Abstract
To investigate the larvicidal activities of novel anthraquinones (1a-1k) against Culex quinquefasciatus mosquito larvae. Novel anthraquinones (1a-1k) derivatives were synthesis via condensation method. The compounds were confirmed through FT-IR spectroscopy, 1H & 13C NMR spectrum, and mass spectral studies. The larvicidal activity of compound 1c was highly active LD50 20.92 µg/mL against Culex quinquefasciatus compared standard permethrin with LD50 25.49 µg/mL. Molecular docking studies were carried out for compound 1c against Odorant-binding protein of Culex quinquefasciatus. The compound 1c (−9.8 Kcal/mol) was a potent larvicide with more binding energy than control permethrin (−9.7 Kcal/mol). Therefore, compound (1c) may be more significant inhibitors of mosquito larvicidal.
Keywords: Anthraquinone, Culex quinquefasciatus, Larvicidal activity, Molecular docking, Odorant-binding protein (OBP)
1. Introduction
Mosquitoes are disreputable as the main vectors for the spread a number of diseases, such as malaria, dengue fever, schistosomiasis Japanese encephalitis, filariasis, and yellow fever (Georges et al., 2008, Govindarajan, 2010). These types of disease reason an affect the economic and social impact in all over the word, especially Culex quinquefasciatus is vectors which are obtained frequently from urban and rural human habitat. Control of mosquito has been used big challenge and currently using most effective mosquito inhibitors such as organophosphates, fenthion, chlorpyrifos, menthoprene, and temephos, then usage of this big challenge in various environmental condition. In this reason, we selected anthraquinones, which is most suitable environmental safe secondary metabolites. Anthraquinones analogues are a large group of pigmented polyketides extensively formed by fungi. Basically, different substituted anthraquinone derivatives are most active in biological systems, such as the addition of methyl (—CH3), carboxyl (—COOH), hydroxyl (—OH), and methoxyl (—OCH3) groups of 9,10-anthracenedione outcomes in a broad-spectrum of medicinal properties (Tutin et al., 1911). Fig. 1 shows important larvicidal active compounds that based on structure, relation of target compounds. Larvicidal activities of benzoquinone (LC50: 90 μg/mL) were evaluated on third-instar larvae of A. aegypti (De Sousa et al., 2010). 2-methoxy-1,4-naphthoquinone (LC50: 0.085 μg/mL) against Estagio larval and isolated from Balsaminaceae (Impatiens glandulifera), (Kim and Ahn, 2017). Naphthalene-1,4-dione (LC50: 1.64 μg/mL) against Culex pipiens pallens, (Jeon et al., 2015), anthracence-9,10-dione (LC50: >25.0 μg/mL) (A. aegypti), tectoquinone LC50: 3.3 µg/mL (A. aegypti), and emodin LC50: 5.3 µg/mL (A. aegypti) (Cheng et al., 2008). However, the above chemicals are problems for usage of environmental factor, such as widespread development of resistance, leading to occurrences of mosquito species, disrupted natural biological control systems, and effects of infection from soil, water, air (Park et al., 2005). Therefore, the above drawback requires the new selective control of mosquito larvae (Yang et al., 2013). Odorant-binding proteins were transporting the odorants to olfactory receptors, which plays in major activities of host-seeking (Bazaes et al., 2013, De March and Golebiowski, 2014, Pechlaner and Oostenbrink, 2015). The above scientific information, we have chosen a hydroxyanthraquinone target against Culex quinquefasciatus and using odorant-binding protein (PDB ID: 3OGN) for molecular docking studies. In the present study, synthesis of new hydroxyanthraquinone Mannich base derivatives for evaluation of larvicidal activity.
Fig. 1.
Some important Larvicidal active compounds.
2. Materials and methods
2.1. Chemistry
The chemicals were obtained from commercially and fully purified all chemicals before using the reactions. The FT-IR Shimadzu 8201PC (4000–400 cm−1), and 1H & 13C NMR spectra of Bruker DRX-300 MHz were used by analysis all newly synthesized compounds. Thin layer chromatography (TLC) technique was used by check purity of the compounds with using silica gel plates.
2.1.1. General method for preparation of 4,11-dihydroxy-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoin dole-5,10-dione (1a).
A mixture of 1,4-dihydroxy anthraquinone (0.0005 mol, 0.1 mg), benzaldehyde (0.002 mol, 0.5 ml) and aniline (0.001 mol, 0.5 ml) are dissolved in ethanol. The mixture was reflexes by 24 h at 60 °C. The final target compound was monitored by TLC. The product was recrystallized in suitable alcohol. The same experimental method was used for preparation of other compounds 1b-k.
2.1.2. 4,11-dihydroxy-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10-dione (1a)
IR (kBr, cm−1): 1714, 1691, 1462, 822, 752; 1H NMR (300 MHz): δ ppm 8.37–8.33 (d, J = 13.74 Hz, 2H), 7.82–7.80 (d, J = 13.74 Hz, 2H), 7.21–7.18 (dd, J = 7.72 Hz, J = 7.75 Hz, 2H), 6.95–6.93 (d, J = 7.75 Hz, 2H), 6.79–6.77 (d, J = 7.75 Hz, 1H), 5.30(s, 2H), 4.61 (s, 4H); 13C NMR (75 MHz): δ 187.9, 152.5, 149.1, 133.0, 132.9, 129.0, 128.2, 126.1, 114.6, 114.1, 121.4, 54.9; EI-MS m/z(rel.int): 358.4 (M+, 26%); Anal C22H15NO4: C,73.94; H,4.23; N,3.92; Found: C, 73.92; H, 4.26; N, 3.91;
2.1.3. 4,11-dihydroxy-2-phenyl-1,3-di((E)-prop-1-en-1-yl)-2,3-dihydro-1H-naphtho[2,3-f]isoin dole-5,10-dione (1b)
IR (kBr, cm−1): 1762, 1689, 1452, 812, 741; 1H NMR (300 MHz): δ 8.38–8.36 (d, J = 13.74 Hz, 2H), 7.82–7.80 (d, J = 13.74 Hz, 2H), 7.21–7.18 (dd, J = 7.72 Hz, J = 7.75 Hz, 2H), 6.95–6.93 (d, J = 7.75 Hz, 2H), 6.77–6.75 (d, J = 7.75 Hz, 1H), 6.10(s, 2H), 5.48–5.46 (q, 2H), 5.36 (s, 2H), 4.49 (s, 2H), 2.12(d, 6H); 13C NMR (75 MHz, δ (ppm)): 187.8, 152.4, 148.9, 133.9, 132.7, 129.1, 128.8, 125.6, 125.0, 124.3, 122.1, 115.8, 115.1, 72.6, 17.5; EI-MS m/z(rel.int): 438.0 [M+, 26%); Anal C28H23NO4: C,76.87; H,5.30; N,3.20; Found: C, 76.88; H, 5.32; N, 3.21;
2.1.4. 4,11-dihydroxy-1-((E)-4-methylpenta-1,3-dien-1-yl)-3-((Z)-4-methylpenta-1,3-dien-1-yl)-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10-dione (1c)
IR (kBr, cm−1): 1744, 1678, 1451, 816, 738; 1H NMR (300 MHz): δ 8.31–8.28 (d, J = 13.04 Hz, 2H), 7.80–7.78 (d, J = 13.04 Hz, 2H), 7.26–7.22 (dd, J = 7.70 Hz, 2H), 6.95–6.93 (d, J = 7.70 Hz, 2H), 6.76–6.75(d, J = 7.71 Hz, 1H), 6.20(s, 2H), 6.10(d, 2H), 5.90(d, 2H), 5.34(s, 2H), 4.69 (s, 2H), 2.14(s, 6H), 1.98(s, 6H); 13C NMR (75 MHz, DMSO‑d6, δ (ppm)): 187.5, 151.6, 148.5, 135.9, 133.2, 132.5, 129.5, 128.9, 128.2, 128.0, 126.3, 126.1, 122.3, 113.9, 114.1, 70.6, 27.6, 20.5 ; EI-MS m/z(rel.int): 518.25 (M+, 35%); Anal C34H31NO4: C,78.88; H,6.04; N, 2.71; Found: C, 78.86; H, 6.08; N, 2.70;
2.1.5. 4,11-dihydroxy-2-phenyl-1,3-di((E)-styryl)-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10 –dione (1d)
IR (kBr, cm−1): 1769, 1674, 1462, 812, 738; 1H NMR(300 MHz): δ 8.32–8.30 (d, J = 13.70 Hz, 2H), 7.86–7.84 (d, J = 13.72 Hz, 2H), 7.42–7.44(m, Ph), 7.26–7.24 (dd, J = 7.70 Hz, 2H), 6.92–6.87 (d, J = 7.70 Hz, 2H), 6.69–6.67 (d, J = 7.70 Hz, 1H), 5.32 (s, 2H), 4.59 (s, 2H), 6.69(s, 2H),6.21(s, 2H); 13C NMR (75 MHz, δ (ppm)): 187.9, 152.5, 149.1, 138.5, 132.9, 132.7, 129.0, 128.5, 128.2, 126.1, 121.4, 114.6, 114.1, 125.9, 71.2, 124.2, 128.9, 54.9; EI-MS m/z(rel.int): 562.21(M+, 45%); 44(18); Anal C38H27NO4: C,81.27; H,4.85; N,2.49; Found: C, 81.25; H, 4.81; N, 2.48;
2.1.6. 1,3-di(furan-2-yl)-4,11-dihydroxy-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10 –dione (1e)
IR (kBr, cm−1): 1725, 1681, 1461, 823, 712; 1H NMR (300 MHz): δ 8.39–8.36 (d, J = 13.74 Hz, 2H), 7.86–7.82 (d, J = 13.74 Hz, 2H), 7.62(d, J = 7.62 Hz, 2H), 7.22–7.20 (dd, J = 7.72 Hz, 2H), 6.94–6.91 (d, J = 7.75 Hz, 2H), 6.71–6.68 (d, J = 7.75 Hz, 1H), 6.25–6.21(d, J = 7.62 Hz, 2H), 6.45–6.42(d, J = 7.62 Hz, 2H), 5.38(s, 2H), 5.36 (s, 2H); 13C NMR(75 MHz, δ (ppm)): 187.4, 151.9, 150.6, 147.9, 141.0, 135.6, 131.9, 128.2, 127.9, 127.8, 122.6, 115.7, 115.2, 109.1, 67.1, 53.7; EI-MS m/z(rel.int): 490(M+, 78%); Anal C30H19NO6: C,73.61; H, 3.91; N, 2.86; Found: C, 73.60; H, 3.90; N, 2.84;
2.1.7. 4,11-dihydroxy-1,2,3-triphenyl-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10-dione (1f)
IR (kBr, cm−1): 1747, 1697, 1455, 809, 744; 1H NMR(300 MHz): δ 8.40–8.36(d, J = 13.72 Hz, 2H), 7.85–7.83 (d, J = 13.72 Hz, 2H), 7.36–7.28(m 10H, Ph), 7.26–7.23 (dd, J = 7.72 Hz, 2H), 6.99–6.97 (d, J = 7.71 Hz, 2H), 6.78–6.75 (d, J = 7.71 Hz, 1H), 5.36 (s, 2H), 5.22 (s, 2H); 13C NMR (75 MHz, δ (ppm)): 187.2, 153.6, 149.8, 146.2, 133.6, 132.2, 132.9, 130.0, 128.9, 125.6, 128.9, 126.2, 122.6, 114.5, 113.8, 71.6; EIMS m/z(rel.int): 510.23(M+, 56%); Anal C34H23NO4: C, 80.14; H,4.55; N,2.75; Found: C, 80.15; H, 4.56; N, 2.76;
2.1.8. 1,3-bis(4-chlorophenyl)-4,11-dihydroxy-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoin do le-5,10-dione (1g)
IR (kBr, cm−1): 1741, 1641, 1462, 821, 765, 659; 1H NMR (300 MHz): δ 8.34–8.30 (d, J = 13.69 Hz, 2H), 7.86–7.81 (d, J = 13.69 Hz, 2H), 7.33–7.30(d, J = 7.69 Hz, 4H), 7.24–7.21(dd, J = 7.72 Hz, 2H), 7.20–7.18(d, J = 7.69 Hz, 4H), 6.97–6.91(d, J = 7.72 Hz, 2H), 6.71–6.69(d, J = 7.72 Hz, 1H), 5.39(s, 2H), 5.12(s, 2H); 13C NMR (75 MHz, δ (ppm)): 187.0, 153.6, 147.8, 140.4, 133.5, 132.9, 132.2, 128.6, 128.0, 127.9, 127.3, 125.2, 121.2, 115.8, 113.7, 71.0; EI-MS m/z(rel.int): 578.23 (M+, 47%); Anal C34H21Cl2NO4: C, 70.60; H, 3.66; N,2.42; Found: C, 71.25; H, 3.67; N, 3.68;
2.1.9. 4,11-dihydroxy-1,3-bis(4-hydroxyphenyl)-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoind ole-5,10-dione (1h)
IR (kBr, cm−1): 3232, 1749, 1680, 1461, 810, 741; 1H NMR(300 MHz): δ 8.43–8.40 (d, J = 13.63 Hz, 2H), 7.81–7.78 (d, J = 13.63 Hz, 2H), 7.24–7.22 (dd, J = 7.74 Hz, J = 7.72 Hz, 2H), 7.10–7.08(d, J = 7.70 Hz, 4H), 6.96–6.94 (d, J = 7.74 Hz, 2H), 6.73–6.70 (d, J = 7.72 Hz, 1H), 6.68–6.63(d, J = 7.70 Hz, 4H), 5.42 (s, 2H), 5.25 (s, 2H); 13C NMR (75 MHz, δ (ppm)): 187.9, 157.2, 152.5, 149.1, 135.9, 132.9, 132.2, 128.9, 128.6, 128.2, 126.0, 122.9, 118.6, 113.9, 114.1, 71.2; EIMS m/z(rel.int): 542.12 (M+, 22%); Anal C34H23NO6: C, 75.41; H,4.28; N,2.59; Found: C, 75.40; H, 4.21; N, 2.55;
2.1.10. 4,11-dihydroxy-1,3-bis(3-nitrophenyl)-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]isoindole-5,10-dione (1i)
IR (kBr, cm−1): 1752, 1684, 1545, 1465, 804, 745; 1H NMR(300 MHz): δ 8.40–8.38 (d, J = 13.78 Hz, 2H), 8.12(d, J = 7.70 Hz, 4H), 7.90–7.87 (d, J = 13.78 Hz, 2H), 7.58–7.56(d, J = 7.70 Hz, 4H), 7.26–7.24 (dd, J = 7.72 Hz, J = 7.75 Hz, 2H), 6.95–6.93 (d, J = 7.75 Hz, 2H), 6.83–6.80 (d, J = 7.75 Hz, 1H), 5.38 (s, 2H), 5.26 (s, 2H); 13C NMR (75 MHz, δ (ppm)): 187.1, 152.5, 149.1, 146.8, 142.9, 135.8, 133.3, 133.0, 131.1, 129.0, 127.9, 126.5, 126.1, 122.9, 121.4, 114.1, 113.9, 68.5; EIMS m/z(rel.int): 600.21 (M+, 36%); Anal C34H21N3O8: C, 68.11; H,3.53; N,7.01; Found: C, 68.17; H, 3.57; N, 7.18;
2.1.11. 1,3-bis(4-(dimethylamino)phenyl)-4,11-dihydroxy-2-phenyl-2,3-dihydro-1H-naphth o [2, 3-f]isoindole-5,10-dione (1j)
IR (kBr, cm−1): 1753, 1681, 1469, 795, 745; 1H NMR(300 MHz): δ 8.37–8.34 (d, J = 13.74 Hz, 2H), 7.82–7.80 (d, J = 13.74 Hz, 2H), 7.23–7.19 (dd, J = 7.72 Hz, 2H), 7.08–7.05(d, J = 7.75 Hz, 4H), 6.95–6.93 (d, J = 7.75 Hz, 2H), 6.79–6.76 (d, J = 7.75 Hz, 1H), 6.68–6.62(d, J = 7.75 Hz, 4H), 5.29(s, 2H), 5.22 (s, 2H), 3.04(s, 12H); 13C NMR (75 MHz, δ (ppm)): 187.9, 152.5, 149.1, 148.9, 133.2, 133.1, 132.9, 128.0, 128.6, 128.2, 125.9, 121.4, 113.8, 114.1, 114.1, 70.2, 42.3; EIMS m/z(rel.int): 596.32 (M+, 20%); Anal C38H33N3O4: C, 76.62; H,5.58; N,7.05; Found: C,76.63; H, 5.59; N, 7.09;
2.1.12. 4,11-dihydroxy-1,3-bis(4-methoxyphenyl)-2-phenyl-2,3-dihydro-1H-naphtho[2,3-f]iso in dole-5,10-dione (1k)
IR (kBr, cm−1): 2821, 1742, 1682, 1469, 812, 740; 1H NMR (300 MHz): δ 8.56–8.41 (d, J = 13.70 Hz, 2H), 7.89–7.86 (d, J = 13.65 Hz, 2H), 7.22–7.19(dd, J = 6.65 Hz, J = 7.71 Hz, 2H), 7.16(4H, CH), 6.90–6.88 (d, J = 7.71 Hz, 2H), 6.75–6.73 (d, J = 7.71 Hz, 1H), 6.69(4H, CH), 5.27(s, 2H), 5.15 (s, 2H), 3.85(s, 6H); 13C NMR (75 MHz, δ (ppm)): 187.1, 159.8, 153.0, 148.9, 136.8, 133.2, 132.8, 129.3, 129.1, 128.1, 127.3, 122.2, 116.1, 114.2, 115.6, 70.5, 55.9; EIMS m/z(rel.int): 570.10 (M+, 30%); Anal C36H27NO6: C, 75.91; H, 4.78; N, 2.46; Found: C, 75.05; H, 4.77; N, 2.45;
2.2. Biological activity
2.2.1. Larvicidal activity
Larvicidal activities of 10, 25, 50 and 100 µg/mL of compounds (1a-1k) were screened as we previously described in publication Idhayadhulla et al., (SathishKumar et al., 2020). Mortality caused by the compounds was evaluated as ratios (%) of the numbers of dead vs. live larvae. The 50% lethal doses (LD50) values of the compounds were calculated using probit analysis and statistically analyzed using SPSS version 16.0 software.
2.2.2. Statistical analysis
Larvicidal activities results were calculated through 3 independent evaluations and Microsoft Excel was used to analysis the standard deviations (SD) of each compound.
2.2.3. Molecular docking
This study was carried out via Autodock vina 1.1.2. (Trott and Olson, 2010), which using to interpret the binding mode of compounds (1c) and permethrin with mosquito odorant protein. Crystal structure of mosquito odorant binding protein (PDB ID: 3OGN) was collected from protein data bank web link. The 3D association of the compound (1c) and permethrin was accomplished through Chem Draw Ultra 12.0 software. The 3OGN protein was fixed at center_x: 18.681, center_y: 49.66, and center_z: 11.409 with size_x: 22, size_y: 20, and size_z: 22 with spacing of 1.0 Å. Discovery studio 2019 program was used for analysis visually of results compared with the least binding affinity value of docking compounds.
3. Result and discussion
3.1. Chemistry
The compound 1a-1k was synthesized from anthraquinone reached with aldehyde and primary amine in the ethanol medium by condensation method. The mixture was reflexed by 24 hr at 60 °C. The final products were obtained yield between 80 and 89 %. The method of preparation was outlined in scheme 1. Structures 1a-1k were definite via FT-IR, 1H and 13C NMR, mass spectral studies, the importance of the IR spectral peak at C—N—C, C O, OH corresponding to the average peak at 1641–1697, 174–1769, and 1451–1469 cm−1 respectively. 1H NMR spectral obtained important proton peaks range between δ 4.49–5.26, and 5.32–5.42 ppm conforming to the protons HC-N, and OH respectively. The 13C NMR carbon peaks were obtained the range of value between at δ 54.9–72.6, 187.0–187.9, and 151.6–153.6 conforming to the C—N—C, C O, and C—OH carbons respectively. All compounds were conformed the mass values according to the molecular ion peak obtain in mass spectral values.
Scheme 1.
Route of synthesis larvicidal active target molecules.
3.2. Larvicidal activity
Larvicidal activity was screened for all synthesized anthraquinones (1a-1k) derivatives against second instar C. quinquefasciatus larvae. Compound 1c exerted more larvicidal activity (LD50 20.92 µg/mL) than other compounds and standard permethrin LD50 25.49 µg/mL. Compounds 1a, 1f, 1h, 1i, and 1k were less active against C. quinquefasciatus with LD50 values of above >100 µg/mL LD50 value. All values are represented in Table 1.
Table 1.
Larvicidal activity of anthraquinone analogues (1a-1k) and permethrin.
| Compounds | Concentration (µg/mL)/Mortality (%) |
LD50 (µg/mL) | |||
|---|---|---|---|---|---|
| 10 | 25 | 50 | 100 | ||
| 1a | – | 0 ± 0.00 | 16 ± 0.27 | 36 ± 0.97 | >100 |
| 1b | 7 ± 0.89 | 23 ± 1.25 | 36 ± 0.96 | 55 ± 0.00 | 85.42 |
| 1c | 41 ± 0.00 | 65 ± 1.31 | 100 ± 1.34 | – | 20.92 |
| 1d | 5 ± 1.76 | 22 ± 1.12 | 43 ± 1.87 | 60 ± 1.61 | 74.17 |
| 1e | 4 ± 1.14 | 15 ± 1.48 | 38 ± 1.46 | 54 ± 0.88 | 86.27 |
| 1f | – | – | 0 ± 0.00 | 20 ± 0.47 | >100 |
| 1g | 22 ± 1.87 | 46 ± 1.21 | 62 ± 0.56 | 88 ± 0.30 | 37.95 |
| 1h | 0 ± 0.00 | 10 ± 1.87 | 20 ± 0.67 | 40 ± 1.89 | >100 |
| 1i | – | – | 21 ± 0.00 | 38 ± 0.09 | >100 |
| 1j | 22 ± 0.95 | 36 ± 1.65 | 57 ± 1.41 | 100 ± 1.14 | 37.59 |
| 1k | – | – | 0 ± 0.00 | 40 ± 1.23 | >100 |
| Permethrin | 23 ± 1.76 | 55 ± 1.23 | 82 ± 1.94 | 100 ± 0.00 | 25.49 |
aValues are the means of three replicates ± SD.
3.3. Docked results with AutoDock Vina
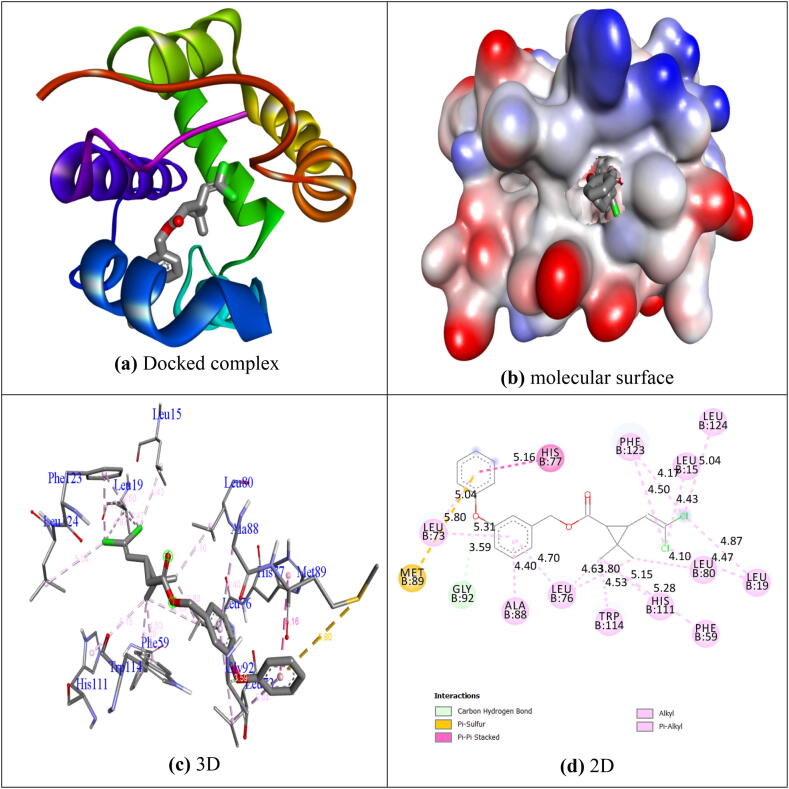
The Autodock Vina program was used to study for compound (1c) and permethrin docking with 3OGN protein. The compound 1c shows significant binding affinity (−9.8 kcal/mol) than permethrin (−9.7 kcal/mol). Stability of protein and ligand was confirmed through the bond distance calculation, which is less than 3.5 Å form H-donor and the H-acceptor of bond distance (Taha et al., 2015). The compound 1c was not conceded for any hydrogen bond in 3OGN. The residues Ala18, Leu19, Leu22, Ala62, Lys63, Val64, Lys75 and Pro81 were complex with hydrophobic connections. The molecular interaction of compound 1c and 3OGN were shown in Fig. 2. The control permethrin was also not conceded any hydrogen bond in 3OGN. The hydrophobic interactions were involved due to the formation of Leu15, Leu19, Phe59, Leu73, Leu76, His77, Leu80, Ala88, Met89, Gly92, His111, Trp114, Phe123, and Leu124. Fig. 3 shows that the molecular interaction of permethrin with 3OGN. Therefore, the compound 1c having remarkable inhibition capability than permethrin in mosquito odorant-binding protein. The results are presented in Table 2.
Fig. 2.
Molecular docked modes of 1c with binding site of 3OGN.
Fig. 3.
Molecular docked modes of permethrin with binding site of 3OGN protein.
Table 2.
Molecular docking interaction of compound 1c and control permethrin.
| Comp. No. | Mosquito odorant-binding protein 3OGN |
||
|---|---|---|---|
| Binding affinity (kcal/mol) | No. of H-bonds | H-bonding residues | |
| 1c | −9.8 | 0 | – |
| Permethrin | −9.7 | 0 | – |
4. Conclusion
Novel anthraquiones (1a-1k) moiety were synthesized and screened for larvicidal activity. The Compound 1c was highly active (LD50 20.92 µg/mL) against second instar C. quinquefasciatus mosquito larvae than permethrin with LD50: 25.49 µg/mL. Molecular docking findings supported the potent larvicidal activity of compound 1c (-9.8 Kcal/mol) compared with Permethrin with binding energy values of (-9.7 Kcal/mol). Therefore, these compounds might serve as a new class of products with larvicidal activity and prospective foundation for emerging ecologically important bioactive compound.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was funded by Researchers Supporting Project number (RSP-2020/27), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bazaes A., Olivares J., Schmachtenberg O. Properties, projections and tuning of teleost olfactory receptor neurons. J. Chem. Ecol. 2013;39:451–464. doi: 10.1007/s10886-013-0268-1. [DOI] [PubMed] [Google Scholar]
- De March C.A., Golebiowski J. A computational microscope focused on the sense of smell. Biochimie. 2014;107:3–10. doi: 10.1016/j.biochi.2014.06.006. [DOI] [PubMed] [Google Scholar]
- De Sousa D.P., Vieira Y.W., Uliana M.P., Melo M.A., Brocksom T.J., Sócrates C.H., Cavalcanti S.C.H. Larvicidal activity of para-Benzoquinones. Parasitol. Res. 2010;107:741–745. doi: 10.1007/s00436-010-1942-7. [DOI] [PubMed] [Google Scholar]
- Georges K., Jayaprakasam B., Dalavoy S.S., Nair M.G. Pestmanaging activities of plant extracts and anthraquinones from Cassia nigricans from Burkina Faso. Bioresour. Technol. 2008;99:2037–2045. doi: 10.1016/j.biortech.2007.02.049. [DOI] [PubMed] [Google Scholar]
- Govindarajan M. Chemical composition and larvicidal activity of leaf essential oil from Clausena anisata (Willd.) Hook. f. ex Benth (Rutaceae) against three mosquito species. Asian Pac. J. Trop. Med. 2010:874–877. [Google Scholar]
- Jeon J.-H., Kim M.-G., Lee H.-S. Larvicidal activity of naturally occurring naphthalenedione and its structurally related analogs against three mosquito species. J. Am. Mosq. Control. Assoc. 2015;31(1):71–76. doi: 10.2987/14-6438.1. [DOI] [PubMed] [Google Scholar]
- Kim S.I., Ahn Y.J. Larvicidal activity of lignans and alkaloid identified in Zanthoxylum piperitum bark toward insecticide-susceptible and wild Culex pipiens pallens and Aedes aegypti. Parasit Vectors. 2017;10(1):221–230. doi: 10.1186/s13071-017-2154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Shin S.C., Kim C.S., Lee H.J., Choi W.S., Ahn Y.J. Larvicidal activity of lignans identified in Phryma leptostachya Var. asiatica roots against tree mosquito species. J. Agric. Food Chem. 2005;53:969–972. doi: 10.1021/jf048208h. [DOI] [PubMed] [Google Scholar]
- Pechlaner M., Oostenbrink C. Multiple binding poses in the hydrophobic cavity of bee odorant binding protein AmelOBP14. J. Chem. Inform. Model. 2015;55:2633–2643. doi: 10.1021/acs.jcim.5b00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SathishKumar C., Selvaraj Keerthana S., Anis A., Ibrahim A.A., SurendraKumar R., Idhayadhulla A. CuII-tyrosinase enzyme catalyst-mediated synthesis of 2- thioxopyrimidine derivatives with potential mosquito larvicidal activity: spectroscopic and computational investigation as well as molecular docking interaction with OBPs of Culex quinquefasciatus. Chem. Sel. 2020:4567–4574. [Google Scholar]
- Cheng S.-S., Huang C.-G., Chen W.-J., Kuo Y.-H., Chang S.-T. Larvicidal activity of tectoquinone isolated from red heartwood-type Cryptomeria japonica against two mosquito species. Bioresource Technol. 2008;99:3617–3622. doi: 10.1016/j.biortech.2007.07.038. [DOI] [PubMed] [Google Scholar]
- Taha, M., Ismail, N.H., Khan, A., Shah, S.A.A., Anwar, A., Halim, S.A., QFatmi, M., Imran, S., Rahim, F., Kha, K.M. 2015. Synthesis of novel derivatives of oxindole, their urease inhibition and molecular docking studies. Bioorg. Med. Chem. Lett. 25, 3285–3289. [DOI] [PubMed]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutin, F., Clewer, H.W.B. XCIX. 1911. The constituents of rhubarb. J. Chem. Soc. Trans. 36, 946–967.
- Yang J.Y., Cho K.S., Chung N.H., Kim C.H., Suh J.W., Lee H.S. Constituents of volatile compounds derived from Melaleuca alternifolia leaf oil and acaricidal toxicities against house dust mites. J. Korean Soc. Appl. Biol. Chem. 2013;56:91–94. [Google Scholar]