Abstract
Crude oil biodegrading microorganism considers the key role for environmental preserving. In this investigation, crude oil biodegrading fungal strains have been isolated in polluted soil of crude-oil at khurais oil ground in Kingdom of Saudi Arabia. Among of 22 fungal isolates, only three isolates reflected potential capability for oil degradation. These isolates were identified and submitted to GenBank as (A1) Aspergillus polyporicola (MT448790), (A2) Aspergillus spelaeus (MT448791) and (A3) Aspergillus niger (MT459302) through internal-transcribed spacer-regions (ITS1&ITS2) for sequencing in molecular marker. Comparing with controls, strain (A1) Aspergillus niger was superior for biodegradation ability (58%) comparing with Aspergillus polyporicola and Aspergillus spelaeus degrading were showed 47 and 51% respectively. Employed CO2 evolution as indicator for petroleum oil biodegradation by the fungal isolates reflected that, Aspergillus niger emission highest CO2 (28.6%) comparing with Aspergillus spelaeus and Aspergillus polyporicola which showed 13% and 12.4% respectively. capability of Aspergillus sp. to tolerate and adapted oil pollutants with successful growth rate on them, indicated that it can be employed as mycoremediation agent for recovering restoring ecosystem when contaminated by crude oil.
Keywords: Crude oil contamination, Mycoremediation, Aspergillus, Internal transcribed spacer region
1. Introduction
The Saudi Kingdom ranks one of the world's top producers of crude oil. Oil spills are an important pollutant to the environment that occurs by leaking oil form tankers or pipelines and consider a real risk to human healthiness because of carcinogenic consequences. Oil spillage is the unintended release into the atmosphere of crude oil resulting in pollution of the ecosystem of any liquid hydrocarbon and endangering public health by contaminating drinking water and damaging natural resources of multiplier effects on the global economy (Obire and Anyanwu, 2009). Mechanical burial, evaporation, dispersion and washing consider traditional methods of remediation of contaminated hydrocarbon soil most commonly used. The key drawbacks of this approach were high shoreline, time consuming and lack of performance. Bioremediation, by contrast, considers the most effective method to be environmentally friendly and cost-effective (Singh et al., 2012). Biodegradation process identified as the use of varied microorganism metabolic capacities to detoxify or remove pollutants from the outputs in the industries of petrol (Medina-Bellver et al., 2005). In an accumulation, Mycoremediation was defined as the process of degradation of organic compounds by fungi. Considering the unique characteristic of most filamentus fungi species with substantial development of oxidative enzyme, organic-acids, chelators and extra-cellular enzymes. Numerous studies indicate that most filamentus fungi species are excellent degraders of hydrocarbons (Gadd and Gadd, 2001, Okoh, 2003, Stamets, 1999, Vanishree, 2014). Certain species of fungi are petroleum resistant to pollution which is capable in eliminating the pollution in soil. Ulfig et al (Ulfig et al., 2003) found, on the basis of previous facts, that Trichophyton ajelloi (who belongs to keratinolytic fungi) provides a powerful tool for associating progress in bioremediation. Alternata fungal strains, aspergillus flavus, curvularia lunata, fusarium solani, mucor racemosum, penicillium notatum and ulocladium atrum were isolated by varied areas in polluted petrol within the Kingdom (Hashem, 2007). This research was performed to isolate and identify the degradation of oil in the strains of fungi through the oil field in Khurais, Saudi Arabia. Biodegrading ability of fungal isolates was assessed.
2. Materials and methods
2.1. Settings of soil samples
Ten different soil samples around 15 g of each were collected from Khurais oil filed (Fig. 1) with an area of 2890-km2, 250 km-Southwest regions of Dhahran and 150kms from East-Northeast’s regions of Riyadh (25.0715 N, 48.0556 E), within the Kingdom in 2020. Samples were then stored in sterile polyethene bags, and stored below −20 °C. Soil suspensions were prepared in 10 mL of DD water, by liquefying one gram of soil.
Fig. 1.
Khurais oil field, Dhahran, Saudi Arabia.
2.2. Identification and isolation
Collected soils of around 10-grams were incubated in 250 mL of Erlenmeyer flasks consists of 100 mL of freshly prepared CDA (Czapak Dox Agar) and PDA (Potato-dextrose agar) media to isolate and purify our interested fungi (Fatima et al., 2019). In-order to detect an ability of fungi isolation to degrade hydrocarbons, pure isolates were grown on Bushnell has media (1 g-K2HPO4; 0.2 g-MgSO4;1g-KH2PO4;0.02 g-CaCl2;1g-NH4NO3;0.05FeCl2/l) supplemented by 1%crude oil as a single source of incubated carbon at 30/l for a week (Adekunle et al., 2007, Mittal and Singh, 2009). Twenty-two fungal isolates were opted to grow on BHS-media consists of 1%crude oil.
2.3. Identification, using molecular technology
According to the manufacturer protocol, fungal complete genomic DNA purified by Fungal DNA mini-kit (Omega Biotech, USA). The internal-transcribed spacer (ITS) was used as a marker for the molecular identification. The primers ITS1 (5′-TCCGTAGGTGAACCTTGCGG-3′) and ITS2 (5′-TCCTCCGCTTATTATGC-3′) were used for this purpose (White et al., 1990). Thermal cycling conditions were built as per the Khan et al (Khan et al., 2019). PCR-amplification was set for 35 cycles and initial denaturation was done for 94 °C-1 min, annealing for 55 °C-2mins, extension at 72 °C-2mins and final extension for 10mins at 72 °C. 1.5% agarose gel was used to run the PCR products and 100 bp ladder to measure the various fragments (Khan et al., 2015). Targeted amplicons were evaluated through EZNA gel extraction kit (Omega Bio-Tek, USA) after documented amplicons by DigiDoc-it and sequenced by Macrogen (Co, ltd, South Korea) through 3730Xl DNA sequences. Further, sequences were then converted to fasta format for an analysis of the alignment. To assess genetic similarity, the NCBI data-base were used for comparison with other gene sequences. MEGA software was used to build the phylogenetic tress of neighbor-joining method (Tamura et al., 2013).
2.4. CO2-evolution estimate
Evolved CO2 was measured with oil after the fungal strains were incubated. Sturm work was conducted to determine the evolution of both volumetric and gravimetric incubation of CO2. Gravimetric research was performed by extracting atmospheric CO2 through the passage of sterile air through a solution 1 M KOH. Bubbling air aerated the fungal activity and simultaneously permitted and CO2 that formed by the fungal activity to melt easily in broth; dissolved CO2 has been estimated by precipitate barium carbonate and CO2 release after adding 0.1 M BaCl2 to 100 mL. Gravimetrically, CO2 release was estimated through measuring the weight of the precipitate. The volumetrically estimated dissolves CO2 in medium which was through titration. 25 mL of filtrate was applied to 0.05 mL of 0.1 N thiosulphate solution following broth filtration. Then, after adding couple of drops of methyl orange indicator, then the solution was titrated against 0.02 M NaOH solution. Converting orange red to yellow in color represent the end point reaction. Titration was continued by adding two drops of indicator phenolphthalein further a pink color was shown.
The following formula was used for the titrate volume which was used for the quantity of CO2 evolved
[From × B × 50 × 1000]; where A = titrant volume NaOH
2.5. Assay on biodegradation of crude-oil
The DCPIP method was used to evaluate the degradation of crude-oil for the fungal isolates (Hanson and Desai, 1993). Around, 100 mL BHS-media involves 0.1% of Tween 80, 1% of crude-oil was mixed with 1 cm 2 of fungal hyphae as the complete source of carbon. Thereafter, redox substratum was added as indicator. After two weeks of incubation at 30oC, using a spectrophotometer (T80, PG, UK) at 420 nm, converting DCPIP into colorless fungal isolate capability for crude oil degradation.
2.6. Analysis with spectrophotometer
After 14-days of incubation the spectrophotometer device was applied to estimate the residual hydro-carbon appears in media. Control was initially obtained through 3 µL of injection into the cuvette and a standard reading was obtained. Sample of 3-µl of each was injected after taking the standard to get the reading. In a capillary column inside spectrophotometer each and every sample was estimated with 3 µL of injected into cuvette and measured at a wavelength of 609 nm.
3. Results and discussion
In this current study, overall, 22 fungal isolates were evaluated for biodegradation of crude oil, only three isolates could be considered a hydrocarbon oxidation of isolates, and showed promising biodegradation pattern with changing Czapek's broth color from blue to colorless showed promise, which were: Aspergillus polyporicola and Aspergillus niger. Aspergillus niger has represented the highest degradation potential among three fungal isolates.
3.1. Molecular markers of ITS1 and ITS2
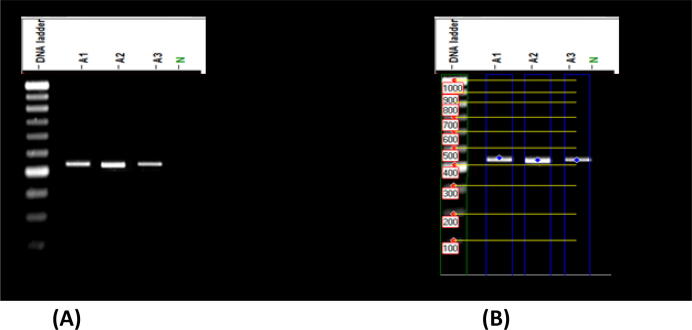
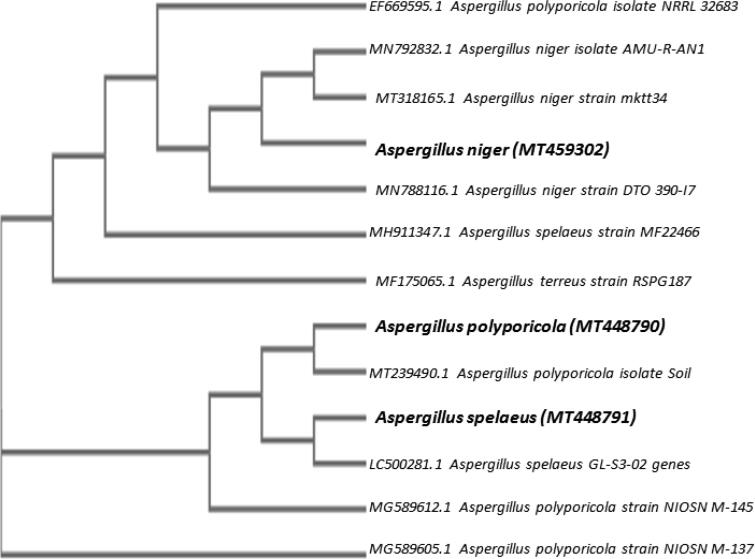
An internal area of transcribed spacers (ITS1 and ITS2) was used as a molecular marker to detect biodegradation of fungal hydrocarbons. Approximately 500 bp of the selected amplicons (Fig. 2) have been sequenced, identified and submitted to GenBank as (A1) Aspergillus polyporicola (MT448790), (A2) Aspergillus spelaeus (MT448791) and (A3) Aspergillus niger (MT459302) with 100% and 100% identity percent. Our findings of identification indicated that isolated fungi belong to the Aspergillus genus. Fig. 3 showed a constructed phylogenetic tree with the highest homology isolates from the database for our fungal insulates.
Fig. 2.
Amplified product of Internal Transcribed Spacer (ITS) for three fungal isolates (A1), (A2), (A3) and negative control (N).
Fig. 3.
Phylogenetic-tree of 3 of the petroleum hydrocarbons biodegradation fungi based on internal transcribed spacer region sequence.
The process of adaptation which isolates were exposed to in the oil polluted soil area can explain the capacity for oil degradation. With agreement to use ITS1 and ITS2 for fungal identification, Pang et al (Pang and Mitchell, 2005) indicated that sequencing fungal rDNA is a valuable aid in the detection of genus-level fungal diversity. In addition, many recent studies identifying oil-degrading fungi through molecular techniques have added more support to our findings for using molecular markers for fungal identification (Hu et al., 2011, Al-Jawhari, 2015, Maddela et al., 2015, Al-Nasrawi, 2012). Apart from this, additional studies (Alwakeel, 2017, Antunes et al., 2011, Pecoraro et al., 2015) added more light to our molecular identification findings. In Jeddah, Saudi Arabia, they characterized Aspergillus, Penicillium, and fusarium genera as dominant member of fungal communities from coastal soil and sea water.
3.2. Assay on biodegradation of crude oil
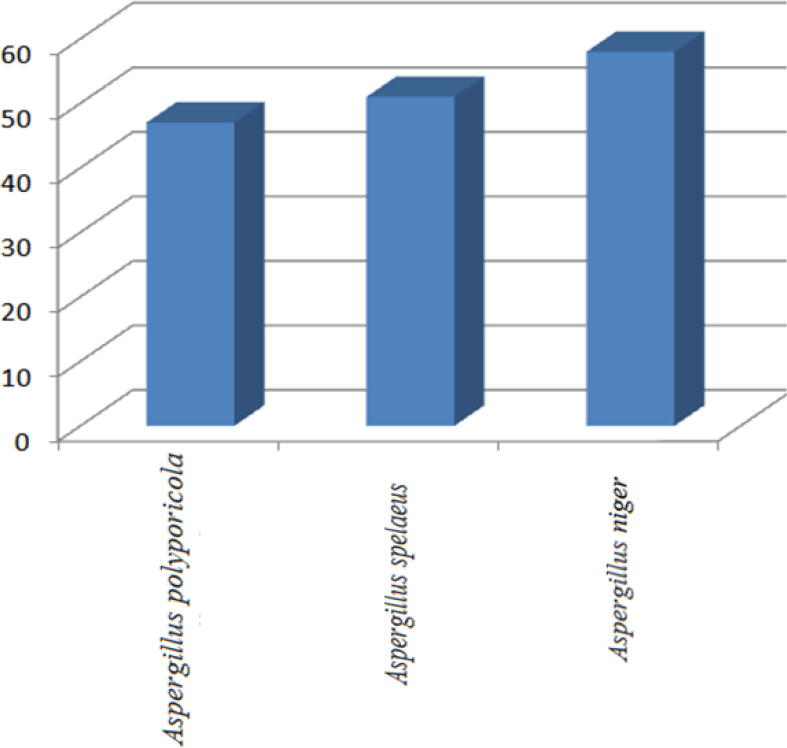
For our fungal isolates, spectrophotometric quantitative estimation of DCPIP (2,6-dichlorophenol indophenol) reflected varied ability to degrade oil. As shown in Table 1 and Fig. 4, Aspergillus niger's strain (A1) represented the highest degradation capability (58%) compared with Aspergillus polyporicola and Aspergillus spelaeus degrading 47% and 51% respectively. Leahy et al (Leahy et al., 2003) applied the same method to evaluate the hydrocarbon degrading capabilities of bacterial isolates in accordance with our findings which relay on spectrophotometric method to detect biodegradation capabilities. Our tests for Aspergillus niger 's superiority as a biodegradable fungal isolate by Gesinde et al (Gesinde et al., 2008) have been further supported. For Aspergillus niger, they cleared distinguishable biodegradation efficiency against four types of oil components (i) durb-oil, (ii) Escravos-light, (iii) Arabian-light and (iv) Bonny-light. In addition, several studies have reported Aspergillus, Penicillium and Fusarium species as effective biodegradable isolates of hydrocarbons (Obire and Anyanwu, 2009, De et al., 2000). In addition, Bartha and Atlas review detected 14 fungal-kinds with recommended potential for degradation of hydrocarbons, in agreement with our findings.
Table 1.
showed distinguishable oil degradation ability for three isolates.
| Species | Degradation % |
|---|---|
| Aspergillus polyporicola | 47 |
| Aspergillus spelaeus | 51 |
| Aspergillus niger | 58 |
Fig. 4.
Petroleum hydrocarbon degradation % of three fungal isolates.
3.3. CO2 evolution resulting from fungal activity on degradation of crude oil
Because of insignificant results, mean volumetric and gravimetric values were recorded for estimated evaluates of CO2. As recorded in Table 2, Aspergillus niger (28.6%) was the highest CO2 emission, followed by Aspergillus spelaeus and Aspergillus polyporicola showing 13% and 12.4% respectively. Use of CO2 evolution as an indicator of bio-degradation of petroleum oil through the fungal isolations also known as the Sturm test (Sturm, 1973). In accordance with our application of carbon dioxide release as an indicator of fungal activity during oil biodegradation, Vanishree et al (Vanishree, 2014) evaluate Aspergillus sp activity for petroleum biodegradation by recording released CO2 during process. Balba et al (Balba et al., 1998) further indicated that total CO2 produced could be considered an efficient index for contaminated soils with hydrocarbons.
Table 2.
CO2 progressed during co-cultivation of fungal isolates with petrol as sole carbon source.
| Fungal isolate |
Comparison of biomass (Dry Weight) after 4-weeks on incubation |
|||
|---|---|---|---|---|
| Controls (g/ l) | Treatments (g /l) | Weight (g /l) | % | |
| Aspergillus polyporicola | 0.591 ± 0.042 | 0.715 ± 0.077 | 0.124 | 12.4 |
| Aspergillus spelaeus | 0.593 ± 0.048 | 0.723 ± 0.093 | 0.13 | 13 |
| Aspergillus niger | 0.596 ± 0.033 | 0.767 ± 0.061 | 0.171 | 28.6 |
4. Conclusion
Our results showed that three fungal isolates reflected promising potential for the biodegrading of the crude oil. These isolates were identified as A1) Aspergillus polyporicola (MT448790), (A2) Aspergillus spelaeus (MT448791) and (A3) Aspergillus niger (MT459302) via ITS1 and ITS2. The biodegradation capacity was detected and supported by the evolution of CO2 as an indicator of biodegradation of petroleum oil.
Footnotes
Peer review under responsibility of King Saud University.
References
- Adekunle I., Adetunji M., Gbadebo A.M., Banjoko O.P. Assessment of groundwater quality in a typical rural settlement in Southwest Nigeria. Int. J. Environ. Res. Public Health. 2007;4:307–318. doi: 10.3390/ijerph200704040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Jawhari I.F.H. Ability of some fungi isolated from a sediment of Suq-Al Shuyukh marshes on biodegradation of crude oil. Int. J. Curr. Microbiol. App. Sci. 2015;4:19–32. [Google Scholar]
- Al-Nasrawi H. Biodegradation of crude oil by fungi isolated from Gulf of Mexico. J. Bioremed. Biodegrad. 2012;3:1–6. [Google Scholar]
- Alwakeel S.S. Molecular identification of fungi isolated from coastal regions of Red Sea, Jeddah, Saudi Arabia. J. Assoc. Arab Univ. Basic Appl. Sci. 2017;24:115–119. [Google Scholar]
- Antunes A., Ngugi D.K., Stingl U. Microbiology of the Red Sea (and other) deep-sea anoxic brine lakes. Environ. Microbiol. Rep. 2011;3:416–433. doi: 10.1111/j.1758-2229.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- Balba M., Al-Awadhi N., Al-Daher R. Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J. Microbiol. Methods. 1998;32:155–164. [Google Scholar]
- De, N., Bello, Y., Saleh, M.J.W., Management. Biodegradation of crude oil by Fusarium sp. and Trichoderma sp. Isolated from oil contaminated soil in different auto mechanic garages. 2000;18.
- Fatima N., Khan M.M., Khan I.A. L-asparaginase produced from soil isolates of Pseudomonas aeruginosa shows potent anti-cancer activity on HeLa cells. Saudi J. Bio.l Sci. 2019;26:1146–1153. doi: 10.1016/j.sjbs.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd G.M., Gadd G.M. Cambridge University Press; 2001. Fungi in Bioremediation. [Google Scholar]
- Gesinde A., Agbo E., Agho M., Dike E.F.C. Bioremediation of some Nigerian and Arabian crude oils by fungal isolates. Int. J. Pure Appl. Sci. 2008;2:37–44. [Google Scholar]
- Hanson K., Desai J.D., Desai A.J. A rapid and simple screening technique for potential crude oil degrading microorganisms. Biotechnol. Tech. 1993;7:745–748. [Google Scholar]
- Hashem, A.J.S., 2007. Bioremediation of petroleum contaminated soils in the Arabian Gulf region: a review, 19.
- Hu H., Van den Brink J., Gruben B., Wösten H., Gu J.-D., De Vries R.J. Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi. Int. Biodeterioration Biodegrad. 2011;65:248–252. [Google Scholar]
- Khan, I.A., Jahan, P., Hasan, Q., Rao, P.J.D., 2019. Research MSC, Reviews. Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. 13:688–94. [DOI] [PubMed]
- Khan I.A., Jahan P., Hasan Q., Rao P. Relationship between PTEN and gestational diabetes in Asian Indians womens. J. Health Specialties. 2015;3:184. [Google Scholar]
- Leahy J.G., Tracy K.D., Eley M.H. Degradation of mixtures of aromatic and chloroaliphatic hydrocarbons by aromatic hydrocarbon-degrading bacteria. FEMS Microbiol. Ecol. 2003;43:271–276. doi: 10.1111/j.1574-6941.2003.tb01067.x. [DOI] [PubMed] [Google Scholar]
- Maddela N.R., Masabanda M., Leiva-Mora M. Novel diesel-oil-degrading bacteria and fungi from the Ecuadorian Amazon rainforest. Water Science & Technology. 2015;71:1554–1561. doi: 10.2166/wst.2015.142. [DOI] [PubMed] [Google Scholar]
- Medina-Bellver J.I., Marín P., Delgado A., Rodríguez-Sánchez A., Reyes E., Ramos J.L. Evidence for in situ crude oil biodegradation after the Prestige oil spill. 2005;7:773–779. doi: 10.1111/j.1462-2920.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- Mittal A., Singh P. Studies on biodegradation of crude oil by Aspergillus niger. The South Pacific J. Natl. Sci. 2009;27:57–60. [Google Scholar]
- Obire O., Anyanwu E.C. Impact of various concentrations of crude oil on fungal populations of soil. Int. J. Sci. Technol. 2009;6:211–218. [Google Scholar]
- Okoh A.I. Biodegradation of Bonny light crude oil in soil microcosm by some bacterial strains isolated from crude oil flow stations saver pits in Nigeria. Afr. J. Biotechnol. 2003;2:104–108. [Google Scholar]
- Pang K.-L., Mitchell J. Molecular approaches for assessing fungal diversity in marine substrata. Botanica Marina. 2005;48:332–347. [Google Scholar]
- Pecoraro L., Girlanda M., Liu Z.-J., Huang L., Perotto S. Molecular analysis of fungi associated with the Mediterranean orchid Ophrys bertolonii Mor. Ann. Microbiol. 2015;65:2001–2007. [Google Scholar]
- Singh S., Kumari B., Mishra S. Springer; 2012. Microbial Degradation of Alkanes. Microbial Degradation of Xenobiotics; pp. 439–469. [Google Scholar]
- Stamets, P., 1999. Helping the ecosystem through mushroom cultivation In: Growing gourmet and medicinal mushroom. Batellet, A.(edition). Ten Speed Press, Berkeley, California.
- Sturm R.N. Biodegradability of nonionic surfactants: screening test for predicting rate and ultimate biodegradation. J. Am. Oil Chem. Soc. 1973;50:159–167. doi: 10.1007/BF02640470. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, SJMb, evolution. MEGA6: molecular evolutionary genetics analysis version 6.0. 2013;30:2725-9. [DOI] [PMC free article] [PubMed]
- Ulfig K., Plaza G., Worsztynowicz A., Manko T., Tien A., Brigmon R. Keratinolytic fungi as indicators of hydrocarbon contamination and bioremediation progress in a petroleum refinery. Polish J. Environ. Stud. 2003;12 [Google Scholar]
- Vanishree M. Biodegradation of petrol using Aspergillus sp. ARRB. 2014;4(6):914–923. doi: 10.9734/ARRB10.9734/ARRB/2014/5630. [DOI] [Google Scholar]
- White T., Bruns T., Lee S., Taylor JJPPAgtm, applications. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols: a guide to methods and applications. 1990. pp. 315–322. [Google Scholar]