms. Preferences of the culturable Labyrinthulomycetes protists for specific substrates and hosts can even extend to saprophytic species associated with detritus types (e.g., Aurantiochytrium mangrovei with fallen mangrove leaves) and parasitic/symbiotic species with specific marine animals (e.g., QPX with the hard clam Mercenaria mercenaria) (26, 68–73). Additionally, our results suggest that some Labyrinthulomycetes ASVs and fungal OTUs may respond to the same episodic factors. Labyrinthulomycetes and fungi have both been reported to be enriched on particulate organic matter, which is patchily distributed in the ocean (9, 18, 19, 33). The short-term blooms in both these Labyrinthulomycetes protists and fungi could indicate either a narrow niche for their preferred substrates, hosts, or other environmental conditions (e.g., bottom-up controls) or, alternately, strong density-dependent selection (top-down controls) by either grazers or viruses. In comparison with other microbial groups sampled at the PICO time series, the patterns in dominant ASVs are more similar to strong summer or winter preferences observed for heterotrophic bacteria and cyanobacteria, while less abundant ASVs exhibit annually reoccurring blooms as previously observed for the fungi (Fig. S8). These observations demonstrate the important but complex patterns of Labyrinthulomycetes in the coastal food webs and highlight the need to consider a broad spectrum of eukaryotic heterotrophs as well as the potential for different factors to structure their distributions in the investigation of marine microbial communities.
FIG 6.
(A to H) Examples of associations between Labyrinthulomycetes and other organisms, including heterotrophic bacteria (A and B), cyanobacteria (C), algae (D to F), and fungi (G and H), which were identified from the weekly time series at Piver’s Island Coastal Observatory (PICO) from 2011 to 2013. The relative abundances of all the illustrated pairs showed significant time-lagged (A and B) or cooccurring (C to H) correlations (P < 5 × 10−23, R = R0). For the second pair (B), the detrended abundances showed significant time-lagged correlations when their polynomial trends at the 25% (P < 0.05, R = 0.182) or 30% (P < 0.01, R = 0.234) span were removed. For other pairs, the detrended correlations were significant (P < 4 × 10−7) when their polynomial trends at the 10% (R = R10), 25% (R = R25), or 30% (R = R30) span were removed. The P values were not corrected for multiple hypothesis testing. Each ASV/OTU is labeled with the most specific available taxonomic assignment and its identification number.
Conclusions.
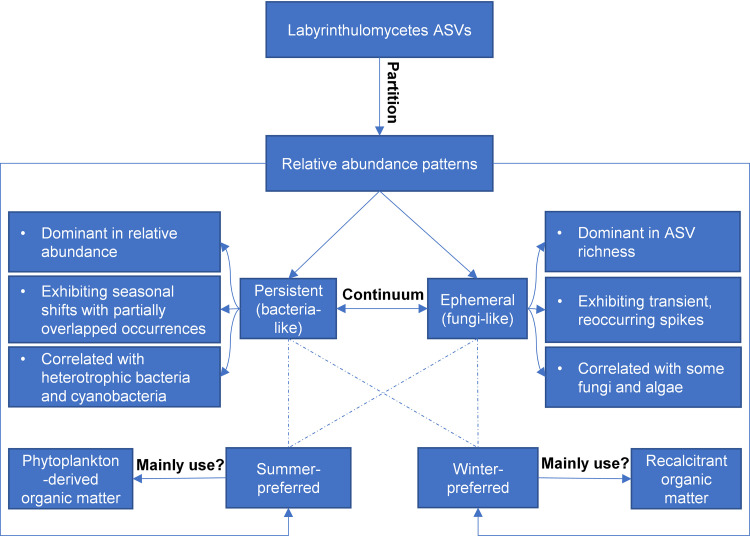
Research on heterotrophic microbes in marine ecosystems has historically largely focused on prokaryotes; however, recent work has revealed the potential importance of understudied, heterotrophic eukaryotes in coastal ecosystems. Here, we utilized a well-characterized weekly coastal time series with available data on bacterioplankton and fungal communities (12, 14) to gain a greater understanding of the similarities and differences in the factors structuring microbial groups in the coastal ocean (Fig. S8). The strong seasonal associations with either summer or winter of the bacterioplankton at this site are observed for only a few persistent Labyrinthulomycetes ASVs, with the majority of the Labyrinthulomycetes taxa exhibiting seasonally associated but brief (weeks to a month) blooms that generally reoccurred in each of the 3 years of the time series. Such obvious and novel partitioning of Labyrinthulomycetes in dynamic patterns suggests distinct ecotypes and multifaceted functions of this protistan group (Fig. 7), which may shed new light on the intricate ecological roles of unicellular heterotrophic eukaryotes in the coastal oceans.
FIG 7.
Conceptual diagram of the bidimensional (persistent versus ephemeral; summer- versus winter-preferred) partitioning of Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs), their relative abundance patterns, and predicted ecological roles in the coastal ocean, based on the weekly observations at the Piver’s Island Coastal Observatory (PICO) from 2011 to 2013.
While most studies of heterotrophic communities focus on bottom-up processes, the constantly changing Labyrinthulomycetes community composition could suggest a strong role for phylotype-specific, density-dependent selection. Moreover, the seasonal associations, persistent occupation, and recurring patterns in this coastal site argue against Labyrinthulomycetes being solely scavengers of detritus “left over” from bacterioplankton metabolism and suggest that they occupy important year-round ecological niches in the coastal system. Thus, the total rates of carbon and nutrient cycling in the coastal ocean likely include significant contributions from a range of heterotrophic eukaryotes, including Labyrinthulomycetes. However, characterizing the responses of Labyrinthulomycetes and other heterotrophic protists to both top-down and bottom-up controls on their community structure remains a subject for future research investigation.
MATERIALS AND METHODS
Environmental samples and metadata.
As previously described, seawater samples were collected weekly at a fixed site (34.7181°N, 76.6707°W) near the Beaufort Inlet in North Carolina (USA) from January 2011 to December 2013 as part of the Piver’s Island Coastal Observatory (PICO), which concurrently measures key environmental variables (14, 74). Roughly, 1 liter of near-surface seawater (1 m depth) was filtered through 0.22-μm Sterivex filter units (Millipore), and the resulting filters were stored at −80°C until DNA was extracted. Methods for determination of surface water temperature, pH, salinity, nutrient concentrations, chlorophyll a, dissolved inorganic carbon, tidal height (mean lower low water), insolation (incoming no-sky solar radiation), and oxygen saturation were described previously (14, 74). The prokaryotic and fungal abundances were determined by flow cytometry and quantitative PCR, respectively (12, 14).
DNA extraction and sequencing.
The DNA was extracted as described previously (14, 75), through physical lysis by bead beating, phenol-chloroform extraction coupled with RNase treatment, and isopropanol precipitation followed by PCR inhibitor removal (Zymo). Partial Labyrinthulomycetes 18S rRNA genes were amplified using primers including Illumina adapters and indexes of LABY-A (5′‐GGGATCGAAGATGATTAG‐3′) and LABY-Y (5′‐CWCRAACTTCCTTCCGGT‐3′) (16–18, 30, 76). The 25-μl PCR mixture contained 1× Qiagen multiplex mastermix, 0.2 μM each primer, and ∼20 ng of DNA template. The PCR program was run as follows: initial denaturation at 95°C for 15 min, followed by 31 cycles of 30 sec at 94°C, 1.5 min at 50°C, and 1.5 min at 72°C, and a final extension at 72°C for 10 min. Duplicate PCR products for each sample were pooled, gel purified using a Qiagen gel extraction kit, and quantified using the Qubit 3 fluorometer. In total, 149 amplicon libraries were pooled and sent to the Duke Center for Genomic and Computational Biology for paired-end (2 × 250 bp) sequencing on the Illumina MiSeq platform.
Quantitative PCR of the Labyrinthulomycetes 18S rRNA genes.
Quantitative PCR was employed to assess the total abundance of the Labyrinthulomycetes 18S rRNA genes per liter of seawater for representative winter (mid-January and mid-February) and summer (mid-July and mid-August) samples. Primers LABY-A and LABY-Y were used with the SYBR premix Ex Taq (TaKaRa, Japan). The 10-μl reaction mixture contained 1× SYBR premix Ex Taq, 0.25 μM each primer, and ca. 5 to 45 ng DNA template. The quantitative PCR program was run on an Eppendorf Mastercycler ep realplex instrument with an initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 5 sec, annealing at 50°C for 30 sec, and elongation and acquisition of fluorescence data at 72°C for 1 min. The melting curve was used to confirm the specificity of the amplification. The standard curve was constructed using known amounts of standard linearized plasmid, a combination of the pGEM-T vector (Promega, USA) and the target gene derived from a cultured thraustochytrid genome. Pearson correlations between the Labyrinthulomycetes 18S rRNA gene abundance and environmental factors were analyzed in SPSS 22.
Processing of the Labyrinthulomycetes 18S rRNA gene sequences.
Raw sequences were demultiplexed and assigned to corresponding libraries using CASAVA software (Illumina). Low-quality sequence ends were trimmed at a Phred quality (Q) of 25 using a 10-bp running window, and only sequences of ≥130 bp after trimming were retained. The paired-end sequences were joined when they had a ≥10-bp overlap with ≤3 mismatches. Then, the Deblur workflow (77) was performed in QIIME 2 (78) to denoise the joined sequences and to resolve amplicon sequence variants (ASVs), using “silva_132_99_18S.fna” (https://www.arb-silva.de/download/archive/qiime/) as the positive filtering database, with the key parameters “sequence trim length,” “mean per nucleotide error,” “indel probability,” and “maximum indel number” set as 380, 0.005, 0.01, and 3, respectively; only ASVs appearing at least 10 times across all libraries were retained. The resulting ASVs were assigned by the BLAST+ consensus taxonomy classifier (79) against the SILVA database, and those not assigned to the class Labyrinthulomycetes were removed from downstream analyses. Libraries were rarified to 11,800 sequences.
Characterization of the Labyrinthulomycetes diversity.
The α-diversity measures, including richness, Pielous’ evenness, and Shannon’s diversity, for ASVs were calculated in USEARCH 10 (80), and the Pearson correlations between the α-diversity measures and environmental factors were analyzed in SPSS 22 (16, 81, 82). The nonmetric multidimensional scaling (NMDS) and redundancy analysis (RDA) were performed using the vegan package in R to visualize patterns and identify potential environmental drivers for the Labyrinthulomycetes community composition, based on the log-transformed ASV abundances and scaled environmental data. The constrained environmental parameters were determined for the RDA model by performing a stepwise selection (Akaike information criterion, 999 permutations per step) using the “step” function and plotted on the NMDS ordination using the “envfit” function. The 100 most abundant ASVs were extracted for soft clustering using the Mfuzz package in R, based on their log2-transformed abundances (83). A heatmap for the dynamic of the log2-transformed abundances of these 100 ASVs across the time series was drawn using the pheatmap package in R, with the ASVs clustered by Ward’s hierarchical agglomerative method (84, 85) and annotated using taxonomic information.
Construction of the Labyrinthulomycetes phylogenetic tree.
The representative sequences for the 100 most abundant ASVs, additional less abundant ASVs that were classified by the BLAST+ consensus taxonomy classifier at the genus level (or at the family level for the poorly resolved Amphitraemidae), and reference sequences downloaded from NCBI GenBank were aligned using MUSCLE (86). The phylogenetic tree was inferred by using the maximum likelihood method based on the general time reversible (GTR) model (87). Initial trees for the heuristic search were obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories, +G, parameter = 0.2399). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 28.73% sites), and all positions with less than 95% site coverage were eliminated. The best‐fit nucleotide substitution model (GTR + G + I) described above was preselected from 24 different models based on the lowest Bayesian information criterion (BIC) and other evaluation scores (Table S5) and then improved by performance of 1,000 bootstrap replicates (87). Evolutionary analyses were conducted in MEGA7 (88) and annotated in iTOL (89).
Correlation analysis between Labyrinthulomycetes and other organisms.
In order to investigate the relationship between Labyrinthulomycetes and other microbial taxa, we obtained the relative abundances of prokaryotic 16S rRNA genes and fungal internal transcribed spacer (ITS) amplicon library data sets from the same time points (12, 14). The synchronous or time-lagged Pearson correlations between the 100 most abundant Labyrinthulomycetes ASVs and the 100 most abundant heterotrophic bacterial, phytoplankton (including cyanobacteria and chloroplasts), and fungal OTUs were examined in SPSS 22. In order to correct for time series autocorrelation, we illustrated typical pairwise examples and detrended these data by subtracting their local polynomial regression fitting trends, which were determined by the R function “loess” at 10%, 25%, and 30% spans, and then recalculated the Pearson correlation of each pair. The P values were not corrected for multiple hypothesis testing, but their precise numbers are presented to indicate if the correlations meet the corrected or conservative threshold (P < 0.05/30000).
Data availability.
The raw sequence reads and environmental metadata that support the findings of this study have been deposited in the NCBI under BioProject numbers PRJNA590600 (Labyrinthulomycetes 18S rRNA gene libraries), PRJNA432592 (fungal ITS libraries) (12), and PRJNA309156 (16S rRNA gene libraries) (14). Other data generated or analyzed during this study are included in this published article and its supplementary information files.
Supplementary Material
ACKNOWLEDGMENTS
The work was partially supported by the National Key R&D Program of China (2016YF0601401) and NSFC (31670044 and 91751115) to G.W., a US-NSF grant (OCE: 14-16665) to D.E.H. and Z.I.J., and a CSC scholarship (201806250109) to N.X. These funders had no role in the study design, data collection, or manuscript preparation.
We acknowledge the entire PICO team for sampling and measuring environmental parameters. We also appreciate Yunxuan Xie’s preparation of the standard plasmid used for qPCR quantification of the Labyrinthulomycetes 18S rRNA genes, Thomas Schultz’s assistance in sharing the qPCR instrument, and Zhao Wang’s helpful discussion of data analysis.
G.W., Z.I.J., N.X., and Y.H. conceived and designed the study. N.X., D.E.H., and Z.I.J. contributed to the data acquisition and analysis. N.X., D.E.H., Z.I.J., and G.W. contributed to the data interpretation. N.X. drafted the manuscript, and all authors were involved in revision of the final version. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, Kirchman DL, Weinbauer MG, Luo T, Chen F, Azam F. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol 8:593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 2.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 4.Santoro AE, Richter RA, Dupont CL. 2019. Planktonic marine archaea. Annu Rev Mar Sci 11:131–158. doi: 10.1146/annurev-marine-121916-063141. [DOI] [PubMed] [Google Scholar]
- 5.Jiao N, Robinson C, Azam F, Thomas H, Baltar F, Dang H, Hardman-Mountford NJ, Johnson M, Kirchman DL, Koch BP, Legendre L, Li C, Liu J, Luo T, Luo Y-W, Mitra A, Romanou A, Tang K, Wang X, Zhang C, Zhang R. 2014. Mechanisms of microbial carbon sequestration in the ocean: future research directions. Biogeosciences 11:5285–5306. doi: 10.5194/bg-11-5285-2014. [DOI] [Google Scholar]
- 6.Zhang Y, Zhao M, Cui Q, Fan W, Qi J, Chen Y, Zhang Y, Gao K, Fan J, Wang G, Yan C, Lu H, Luo Y, Zhang Z, Zheng Q, Xiao W, Jiao N. 2017. Processes of coastal ecosystem carbon sequestration and approaches for increasing carbon sink. Sci China Earth Sci 60:809–820. doi: 10.1007/s11430-016-9010-9. [DOI] [Google Scholar]
- 7.Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 8.Pernice MC, Forn I, Gomes A, Lara E, Alonso-Sáez L, Arrieta JM, del Carmen Garcia F, Hernando-Morales V, MacKenzie R, Mestre M, Sintes E, Teira E, Valencia J, Varela MM, Vaqué D, Duarte CM, Gasol JM, Massana R. 2015. Global abundance of planktonic heterotrophic protists in the deep ocean. ISME J 9:782–792. doi: 10.1038/ismej.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bochdansky AB, Clouse MA, Herndl GJ. 2017. Eukaryotic microbes, principally fungi and Labyrinthulomycetes, dominate biomass on bathypelagic marine snow. ISME J 11:362–373. doi: 10.1038/ismej.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarkar M, Countway PD, Du Yoo Y, Daniels E, Poulton NJ, Palenik B. 2018. Temporal dynamics of eukaryotic microbial diversity at a coastal Pacific site. ISME J 12:2278–2291. doi: 10.1038/s41396-018-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rachik S, Christaki U, Li LL, Genitsaris S, Breton E, Monchy S. 2018. Diversity and potential activity patterns of planktonic eukaryotic microbes in a mesoeutrophic coastal area (eastern English Channel). PLoS One 13:e0196987. doi: 10.1371/journal.pone.0196987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan Y, Xie N, Song Z, Ward CS, Yung C-M, Hunt DE, Johnson ZI, Wang G. 2018. A high-resolution time-series reveals seasonal patterns of planktonic fungi at a temperate coastal ocean site (Beaufort, North Carolina, USA). Appl Environ Microbiol 84:e00967. doi: 10.1128/AEM.00967-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. 2015. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347:1257594. doi: 10.1126/science.1257594. [DOI] [PubMed] [Google Scholar]
- 14.Ward CS, Yung C-M, Davis KM, Blinebry SK, Williams TC, Johnson ZI, Hunt DE. 2017. Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J 11:1412–1422. doi: 10.1038/ismej.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan J, Campo J, Keeling PJ. 2017. Reference tree and environmental sequence diversity of Labyrinthulomycetes. J Eukaryot Microbiol 64:88–96. doi: 10.1111/jeu.12342. [DOI] [PubMed] [Google Scholar]
- 16.Xie N, Sen B, Song Z, Zhao Y, Chen Z, Shi W, Zhang Y, Zhang J, Johnson ZI, Wang G. 2018. High phylogenetic diversity and abundance pattern of Labyrinthulomycete protists in the coastal waters of the Bohai Sea. Environ Microbiol 20:3042–3056. doi: 10.1111/1462-2920.14341. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Singh P, Liang Y, Li J, Xie N, Song Z, Daroch M, Leng K, Johnson ZI, Wang G. 2017. Abundance and molecular diversity of thraustochytrids in coastal waters of southern China. FEMS Microbiol Ecol 93:fix070. doi: 10.1093/femsec/fix070. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Wang X, Liu X, Jiao N, Wang G. 2013. Abundance and novel lineages of thraustochytrids in Hawaiian waters. Microb Ecol 66:823–830. doi: 10.1007/s00248-013-0275-3. [DOI] [PubMed] [Google Scholar]
- 19.Naganuma T, Kimura H, Karimoto R, Pimenov NV. 2006. Abundance of planktonic thraustochytrids and bacteria and the concentration of particulate ATP in the Greenland and Norwegian Seas. Polar Biosci 20:37–45. [Google Scholar]
- 20.Kimura H, Fukuba T, Naganuma T. 1999. Biomass of thraustochytrid protoctists in coastal water. Mar Ecol Prog Ser 189:27–33. doi: 10.3354/meps189027. [DOI] [Google Scholar]
- 21.Ueda M, Nomura Y, Doi K, Nakajima M, Honda D. 2015. Seasonal dynamics of culturable thraustochytrids (Labyrinthulomycetes, Stramenopiles) in estuarine and coastal waters. Aquat Microb Ecol 74:187–204. doi: 10.3354/ame01736. [DOI] [Google Scholar]
- 22.Nakai R, Naganuma T. 2015. Diversity and ecology of thraustochytrid protists in the marine environment, p 331–346. In Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, Not F (ed), Marine protists. Springer Japan, Tokyo, Japan. [Google Scholar]
- 23.Raghukumar S. 2002. Ecology of the marine protists, the Labyrinthulomycetes (thraustochytrids and labyrinthulids). Eur J Protistol 38:127–145. doi: 10.1078/0932-4739-00832. [DOI] [Google Scholar]
- 24.Raghukumar S. 1992. Bacterivory: a novel dual role for thraustochytrids in the sea. Mar Biol 113:165–169. doi: 10.1007/BF00367650. [DOI] [Google Scholar]
- 25.Wong MK, Vrijmoed LL, Au DW. 2005. Abundance of thraustochytrids on fallen decaying leaves of Kandelia candel and mangrove sediments in Futian National Nature Reserve, China. Bot Mar 48:374–378. doi: 10.1515/bot.2005.050. [DOI] [Google Scholar]
- 26.Rubin E, Tanguy A, Pales Espinosa E, Allam B. 2017. Differential gene expression in five isolates of the clam pathogen, Quahog parasite unknown (QPX). J Eukaryot Microbiol 64:647–654. doi: 10.1111/jeu.12400. [DOI] [PubMed] [Google Scholar]
- 27.Hamamoto Y, Honda D. 2019. Nutritional intake of Aplanochytrium (Labyrinthulea, Stramenopiles) from living diatoms revealed by culture experiments suggesting the new prey-predator interactions in the grazing food web of the marine ecosystem. PLoS One 14:e0208941. doi: 10.1371/journal.pone.0208941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghukumar S, Damare VS. 2011. Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Bot Mar 54:3–11. doi: 10.1515/bot.2011.008. [DOI] [Google Scholar]
- 29.Damare V, Raghukumar S. 2008. Abundance of thraustochytrids and bacteria in the equatorial Indian Ocean, in relation to transparent exopolymeric particles (TEPs). FEMS Microbiol Ecol 65:40–49. doi: 10.1111/j.1574-6941.2008.00500.x. [DOI] [PubMed] [Google Scholar]
- 30.Bai M, Sen B, Wang Q, Xie Y, He Y, Wang G. 2019. Molecular detection and spatiotemporal characterization of Labyrinthulomycete protist diversity in the coastal waters along the Pearl River Delta. Microb Ecol 77:394–405. doi: 10.1007/s00248-018-1235-8. [DOI] [PubMed] [Google Scholar]
- 31.Raghukumar S, Raghukumar C, Rajendran A. 1990. Abundance of thraustochytrid fungi in the Arabian Sea. Estuar Coast Shelf Sci 31:351–358. doi: 10.1016/0272-7714(90)90109-5. [DOI] [Google Scholar]
- 32.Bongiorni L, Dini F. 2002. Distribution and abundance of thraustochytrids in different Mediterranean coastal habitats. Aquat Microb Ecol 30:49–56. doi: 10.3354/ame030049. [DOI] [Google Scholar]
- 33.Kimura H, Sato M, Sugiyama C, Naganuma T. 2001. Coupling of thraustochytrids and POM, and of bacterio- and phytoplankton in a semi-enclosed coastal area: implication for different substrate preference by the planktonic decomposers. Aquat Microb Ecol 25:293–300. doi: 10.3354/ame025293. [DOI] [Google Scholar]
- 34.Kimura H, Naganuma T. 2001. Thraustochytrids: a neglected agent of the marine microbial food chain. Aquat Ecosyst Health 4:13–18. doi: 10.1080/146349801753569243. [DOI] [Google Scholar]
- 35.Naganuma T, Takasugi H, Kimura H. 1998. Abundance of thraustochytrids in coastal plankton. Mar Ecol Prog Ser 162:105–110. doi: 10.3354/meps162105. [DOI] [Google Scholar]
- 36.Duan Y, Sen B, Xie N, Paterson JS, Chen Z, Wang G. 2018. Flow cytometry for rapid enumeration and biomass quantification of thraustochytrids in coastal seawaters. Microbes Environ 33:195–204. doi: 10.1264/jsme2.ME17162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghukumar S, Ramaiah N, Raghukumar C. 2001. Dynamics of thraustochytrid protists in the water column of the Arabian Sea. Aquat Microb Ecol 24:175–186. doi: 10.3354/ame024175. [DOI] [Google Scholar]
- 38.Takao Y, Tomaru Y, Nagasaki K, Honda D. 2015. Ecological dynamics of two distinct viruses infecting marine eukaryotic decomposer thraustochytrids (Labyrinthulomycetes, Stramenopiles). PLoS One 10:e0133395. doi: 10.1371/journal.pone.0133395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chafee M, Fernàndez-Guerra A, Buttigieg PL, Gerdts G, Eren AM, Teeling H, Amann RI. 2018. Recurrent patterns of microdiversity in a temperate coastal marine environment. ISME J 12:237–252. doi: 10.1038/ismej.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chase AB, Karaoz U, Brodie EL, Gomez-Lunar Z, Martiny AC, Martiny JBH. 2017. Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio 8:e01809-17. doi: 10.1128/mBio.01809-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin AA, Martiny AC. 2017. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ Microbiol Rep 9:55–70. doi: 10.1111/1758-2229.12523. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JD, Cunliffe M. 2016. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J 10:2118–2128. doi: 10.1038/ismej.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Platero AM, Cleary B, Kauffman K, Preheim SP, McGillicuddy DJ, Alm EJ, Polz MF. 2018. High resolution time series reveals cohesive but short-lived communities in coastal plankton. Nat Commun 9:266. doi: 10.1038/s41467-017-02571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghukumar S, Sathe-Pathak V, Sharma S, Raghukumar C. 1995. Thraustochytrid and fungal component of marine detritus. III. Field studies on decomposition of leaves of the mangrove Rhizophora apiculata. Aquat Microb Ecol 9:117–125. doi: 10.3354/ame009117. [DOI] [Google Scholar]
- 45.Liu X, Sen B, Zhao Y, Bai M, He Y, Xie Y, Li J, Wang G. 2019. Gradients of three coastal environments off the South China Sea and their impacts on the dynamics of heterotrophic microbial communities. Sci Total Environ 659:499–506. doi: 10.1016/j.scitotenv.2018.12.405. [DOI] [PubMed] [Google Scholar]
- 46.Hicks N, Liu X, Gregory R, Kenny J, Lucaci A, Lenzi L, Paterson DM, Duncan KR. 2018. Temperature driven changes in benthic bacterial diversity influences biogeochemical cycling in coastal sediments. Front Microbiol 9:1730. doi: 10.3389/fmicb.2018.01730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Currie AR, Tait K, Parry H, de Francisco-Mora B, Hicks N, Osborn AM, Widdicombe S, Stahl H. 2017. Marine microbial gene abundance and community composition in response to ocean acidification and elevated temperature in two contrasting coastal marine sediments. Front Microbiol 8:1599. doi: 10.3389/fmicb.2017.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell AM, Fleisher J, Sinigalliano C, White JR, Lopez JV. 2015. Dynamics of marine bacterial community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast Florida. Microbiologyopen 4:390–408. doi: 10.1002/mbo3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, Sen B, Zhou S, Xie N, Zhang Y, Zhang J, Wang G. 2017. Distinct seasonal patterns of bacterioplankton abundance and dominance of phyla α-Proteobacteria and Cyanobacteria in Qinhuangdao coastal waters off the Bohai Sea. Front Microbiol 8:1579. doi: 10.3389/fmicb.2017.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefanidou N, Genitsaris S, Lopez-Bautista J, Sommer U, Moustaka-Gouni M. 2018. Unicellular eukaryotic community response to temperature and salinity variation in mesocosm experiments. Front Microbiol 9:2444–2444. doi: 10.3389/fmicb.2018.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta A, Singh D, Byreddy AR, Thyagarajan T, Sonkar SP, Mathur AS, Tuli DK, Barrow CJ, Puri M. 2016. Exploring omega-3 fatty acids, enzymes and biodiesel producing thraustochytrids from Australian and Indian marine biodiversity. Biotechnol J 11:345–355. doi: 10.1002/biot.201500279. [DOI] [PubMed] [Google Scholar]
- 52.Byreddy AR. 2016. Thraustochytrids as an alternative source of omega-3 fatty acids, carotenoids and enzymes. Lipid Technol 28:68–70. doi: 10.1002/lite.201600019. [DOI] [Google Scholar]
- 53.Taoka Y, Nagano N, Okita Y, Izumida H, Sugimoto S, Hayashi M. 2009. Extracellular enzymes produced by marine eukaryotes, thraustochytrids. Biosci Biotechnol Biochem 73:180–182. doi: 10.1271/bbb.80416. [DOI] [PubMed] [Google Scholar]
- 54.Bremer GB, Talbot G. 1995. Cellulolytic enzyme activity in the marine protist Schizochytrium aggregatum. Bot Mar 38:37–42. [Google Scholar]
- 55.Sathe-Pathak V, Raghukumar S, Raghukumar C, Sharma S. 1993. Thraustochytrid and fungal component of marine detritus. I. Field studies on decomposition of the brown alga Sargassum cinereum J. Ag. Indian J Mar Sci 22:159–167. [Google Scholar]
- 56.Damare V, Raghukumar S. 2010. Association of the stramenopilan protists, the aplanochytrids, with zooplankton of the equatorial Indian Ocean. Mar Ecol Prog Ser 399:53–68. doi: 10.3354/meps08277. [DOI] [Google Scholar]
- 57.Nagano N, Matsui S, Kuramura T, Taoka Y, Honda D, Hayashi M. 2011. The distribution of extracellular cellulase activity in marine eukaryotes, thraustochytrids. Mar Biotechnol (NY) 13:133–136. doi: 10.1007/s10126-010-9297-8. [DOI] [PubMed] [Google Scholar]
- 58.Fossier Marchan L, Lee Chang KJ, Nichols PD, Mitchell WJ, Polglase JL, Gutierrez T. 2018. Taxonomy, ecology and biotechnological applications of thraustochytrids: a review. Biotechnol Adv 36:26–46. doi: 10.1016/j.biotechadv.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Hassett BT, Gradinger R. 2018. New species of saprobic Labyrinthulea (=Labyrinthulomycota) and the erection of a gen. nov. to resolve molecular polyphyly within the aplanochytrids. J Eukaryot Microbiol 65:475–483. doi: 10.1111/jeu.12494. [DOI] [PubMed] [Google Scholar]
- 60.Collado-Mercado E, Radway JC, Collier JL. 2010. Novel uncultivated Labyrinthulomycetes revealed by 18S rDNA sequences from seawater and sediment samples. Aquat Microb Ecol 58:215–228. doi: 10.3354/ame01361. [DOI] [Google Scholar]
- 61.Damare V, Raghukumar S. 2006. Morphology and physiology of the marine straminipilan fungi, the aplanochytrids isolated from the equatorial Indian Ocean. Indian J Mar Sci 35:326–340. [Google Scholar]
- 62.Wang Y, Sen B, He Y, Xie N, Wang G. 2018. Spatiotemporal distribution and assemblages of planktonic fungi in the coastal waters of the Bohai Sea. Front Microbiol 9:584. doi: 10.3389/fmicb.2018.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bongiorni L, Pusceddu A, Danovaro R. 2005. Enzymatic activities of epiphytic and benthic thraustochytrids involved in organic matter degradation. Aquat Microb Ecol 41:299–305. doi: 10.3354/ame041299. [DOI] [Google Scholar]
- 64.Raghukumar S, Sharma S, Raghukumar C, Sathe-Pathak V, Chandramohan D. 1994. Thraustochytrid and fungal component of marine detritus. IV. Laboratory studies on decomposition of leaves of the mangrove Rhizophora apiculata Blume. J Exp Mar Bio Ecol 183:113–131. doi: 10.1016/0022-0981(94)90160-0. [DOI] [Google Scholar]
- 65.Sharma S, Raghukumar C, Raghukumar S, Sathe-Pathak V, Chandramohan D. 1994. Thraustochytrid and fungal component of marine detritus. II. Laboratory studies on decomposition of the brown alga Sargassum cinereum J. Ag. J Exp Mar Bio Ecol 175:227–242. doi: 10.1016/0022-0981(94)90028-0. [DOI] [Google Scholar]
- 66.Coleman NK, Vestal JR. 1987. An epifluorescent microscopy study of enzymatic hydrolysis of fluorescein diacetate associated with the ectoplasmic net elements of the protist Thraustochytrium striatum. Can J Microbiol 33:841–843. doi: 10.1139/m87-147. [DOI] [Google Scholar]
- 67.Ralph PJ, Short FT. 2002. Impact of the wasting disease pathogen, Labyrinthula zosterae, on the photobiology of eelgrass Zostera marina. Mar Ecol Prog Ser 226:265–271. doi: 10.3354/meps226265. [DOI] [Google Scholar]
- 68.Fan K, Vrijmoed L, Jones E. 2002. Physiological studies of subtropical mangrove thraustochytrids. Bot Mar 45:50–57. doi: 10.1515/BOT.2002.006. [DOI] [Google Scholar]
- 69.Fan K, Vrijmoed L, Jones E. 2002. Zoospore chemotaxis of mangrove thraustochytrids from Hong Kong. Mycologia 94:569–578. doi: 10.1080/15572536.2003.11833185. [DOI] [PubMed] [Google Scholar]
- 70.Raghu-Kumar S. 1988. Schizochytrium mangrovei sp. nov., a thraustochytrid from mangroves in India. Trans Br Mycol Soc 90:627–631. doi: 10.1016/S0007-1536(88)80068-8. [DOI] [Google Scholar]
- 71.Harel M, Ben-Dov E, Rasoulouniriana D, Siboni N, Kramarsky-Winter E, Loya Y, Barak Z, Wiesman Z, Kushmaro A. 2008. A new thraustochytrid, strain Fng1, isolated from the surface mucus of the hermatypic coral Fungia granulosa. FEMS Microbiol Ecol 64:378–387. doi: 10.1111/j.1574-6941.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 72.Mo C, Douek J, Rinkevich B. 2002. Development of a PCR strategy for thraustochytrid identification based on 18S rDNA sequence. Mar Biol 140:883–889. doi: 10.1007/s00227-002-0778-9. [DOI] [Google Scholar]
- 73.Raghu-Kumar S. 1988. Detection of the thraustochytrid protist Ulkania visurgensis in a hydroid, using immunofluorescence. Mar Biol 97:253–258. doi: 10.1007/BF00391310. [DOI] [Google Scholar]
- 74.Johnson ZI, Wheeler BJ, Blinebry SK, Carlson CM, Ward CS, Hunt DE. 2013. Dramatic variability of the carbonate system at a temperate coastal ocean site (Beaufort, North Carolina, USA) is regulated by physical and biogeochemical processes on multiple timescales. PLoS One 8:e85117. doi: 10.1371/journal.pone.0085117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Massana R, Murray AE, Preston CM, DeLong EF. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol 63:50–56. doi: 10.1128/AEM.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stokes NA, Calvo LMR, Reece KS, Burreson EM. 2002. Molecular diagnostics, field validation, and phylogenetic analysis of Quahog parasite unknown (QPX), a pathogen of the hard clam Mercenaria mercenaria. Dis Aquat Organ 52:233–247. doi: 10.3354/dao052233. [DOI] [PubMed] [Google Scholar]
- 77.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 81.Jiang L, Song M, Yang L, Zhang D, Sun Y, Shen Z, Luo C, Zhang G. 2016. Exploring the influence of environmental factors on bacterial communities within the rhizosphere of the Cu-tolerant plant, Elsholtzia splendens. Sci Rep 6:36302. doi: 10.1038/srep36302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Na X, Xu T, Li M, Zhou Z, Ma S, Wang J, He J, Jiao B, Ma F. 2018. Variations of bacterial community diversity within the rhizosphere of three phylogenetically related perennial shrub plant species across environmental gradients. Front Microbiol 9:709. doi: 10.3389/fmicb.2018.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar L, E Futschik M. 2007. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ward JH. 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 85.Murtagh F, Legendre P. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Classif 31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 86.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nei M, Kumar S. 2001. Molecular evolution and phylogenetics. Oxford University Press, Oxford. [Google Scholar]
- 88.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads and environmental metadata that support the findings of this study have been deposited in the NCBI under BioProject numbers PRJNA590600 (Labyrinthulomycetes 18S rRNA gene libraries), PRJNA432592 (fungal ITS libraries) (12), and PRJNA309156 (16S rRNA gene libraries) (14). Other data generated or analyzed during this study are included in this published article and its supplementary information files.