Abstract
Objectives
The common data elements (CDE) project was developed by the National Institute of Neurological Disorders and Stroke (NINDS) to provide clinical researchers with tools to improve data quality and allow for harmonization of data collected in different research studies. CDEs have been created for several neurological diseases; the aim of this project was to develop CDEs specifically curated for mitochondrial disease (Mito) to enhance clinical research.
Methods
Nine working groups (WGs), composed of international mitochondrial disease experts, provided recommendations for Mito clinical research. They initially reviewed existing NINDS CDEs and instruments, and developed new data elements or instruments when needed. Recommendations were organized, internally reviewed by the Mito WGs, and posted online for external public comment for a period of eight weeks. The final version was again reviewed by all WGs and the NINDS CDE team prior to posting for public use.
Results
The NINDS Mito CDEs and supporting documents are publicly available on the NINDS CDE website (https://commondataelements.ninds.nih.gov/), organized into domain categories such as Participant/Subject Characteristics, Assessments, and Examinations.
Conclusion
We developed a comprehensive set of CDE recommendations, data definitions, case report forms (CRFs), and guidelines for use in Mito clinical research. The widespread use of CDEs is intended to enhance Mito clinical research endeavors, including natural history studies, clinical trial design, and data sharing. Ongoing international collaboration will facilitate regular review, updates and online publication of Mito CDEs, and support improved consistency of data collection and reporting.
Introduction
The Common Data Element (CDE) project began in 2005 as part of an initiative by the National Institute of Neurological Disorders and Stroke (NINDS) to assist NINDS-funded investigators in the collection of neuroscientific clinical trial research data in a standardized and consistent fashion (Grinnon et al 2012). The CDEs are content standards that can be applied to various data collection models and are intended to be dynamic and evolve over time, as indicated by research advances. The CDE project is not a database—rather it is a collection of metadata and data standards, used to facilitate sharing and combination of data across studies as a means of data comparison and analysis. Its goal is to develop common definitions for clinical research data as well as the creation of standardized case report forms (CRFs) and instruments. The goals of the NINDS CDE Project are to: 1) Disseminate standards for data collection from participants enrolled in neurological disease studies; 2) Create easily accessible tools for investigators to collect study data. These tools should be especially helpful to new investigators and others working with limited budgets; 3) Encourage focused and simplified data collection to reduce burden on investigators and practice-based clinicians to facilitate their participation in clinical research; 4) Improve data quality while controlling cost by providing uniform data descriptions and tools across NINDS-funded clinical studies (Grinnon et al 2012). As the CDEs were being developed, the number of clinical trials for patients with mitochondrial disease also rose, highlighting the value and urgency of such tools to be developed.
To date, the NINDS CDE database has developed metadata with data standards and instruments for 18 neurological diseases. In the case of mitochondrial disease, CDE validation can be complex due to the broad array of mechanisms causing mitochondrial diseases and dysfunction. This paper reviews the process by which the Mito WGs, an international group of mitochondrial disease experts, developed CDEs to be used in the field of mitochondrial disease research. The draft Mito CDEs were reviewed by the external mitochondrial disease research community prior to finalization and posted to the NINDS CDE website in 2015. The WG participant rosters may be found on the NINDS CDE web page (https://commondataelements.ninds.nih.gov/).
Background
Brief description of mitochondrial diseases
Mitochondrial diseases (also known as disorders of oxidative phosphorylation (OXPHOS), mitochondrial respiratory chain diseases, mitochondrial cytopathies, and mitochondriopathies) are a group of disorders caused by genetic defects that directly or indirectly affect the OXPHOS system, the major energy generating pathway in cells (Chinnery 2014; DiMauro and Schon 2003). The prevalence of mitochondrial disease is difficult to establish for many reasons, including their clinical and genetic heterogeneity, challenges in establishing a precise genetic diagnosis, and complexities in patient ascertainment and referral. The prevalence of all pathogenic mutations in both nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) is at least 1:4300 (Gorman et al 2015). Approximately 15% (DiMauro and Schon 2003) of mitochondrial disorders are caused by inherited germline mutations in the mtDNA. Mitochondrial disorders can also be caused by mutations in over 250 nDNA genes (Gorman et al 2016) and dysfunction can be acquired due to adverse environmental effects of drugs and infections (Niyazov et al 2016). Those disorders resulting from inherited nDNA or mtDNA gene mutations that have an effect on the structure or function of the OXPHOS system are termed “primary mitochondrial diseases” (Parikh et al 2015). Some mitochondrial disorders affect a single organ, but many involve multiple organ systems and can present with a bewildering array of multisystem phenotypes, including neurologic symptoms and myopathies, visual and hearing loss, as well as cardiac, endocrine, gastrointestinal, hepatic, and/or renal dysfunction (Chinnery 2014). Some well-characterized multisystem clinical syndromes have now been recognized as being mitochondrial diseases. Primary inherited mitochondrial diseases encompass hundreds of individual genetic disorders that are heterogeneous and frequently multisystemic, yet share common disease mechanisms and overlapping clinical phenotypes. One challenge, not necessarily unique to mitochondrial disorders, is that there may be great variation between individuals with the same mutation. With respect to mtDNA mutations (e.g., Mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) mutation), variation may depend on the percentage heteroplasmy (mutation load) in any given tissue and the number of tissues harboring the mutation, adding to the wide phenotypic spectrum and variable disease severity. A comprehensive overview can be found on the North American Mitochondrial Disease Consortium (NAMDC) website (https://www.rarediseasesnetwork.org/cms/namdc/Learn-More/Disorder-Definitions).
Marked variability remains in the diagnostic approaches, treatment, and management of mitochondrial diseases (Parikh et al 2015). Conducting large-scale clinical trials for patients with mitochondrial diseases is difficult, given their extreme clinical variability, biochemical and genetic heterogeneity, and the rarity of each etiology or subtype. Despite all of these constraints, several clinical trials for mitochondrial diseases are underway, none of which use any mitochondrial disease-specific curated or validated outcome measure for this specific patient group.
Materials and methods
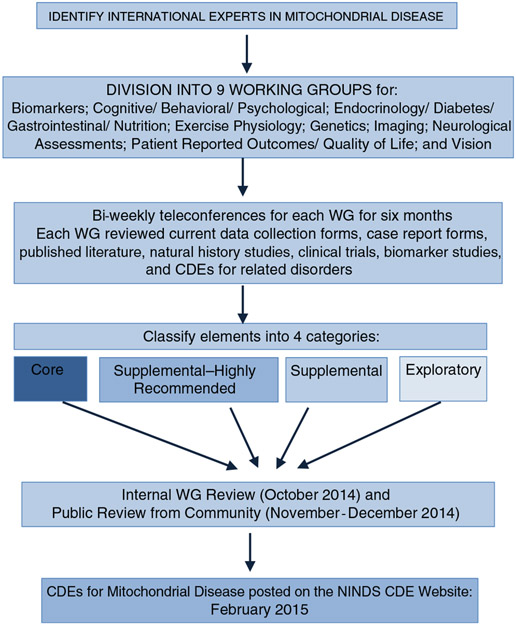
Developing CDEs for mitochondrial disorders was challenging due to the heterogeneity of potential disease symptoms that may develop. CDEs reviewed and selected for this project focused on features within and outside the domain of clinical neurology, spanning almost all organ systems. Selected CDEs would benefit the majority of individuals with mitochondrial disorders and would be linked to appropriate measures to objectively assess disease severity and progression. Therefore, the Mitochondrial Disease CDE WG was divided into nine subgroups to focus on identifying and defining data elements in the following domains: Biomarkers; Cognitive/Behavioral/Psychological; Endocrinology/Diabetes/Gastrointestinal/Nutrition; Exercise Physiology; Genetics; Imaging; Neurological Assessments; Patient Reported Outcomes/Quality of Life; and Vision. Bi-weekly teleconferences for each WG for a period of six months were held until project completion. In order to remain consistent with the overall CDE format, the WG recommendations were classified into one of four categories:
Core: A data element for recording essential information applicable to any mitochondrial disease study including therapeutic areas and study designs. Consistent with all NINDS disease-specific CDE sets, the Core Mito CDEs are a small subset of all available CDEs that are the most specific and valuable for all Mito studies.
Supplemental–highly recommended: A data element which is recommended for use whenever applicable, based on certain conditions or clinical study designs. In most cases, these have been used and validated with strong psychometrics for use in mitochondrial disease, and are considered essential for clinical research studies by experts in the field.
Supplemental: A data element which has some evidence of validity and is commonly collected in clinical studies in mitochondrial disease. Use depends upon the study design, protocol or type of research involved. Exploratory: A data element that could be emerging or that requires further validation in target populations, but may fill current gaps in the CDEs and/or substitute for an existing CDE with additional evidence.
Pre-existing individual CDEs selected for other clinically similar disease phenotypes were included and other appropriate instruments were added when available. All were critically evaluated for appropriateness to mitochondrial disorders even if historical reliability and validity was accepted for other diseases. Statistical analysis was not performed at this time, as this is not applicable for the development and initial description of the Mito CDEs. In the future, statistical analyses will help determine if these CDEs can be specifically validated for mitochondrial diseases. The draft Mito CDEs were posted on the NINDS CDE website for public review from November 20, 2014 to January 16, 2015. The final Version 1.0 Mito CDEs were posted on February 25, 2015, following the incorporation of comments received from public review from the mitochondrial clinical research community. The process describing the formation of the CDEs is available on the CDE website at https://www.commondataelements.ninds.nih.gov/CDEStandard.aspx. Figure 1 illustrates the mitochondrial CDE development process.
Fig. 1.
Flowchart illustrating the mitochondrial CDE development process
Results
The nine WGs reviewed a total of 153 CRFs and instruments (56 for Cognitive/Behavioral/Psychological outcomes, 17 for Neurological Assessments, 28 for Patient Reported Outcomes/QoL, 41 for Exercise Physiology, and 11 for Endocrinology/Diabetes/GI/Nutrition). This resulted in a library of 120 CRF and instruments recommendations divided into four domains including: 1) participant history and family history, 2) participant characteristics, 3) assessment and examinations, and 4) outcomes and endpoints. Recommendations from the Mito CDE project posted on the NINDS CDE website contain a summary of each instrument, its recommended use, and comparative strengths and weaknesses. Although the CDEs have been developed for the unique purpose of advancing mitochondrial medicine clinical research, several of the CRFs reviewed include ones used on a clinical basis for patient care purposes. The WGs did not develop the CDEs with the intention of utilizing these in clinical care, and further consideration about care guidelines (i.e., diagnostic algorithm, nutrition/exercise guidelines, etc.) would exceed the scope of this paper.
Core elements for the mitochondrial disorders included one general core element common to all other disorders reviewed for CDEs, namely the demographic patient/participant characteristics. The WGs did not find any element that was strongly representative of mitochondrial disorders, and thus no additional core elements specifically required for all mitochondrial disorders were selected. Of the remaining recommended categories, 17% of the elements assessed were categorized as supplemental-highly recommended, 75% as supplemental, and 8% as exploratory. The Highlight Summary Document is available in table format on the CDE webpage (https://www.commondataelements.ninds.nih.gov/Doc/MITO/Mitochondrial_Disease_CDE_Highlight_Summary.pdf). There were no core elements recommended due to lack of validation in mitochondrial disease cohorts. The Supplemental–highly recommended instruments (for specific disease conditions or types of study) included: Anthropometrics-Vital Signs, Apathy Evaluation Scale, Automated Self-administered 24-hour Dietary Recall (ASA 24), Barry Albright Dystonia Scale (BADS), Behavior Rating Inventory of Executive Function (BRIEF), Behavior Rating Inventory of Executive Function-Preschool Version (BRIEF-P), Conners 3, Diabetes-Related Medical History, Echocardiogram, Genetic Testing Short Form, Genetic Testing Clinical Diagnostics, Laboratory Tests and Non-Imaging Diagnostics (Diabetes), Maximal Exercise Test, Modified Hammersmith Functional Motor Scale (MHFMS-SMA, MHFMS), Peabody Development Motor Scale II, Pediatric Quality of Life Inventory (PEDSQL), Pulmonary Function, Scale for the Assessment and Rating of Ataxia (SARA), Sub-Maximal Exercise Test, Test of Variable Attention (TOVA), The Borg Scale of Perceived Exertion, Vineland Adaptive Behavior Scales, 2nd Ed., World Health Organization Quality of Life Assessment (WHOQOL). Table 1 summarizes the CDEs based on working group category and level of recommended use (core, supplemental–highly recommended, supplemental, exploratory).
Table 1.
Common data elements based on Working Group and level of recommendation (core, supplemental–highly recommended, supplemental, exploratory)
| Domain | Instrument or CDEs |
|||
|---|---|---|---|---|
| Core | Supplemental-highly recommended | Supplemental | Exploratory | |
| Demographics | General core (e.g., gender, birth date, race) | |||
| General health history | Diabetes-related medical history | Dietary supplements, reproductive and hormonal history | ||
| Physical examinations | Automated Self-Administered 24-hour Dietary Recall (ASA 24) | Mitochondrial and gastrointestinal diseases assessment | ||
| Imaging diagnostics | Brain magnetic resonance imaging (MRI), brain perfusion magnetic resonance imaging, cardiac magnetic resonance imaging (MRI), imaging–mitochondrial disease, electrocardiogram (ECG), imaging guidelines and definition | Phosphorus magnetic resonance spectroscopy (31PMRS), two dimensional speckle tracking echocardiography imaging | ||
| Laboratory tests/biomarkers | Laboratory tests and non-imaging diagnostics (diabetes) | Biomarkers guidelines mitochondrial disease, biomarkers in mitochondrial disease | ||
| Genetics | Genetic Testing Short Form, genetics testing clinical diagnostics | Genetic Testing Short Form, Genetics Testing Clinical Diagnostics | ||
| Non-imaging diagnostics | Echocardiogram | Holter examination | ||
| Vision | Vision Mitochondria Disease OCT Guidelines, Vision Mitochondrial Disease Test Guidelines, Vision Mitochondrial Disease Visual Fields Guidelines | |||
| Vital signs and other body measures | Vital Signs | |||
| Academic achievement | American National Adult Reading Test (AmNART) | |||
| Adaptive | Vineland Adaptive Behavior Scales | Adaptive Behavior Assessment Scale (ABAS-II), Delis Kaplan Executive Functioning System (DKEFS), 2nd Ed. (Vineland-II) | ||
| Attention | Conners III | Delis Kaplan Executive Function System (DKEFS), National Institutes of Health (NIH) Toolbox, Test of Variable Attention (TOVA) | ||
| Ataxia and performance measures | Scale for the Assessment and Rating of Ataxia | International Cooperative Ataxia Rating Scale (ICARS) | ||
| Cognitive | Automated Neuropsychological Assessment Metrics (ANAM), Axon Sports Computerized Cognitive Assessment, Brief Test of Adult Cognition by Telephone (BTACT), Cambridge Cognitive Assessment Revised (CAMCOG-R) | |||
| Dementia | Informant Questionnaire on Cognitive Decline in the Elderly (IQCODE), Montreal Cognitive Assessment (MoCA) | |||
| Emotional/behavioral | Apathy Evaluation Scale | ALS Depression Inventory (ADI-12), Apathy Scale (AS), Autism Diagnostic Interview (ADI), Autism Diagnostic Observation Schedule (ADOS), Beck Anxiety Inventory (BAI), Brief Infant Toddler Social Emotional Assessment (BITSEA), Brief Psychiatric Rating Scale (BPRS), Cambridge Behavioral Inventory – Revised, Center for Epidemiological Studies – Depression Scale (CES-D), Child Behavior Checklist (CBCL), Child Depression Inventory (CDI), Children Depression Inventory – 2 (CDI-2), Columbia Suicide Severity Rating Scale (CSSRS), Diagnostic and Statistical Manual of Mental Disorders (DSM-5), Generalized Anxiety Disorder (GAD-7), Hamilton Anxiety Rating Scale (HAM-A), Hamilton Depression Rating Scale (HDRS), Irritability Scale, Modified Overt Aggression Scale (MOAS), Montgomery- Åsberg Depression Scale (MADRS), Mood Disorder Questionnaire (MDQ), Problem Behaviours Assessment HD – Short Version (PBA-S), Social Responsiveness Scale (SRS) | ||
| Executive functioning | Adaptive Behavior Assessment System-II (ABAS-II) *, Behavior Rating Inventory of Executive Function (BRIEF) *, Behavior Rating Inventory of Executive Function – Preschool Version (BRIEF-P) * | |||
| Motor/physical function | Borg Rating of Perceived Exertion (RPE) Scale, Modified Hammersmith Functional Motor Scale for Children with Spinal Muscular Atrophy (MHFMS-SMA/MHFMS-Extend), Peabody Developmental Motor Scale II (PDMS-2), Barry Albright Dystonia Scale (BADS) | 6 Minute Walk Test, Physical Activity Questionnaire for Adolescents (PAQ-A), Beery-Buktenica Developmental Test of Visual-Motor Integration (Beery VMI), Burke-Fahn-Marsden Movement Scale (BFMMS), Newcastle Mitochondrial Disease Adult Scale, Newcastle Pediatric Mitochondrial Disease Scale (NPMDS), Unified Dystonia Rating Scale (UDRS) | 2 Minute Walk Test, Alberta Infant Motor Scale, Gross Motor Function Measure (GMFM-88, GMFM-66), Motor Function Measure (MFM), Physical and Neurological Examination for Subtle Signs (PANESS), International Pediatric Mitochondrial Disease Score (IPMDS) | |
| Intellectual functioning | Kaufman Assessment Battery for Children, Leiter International Performance Scale-3 (Leiter-3), Reynolds Intellectual Assessment Scale (RIAS), Wechsler Abbreviated Scale of Intelligence Scale – Fourth Edition (WAIS-IV), Wechsler Intelligence Scale for Children-IV (WISC-IV), Wechsler Preschool and Primary Scale of Intelligence (WPPSI-III) | |||
| Language | Boston Diagnostic Aphasia Exam (BDAE-III), Boston Naming Test (BNT) – 30-item version, Clinical Evaluation of Language Fundamentals (CLEF-4), Comprehensive Assessment of Spoken Language (CASL), Expressive One-Word Picture Vocabulary Test, New Reynell Developmental Language Scale (NRDLS), Peabody Picture Vocabulary Test 4th Edition (PPVT-4), Preschool Language Scales-Fifth Edition (PLS-5) | |||
| Memory | Amyotrophic Lateral Sclerosis Depression Inventory-12 (ADI-12) * | Memory Assessment Scale, Summary of Recommendations for Global Outcome, Wechsler Memory Scale IV (WMS-IV) | ||
| Muscle strength testing | Manual Muscle Testing-Using the Medical Research Council Muscle Grading Scale, Maximum Voluntary Isometric Contraction Testing (MVICT) | |||
| Neuropsychological testing | Bayley Scales of Infant Development (Bayley III, BSID) | |||
| Pulmonary function testing/respiratory status | Pulmonary function | |||
| Quality of life | Pediatric Quality of Life Inventory (PEDSQL), World Health Organization Quality of Life Assessment (WHOQOL-BREF) | Child and Adolescent Scale of Environment (CASE), Child and Adolescent Scale of Participation, Craig Handicap and Assessment Reporting Technique (CHART-SF) – Interview version, Craig Handicap and Assessment Reporting Technique (CHART-SF) Paper version, Quality of Life in Neurological Disorders (Neuro-QOL), Short Form 36-Item Health Survey (SF-36) | EurQol-5 Dimension Questionnaire, Family Strain Questionnaire Assessment of Preschooled Children’s Participation), Pediatric Quality of Life Inventory Multidimensional Fatigue Scale | |
| Visual-spatial processing | Judgment of Line Orientation (Benton JLO) | |||
Navigating the NINDS Mito CDE website
The Mito CDEs are available at the NINDS CDE Mito Website. A brief summary of the project, along with documents to assist in starting a study, are presented on the Data Standards tab of the Mito disease page. The Mito CDE recommendations are grouped by Domains and Sub-Domains below this introductory information. CRFs and their accompanying CDEs (listed as “CDE Details” on the website) can be downloaded as needed. Users are also able to learn more about the CDE project by navigating to the Learn menu at the top right of the page, where tutorials, a project overview and definitions are available (https://www.commondataelements.ninds.nih.gov/MITO.aspx#tab=Data_Standards).
WG results summary
Biomarkers
The Mito Biomarkers WG found that none of the reported biomarkers are consistently altered, nor have been used to assess mitochondrial disease in every study. For example, elevated lactic acid, commonly considered to be a biomarker of mitochondrial disease, is not consistently elevated in blood across all mitochondrial diseases, especially when only a single organ is involved (e.g., the eye in Leber’s hereditary optic neuropathy). Conversely, lactate levels in bold are frequently elevated in many pediatric mitochondrial diseases where the oxidative phosphorylation deficiency is often both severe and widespread. The WG also considered some parameters that are absolutely essential, but in only one disease. For example, thymidine and deoxyuridine levels along with thymidine phosphorylase enzymology are only useful in Mitochondrial neurogastrointestinal encephalopathy (MNGIE) syndrome, a very rare genetic disease caused by thymidine phosphorylase deficiency. The WG found the task of defining an extensive list of “specific” parameters difficult because of the high, and steadily increasing, number of different causes of mitochondrial diseases as well as a growing list of new biomarkers (e.g. FGF-21, GDF-15) (Suomalainen 2013; Yatsuga et al 2015). As a result, all biomarkers recommended by the WG are classified as Supplemental and were fully defined in a guidelines document to assist researchers in their studies.
Cognitive/behavioral/psychological outcomes
A broad range of clinical phenotypes is observed in both children and adults with mitochondrial disorders, with symptoms that may progressively develop and wax and wane in severity over the course of the entire lifespan. As a result, the instruments recommended vary according to age and the specific type of disorder. Furthermore, the scoring and use of the instruments may vary based on the clinical history. Modification of the scoring system for some of the instruments may be recommended to achieve higher sensitivity of symptom assessment (i.e., use of raw scores vs. standard scores). The WG also suggested considering the intended use of the instrument in any given study, such as a natural history to study changes over time in a clinical trial. The WG recommends selecting individual components of the larger tools to study in clinical trials. An example of this would be the use of memory or executive functioning subsets of the NIH Toolbox instead of using the whole instrument. While choosing instrument recommendations, the group did not encounter any tools unique to individuals with mitochondrial disorders and therefore there are no validated tools available for this population of patients.
Endocrinology/diabetes/gastrointestinal/nutrition
Diabetes mellitus, abnormal growth, gastrointestinal (GI) problems and nutrition-related concerns are important features of mitochondrial disease. This WG developed instruments focused on diabetes mellitus, anthropometric measurements, GI symptoms and nutritional assessment, including diet and use of dietary supplements. The WG acknowledged that the diversity of current approaches to treatment of mitochondrial disease in general and particular subtypes in particular, including nutrition and supplements, leads to difficulties in standardized documentation to enable ready evaluation of their potential impact. Given their importance to the overall health of affected individuals, careful assessment and study of endocrine and GI-related health conditions was recommended. The Office of Dietary Supplements at NIH organized a community meeting and subsequent working group dedicated to further investigation of the impact of nutritional interventions in primary mitochondrial diseases (Camp et al 2016).
Exercise physiology
The exercise physiology WG focused on the areas of exercise intolerance, endurance, and exercise recommendations. Exercise intolerance is one of the most prevalent symptoms of mitochondrial disease, especially in adults with mitochondrial myopathy. The WG did not address clinical recommendations on exercise as a treatment, but rather focused on recommended instruments to measure fatigue, exercise intolerance, and ability to exercise. The Borg Scale of Perceived Exertion, The Newcastle Pediatric Mitochondrial Disease Scale (NPMDS), echocardiogram, electrocardiogram, maximum and submaximum exercise testing, and pulmonary function testing were recommended as supplemental–highly recommended tests. The 6-minute walk test is widely used in clinical trials as a reflection of overall exercise capacity, and was considered to be a Supplemental test for mitochondrial disease evaluation as determined by evidence available to the exercise physiology WG.
Genetics
Inclusion in clinical trials increasingly requires the establishment of a known pathogenic mutation in a mitochondrial disease-associated gene using standard criteria. Thus, establishing an accurate molecular diagnosis is important for clinical trial participation. The genetics WG made a clear distinction between genetic testing for clinical purposes (i.e., in a CLIA-certified diagnostic laboratory) versus on a research basis. Genetic testing should be performed in an experienced laboratory with clearly defined analytic methodology. Depending on clinical presentation and exact methodology used, such analyses may include next generation sequencing of relevant nuclear genes in the form of gene panels and/or the exome, genome-wide SNP microarray analysis to detect nuclear chromosomal copy number alterations, and mitochondrial genome next generation sequencing to detect low level heteroplasmy for mitochondrial point mutations or small copy number alterations, real-time quantitative PCR to detect large mtDNA deletions and duplications, and/or quantitative PCR to measure mtDNA genome content in relevant tissues. The specific DNA variant identified should be documented relative to a reference sequence and using established ACMG guidelines (Richards et al 2015). If available, previous publications reporting pathogenicity of the variant should be cited (MacArthur et al 2014). Often this is performed through literature or database searches, using online tools such as MITOMAP (Lott et al 2013) and ClinVar (Harrison et al 2016). If the variant is novel, and prediction programs such as PolyPhen and SIFT are used, the specific versions and tools should be cited because, occasionally, multiple programs may issue conflicting predictions. Segregation studies within the family should be performed to confirm expected inheritance patterns. Biochemical studies should be performed in subjects’ blood, urine, cells, and/or tissue to confirm mitochondrial dysfunction type and degree, as needed or appropriate. In certain instances, in vitro cellular and/or animal model experiments might be needed to further evaluate the pathogenicity of novel variants of unknown significance.
Imaging
The imaging WG aimed to establish a data form that would be all-inclusive, ideally serving as a reference guide for what imaging data points might be useful to collect when researching mitochondrial disorders. While recommending many existing imaging CDEs, the WG highlighted several variables in the Mito imaging CRF that are characteristic of, although not exclusive, to mitochondrial disorders. The most notable elements are the involvement of deep gray nuclei, white matter tracts, and myelination pattern.
Neurological assessments
The spectrum of clinical manifestations of mitochondrial diseases spans every component of the nervous system (i.e., brain, spinal cord, peripheral nerves, autonomic nervous system, and muscle). The scales to capture a specific neurological disability vary in regard to their universal acceptance, sensitivity, complexity, and time-for-completion. As the CDEs are a project of the NINDS, and several other neurological diseases already had completed CDEs available for review, the Mito CDE WGs had the benefit of reviewing CDEs for clinically overlapping disorders including: Friedreich ataxia, epilepsy, amyotrophic lateral sclerosis, multiple sclerosis, Duchenne muscular dystrophy, stroke, spinal cord injury, and Huntington and Parkinson diseases. As many of the progressive neurological disorders have overlapping clinical symptoms, as well as secondary mitochondrial dysfunction contributing to their pathophysiology, many of the CRFs from these other neurological diseases were potentially applicable as Mito CDEs.
The WG recommended that the choice of scales for a specific patient or study should include those that best measure function for the identified disability, and possibly scales that would measure function from the pre-symptomatic state for expected disabilities that could be predicted to develop by the patient’s clinical diagnosis or genotype. The WG aimed to provide a battery of neurological tests that would capture small changes in cognitive function, development, motor weakness (muscle and nerve), coordination, and movement disorders (e.g., ataxia, dystonia).
Patient reported outcomes/quality of life (PRO/QoL)
The PRO/QoL WG found no QoL CDEs to be essential, and thus no instruments were classified as core. The recommended instruments had no differential application to subtypes of mitochondrial disease. However, the scales are often dependent on subject age for administration, and some are better suited for subjects with higher cognitive skills. Among several available QoL scales, the WG determined two that should be supplemental–highly recommended: the World Health Organization Quality of Life Assessment (WHOQOL) and the Pediatric Quality of Life Inventory (PEDSQL). Due to overall limitations, lack of a validated instrument, and the heterogeneity of mitochondrial diseases, it was difficult to define universal CDEs for QoL within mitochondrial diseases.
Vision
The vision WG recommendations were made to be applicable to all types of mitochondrial disease. Adequate training of physicians and technicians performing various ophthalmological tests with ongoing quality control were deemed essential. Several platforms were identified that are available for visual field perimetry and optical coherence tomography (OCT) imaging. The chosen tests will largely depend on the preference of the investigators and the specific facilities available in their respective study centers. The WG emphasized the need to ensure that the same platform and acquisition protocol are used across all the centers involved in a given study to allow for direct comparison and/or grouping of data at study conclusion. For visual electrophysiology, it was deemed imperative that testing be performed to incorporate the International Society for Clinical Electrophysiology of Vision (ISCEV) standards.
Discussion
The development of CDEs to facilitate mitochondrial disease research is timely, in view of the improvements and implementation of widespread genetic testing that has led to genetically-defined mitochondrial diseases and large patient cohorts. In addition, there has been a recent increase in candidate therapies proposed for these currently incurable disorders, with promising results in several preclinical studies in cell and animal models (Rahman 2015; Nightingale et al 2016). The CDE project is an international collaborative effort involving the many key stakeholders in mitochondrial medicine, including clinicians, translational and basic researchers, industry partners, patient advocacy groups, and the NIH. The breadth of our collaborators is a testament to the interest and need for these shared research tools to move the mitochondrial medicine field forward.
The WGs reviewed a total of 153 CRFs and instruments for the possible inclusion in the Mito CDEs. Interestingly, and perhaps not surprisingly, the WGs did not find a single core data element; demonstrating once again the challenges for clinical research in mitochondrial diseases.
The CDEs were categorized based on their prior use in both mitochondrial diseases or similar disorders and whether they were scientifically robust and clinically significant. Together, these CDEs cover almost the entire disease spectrum of these complex multi-systemic disorders, and have been developed with the intention of providing a publicly available resource to facilitate the design of protocols for any clinical study relating to mitochondrial disease. Some caveats should be noted. The Mito CDEs are suggested guidelines rather than definitive requirements for future study protocols and are not intended for use in clinical patient care. Clinical study design should take into account the specific diseases (considering both genotype and phenotype) under study, age group of affected patients and nature of the intervention, and incorporate the most relevant CDEs, in addition to any other outcome measures that the researchers consider appropriate. The Mito CDE recommendations are based on current knowledge of a rapidly expanding and changing group of heterogeneous disorders, and it is anticipated that, while having a stable set of essential elements, these will be dynamic and need to be updated as the field advances and specific instruments become validated for use in mitochondrial disease. The NINDS has developed oversight committees that will review feedback from the community and adjust the CDEs periodically, as needed.
The availability of a global set of CDEs addressing many of the multi-systemic features should help serve as a starting place to harmonize data collection and ultimately enable the combination and comparison of outcomes of clinical studies in mitochondrial diseases. Given the diversity and spectrum of clinical manifestations seen in these disorders, selection of outcome measures for clinical research presents a unique challenge. These difficulties are further compounded by the rarity of each individual genetic entity of this group of disorders, challenges to robust study design, and need for substantive funding. It is thus imperative that patient support groups, together with mitochondrial disease clinician networks and research consortia, partner with industry and the FDA to validate these measures, recently discussed at a Critical Path Innovation Meeting (CPIM) (https://ods.od.nih.gov/attachments/CriticalPathInnovationMeetingSummary.pdf). The international collaboration on this CDE project has continued to be fruitful, with projects ongoing to harmonize global patient registries, to validate patient-centered outcome measures, and to create mitochondrial disease specific outcome measures inspired by the CDEs reviewed in this project.
Acknowledgements
The views expressed here are those of the authors and do not represent those of the NIH, NINDS or the US Government. Logistic support for this project was provided in part through NIH Contract HHSN271201200034C. The development of the NINDS Mito CDEs was made possible thanks to the great investment of time and effort of WG members and the members of the NINDS CDE Project team participating from 2013 to 2015.
MITO Working Group Members
Biomarkers WG
John Shoffner, MD (Chair), Atlanta, GA, USA
Mark Bamberger, PhD, Stealth Peptides, Inc, Newton Centre, MA, USA
Yasutoshi Koga, MD, PhD, Kurume University, Fukuoka-ken, Japan
Anne Lombès, MD, PhD, INSERM, Paris, France
Fernando Scaglia, MD, Baylor College of Medicine, Houston, TX, USA
Cognitive/Behavioral/Psychological Outcomes WG
Lisa Emrick, MD (Chair), Baylor College of Medicine/Texans Children’s Hospital, Houston, TX, USA
Rebecca Vaurio, PhD, Kennedy Krieger Institute, Baltimore, MD, USA
Endocrinology/Diabetes/GI/Nutrition WG
Shana McCormack, MD (Chair), The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
J.E. Abdenur, MD, CHOC Children’s Hospital, Orange, CA, USA
Kristin Fiorino, MD, Children’s Hospital of Philadelphia, Philadelphia, PA, USA (ad hoc member)
Bret Goodpaster, PhD, Sanford, Burnham Medical Research Institute, Orlando, FL, USA
Robert O. Heuckeroth, MD PhD, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Kierstin Keller, CGC, Children’s Hospital of Philadelphia, Philadelphia, PA, USA (ad hoc member)
Robert McFarland, DoH, HEFCE, Newcastle University, UK
Philip Morgan, MD, Mitochondrial Research Guild, Seattle, WA, USA
Fernando Scaglia, MD, Baylor College of Medicine, Houston, TX, USA
Charles A. Stanley, MD, Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Steven M. Willi, MD, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Exercise Physiology WG
Tanja Taivassalo, PhD (Chair), McGill University, Montreal, QC, Canada
Amy Goldstein, MD, Children’s Hospital of Pittsburgh, Pittsburgh, PA, USA
Ronald Haller, MD, Southwestern Medical Center, Dallas, TX, USA
Mark Tarnopolsky, MD, PhD, McMaster University, Hamilton, Ontario, Canada
Karim Wahbi, MD, PhD, Institute of Myology, Paris, France
Genetics WG
David Dimmock, MD (Co-Chair), Medical College of Wisconsin, Milwaukee, Wisconsin, USA
Marn J. Falk, MD (Co-Chair), The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Xiaowu Gai, PhD, Ocular Genomics Institute
Michio Hirano, MD, Columbia University Medical Center
Anne Lombès, MD, PhD, INSERM, Paris, France
Shamima Rahman, FRCP, FRCPCH, PhD, UCL Great Ormond Street Institute of Child Health, London, UK
Fernando Scaglia, MD, Baylor College of Medicine, Houston, TX, USA
Curt Scharfe, MD, PhD, Stanford University School of Medicine, Stanford, CA, USA
David Thorburn, PhD, FHGSA, FFSC (RCPA), Murdoch Children’s Research Institute, Victoria, Australia
Stephan Züchner, MD, PhD, Dr. John T. Macdonald Foundation, Miami, FL, USA
Imaging WG
David M. Mirsky, MD (Chair), Children’s Hospital Colorado, Aurora, CO, USA
Abigail Collins, MD, Children’s Hospital Colorado, Aurora, CO, USA
Andrea L. Gropman, MD, Children’s National Medical Center, Washington, DC, USA
Edward Yang, MD, Boston Children’s Hospital, Boston, MA, USA
Neurological Assessments WG
Bruce Cohen, MD, FAAN (Co-Chair), Akron Children’s Hospital, Akron, OH, USA
Ingrid Tein, MD, FRCP (Co-Chair), University of Toronto, ON, Canada
Laurence Bindoff, MD, PhD, University of Bergen and Haukeland
Michio Hirano, MD, Columbia University Medical Center
Matthew Klein, MD, MS, FACS, Edison Pharmaceuticals
Thomas Klopstock, MD, Friedrich-Baur-Institute, Munich, Germany
Saskia Koene, MD, Radboud University Nijmegen Medical Centre, Netherlands
Pascal Laforêt, MD, PhD, Groupe Hospitalier Pitié-Salpêtrière Assistance Publique-Hôpitaux de Paris, Paris, France
Margherita Milone, MD, PhD, Mayo School of Graduate Medical Education, Rochester, MN, USA
Jan Smeitink, MD, PhD, Radboud University Nijmegen Medical Centre, Netherlands
Patient-Reported Outcomes QoL WG
Mary Kay Koenig, MD (Co-Chair), University of Texas Health Science Center, Houston, TX, USA
Sumit Parikh, MD (Co-Chair), Cleveland Clinic, Cleveland, OH, USA
Cristy Balcells, RN MSN, Mito Action, Boston, MA, USA
Michio Hirano, MD, Columbia University Medical Center
Amel Karaa, MD, Massachusetts General Hospital, Boston, MA, USA
Robert McFarland, DoH, HEFCE, Newcastle University, UK
Russel Saneto, DO, Seattle Children’s Hospital, Seattle, WA, USA
Peter W. Stacpoole, MD, PhD, University of Florida, Gainesville, FL, USA
Philip Yeske, PhD, United Mitochondrial Disease Foundation
Vision WG
Patrick Yu-Wai-Man, MD, PhD, FRCPath, FRCOphth (Chair), Newcastle University and Moorfields Eye Hospital, UK
Piero Barboni, MD, University of Bologna, Bologna, Italy
Valerio Carelli, MD, PhD, University of Bologna School of Medicine, Bologna, Italy
Patrick Chinnery, MD, PhD, FRCPath, FRCP, University of Cambridge, UK
Graham E. Holder, PhD, Moorfields Eye Hospital, London, UK
John L. Keltner, MD, UC Davis School of Medicine, Sacramento, CA, USA
Tony Moore, MA, FRCS, FCROphth, FMedSci, Ophthalmology UCSF School of Medicine, San Francisco, CA
Alfredo A. Sadun, MD, PhD, Doheny Eye Institute/UCLA, Los Angeles, CA, USA
Claire Sheldon, MD, PhD, University of British Columbia, Vancouver, Canada
Marcela Votruba, MD, PhD, FRCOphth, Cardiff University, UK
NINDS CDE Team
Joanne Odenkirchen, MPH, NINDS CDE Project Officer, National Institutes of Health/National Institute of Neurological Disorders and Stroke, (NIH/NINDS), Bethesda, MD, USA
Vernon Anderson, PhD
Kathryn Camp MS, RD, Consultant to the NIH/Office of Dietary Supplements (ODS), Bethesda, MD, USA
William C. Copeland, PhD, National Institute of Environmental Health Sciences (NIEHS), Research Triangle Park, NC, USA
Kerry Goetz, MS, National Eye Institute (NEI)/NIH, Bethesda, MD, USA
Katrina A Gwinn, MD, NINDS-NIH, Bethesda, MD, USA
Petra Kaufmann, MD, MSc, National Center for Advancing Translational Sciences (NCATS)/ NIH, Bethesda, MD, USA
Danuta Krotoski, PhD, Eunice Kennedy Shriver NICHD/NIH, Bethesda, MD, USA
Lynne A. Wolfe, MS, PNP, BC, Undiagnosed Disease Program, NHGRI/NIH, Bethesda, MD, USA
Steven Zullo, PhD, NIH/NIBIB, Bethesda, MD, USA
Sherita Ala’i-Hansen, MS, The Emmes Corporation, Rockville, MD, USA
Muniza K. Sheikh, MS, MBA, The Emmes Corporation, Rockville, MD, USA
Footnotes
Conflict of interest John Shoffner has received research funds from the Department of Defense and owns stock in Medical Neurogenetics, LLC.
David Dimmock is a consultant to Audentes Therapeutics, Biomarin, Ilumina and Complete Genomics and involved in clinical trials with Genzyme/Sanofi, Biomarin, Shire, Cytonet.
Marni Falk has been a consultant to Mitokyne; received research funds from Cardero Therapeutics, Mitobridge, Raptor Pharmaceuticals, RiboNova, Stealth BioTherapeutics, and Vitaflo International; was a former scientific advisory board member for Perlstein Laboratory, Inc., and is involved in clinical trials with REATA pharmaceuticals.
Amel Karaa, Shamima Rahman, Anne Lombès, Patrick Yu-Wai-Man, Muniza K. Sheikh, Sherita Alai-Hansen, Bruce H. Cohen, Lisa Emrick, Shana McCormack, David Mirsky, Tony Moore, Sumit Parikh, Tanja Taivassalo, Mark Tarnopolsky, Ingrid Tein, Joanne C. Odenkirchen, and Amy Goldstein have no conflict of interest.
Studies with human or animal This article does not contain any studies with human or animal subjects performed by the any of the authors.
References
- Camp KM, Krotoski D, Parisi MA, Gwinn KA, Cohen BH, Cox CS, Enns GM, Falk MJ, Goldstein AC, Gopal-Srivastava R, Gorman GS, Hersh SP, Hirano M, Hoffman FA, Karaa A, MacLeod EL, McFarland R, Mohan C, Mulberg AE, Odenkirchen JC, Parikh S, Rutherford PJ, Suggs-Anderson SK, Tang WH, Vockley J, Wolfe LA, Yannicelli S, Yeske PE, Coates PM (2016) Nutritional interventions in primary mitochondrial disorders: developing an evidence base. Mol Genet Metab 119(3): 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery PF (2014) Mitochondrial disorders overview In: Pagon PA, Adam MP, Ardinger HH et al. (eds) Gene reviews. University of Washington, Seattle [Google Scholar]
- DiMauro S, Schon E (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348:2656–2668 [DOI] [PubMed] [Google Scholar]
- Gorman GS, Schaefer AM, Ng Y et al. (2015) Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol 77:753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, Suomalainen A, Thorburn DR, Zeviani M, Turnbull DM (2016) Mitochondrial diseases. Nat Rev Dis Primers. doi: 10.1038/nrdp.2016.80 [DOI] [PubMed] [Google Scholar]
- Grinnon ST, Miller K, Marler JR, Lu Y, Stout A, Odenkirchen J, Kunitz S (2012) National Institute of Neurological Disorders and Stroke Common Data Element Project - approach and methods. Clin Trials 9:322–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SM, Riggs ER, Maglott DR, Lee JM, Azzariti DR, Niehaus A, Ramos EM, Martin CL, Landrum MJ, Rehm HL (2016) Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet 89:8.16.1–8.16.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC (2013) mtDNA variation and analysis using mitomap and mitomaster. Curr Protoc Bioinformatics 44: 1.23.1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP (2014) Guidelines for investigating causality of sequence variants in human disease. Nature 24 508(7497):469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale H, Pfeffer G, Bargiela D, Horvath R, Chinnery PF (2016) Emerging therapies for mitochondrial disorders. Brain 139:1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyazov D, Kahler S, Frye R (2016) Primary mitochondrial disease and secondary mitochondrial dysfunction: importance of distinction for diagnosis and treatment. Mol Syndromol 7:122–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh S, Goldstein A, Koenig MK et al. (2015) Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med 17:689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S (2015) Emerging aspects of treatment in mitochondrial disorders. J Inherit Metab Dis 38:641–653 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A (2013) Fibroblast growth factor 21: a novel biomarker for human muscle-manifesting mitochondrial disorders. Expert Opin Med Diagn 7:313–317 [DOI] [PubMed] [Google Scholar]
- Yatsuga S, Fujita Y, Ishii A et al. (2015) Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann Neurol 7:814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]