Abstract
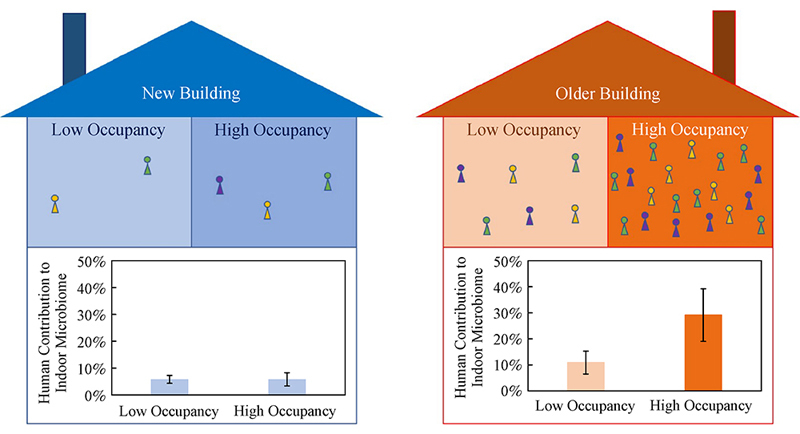
Educational facilities serve as community hubs and consequently hotspots for exposure to pathogenic microorganisms. Therefore, it is of critical importance to understand processes shaping the indoor microbiomes in educational facilities to protect public health by reducing potential exposure risks of students and the broader community. In this study, the indoor surface bacterial microbiomes were characterized in two multifunctional university buildings with contrasting levels of human occupancy, of which one was recently constructed with minimal human occupancy while the other had been in full operation for six years. Higher levels of human occupancy in the older building were shown to result in greater microbial abundance in the indoor environment and greater proportion of the indoor surface bacterial microbiomes contributed from human-associated microbiota, particularly the skin microbiota. It was further revealed that human-associated microbiota had greater influence on the indoor surface bacterial microbiomes in areas of high occupancy than areas of low occupancy. Consistent with minimal impact from human occupancy in a new construction, the indoor microbiomes in the new building exhibited significantly lower influence from human-associated microbiota than in the older building, with microbial taxa originating from soil and plants representing the dominant constituents of the indoor surface bacterial microbiomes. In contrast, microbial taxa in the older building with extensive human occupancy were represented by constituents of the human microbiota, likely from occupants. These findings provide insights into processes shaping the indoor microbiomes which will aid the development of effective strategies to control microbial exposure risks of occupants in educational facilities.
Keywords: Built environment, Indoor microbiome, Occupant, Building, Sequencing
Acknowledgements
This work was supported in part by US National Science Foundation awards 1952140 and 2026719. Any opinions, findings, recommendations, and conclusions in this paper are those of the authors, and do not necessarily reflect the views of NSF and the University of Tennessee, Knoxville of USA.
Footnotes
Highlights
• Exposure to indoor microbiomes is a public health concern in educational facilities.
• Indoor microbiomes were characterized in two multifunctional university buildings.
• Human occupancy had significant impact on the composition of indoor microbiomes.
• The skin microbiota of occupants represented important sources of indoor microbiomes.
Contributor Information
Shuai Li, Email: sli48@utk.edu.
Qiang He, Email: qianghe@utk.edu.
References
- Adams R I, Bateman A C, Bik H M, Meadow J F. Microbiota of the indoor environment: A meta-analysis. Microbiome. 2015;3:49. doi: 10.1186/s40168-015-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Atrouni A, Joly-Guillou M L, Hamze M, Kempf M. Reservoirs of non-baumannii Acinetobacter species. Frontiers in Microbiology. 2016;7:49. doi: 10.3389/fmicb.2016.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J G, Marr L C. Recognizing and controlling airborne transmission of SARS-CoV-2 in indoor environments. Indoor Air. 2020;30(4):557–558. doi: 10.1111/ina.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S Q, Berg G. Stenotrophomonas maltophilia. Trends in Microbiology. 2018;26(7):637–638. doi: 10.1016/j.tim.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Baird-Parker A C. The staphylococci: An introduction. Journal of Applied Bacteriology. 1990;69:1S–8S. doi: 10.1111/j.1365-2672.1990.tb01793.x. [DOI] [PubMed] [Google Scholar]
- Bell K S, Philp J C, Aw D W J, Christofi N. The genus Rhodococcus. Journal of Applied Microbiology. 1998;85(2):195–210. doi: 10.1046/j.1365-2672.1998.00525.x. [DOI] [PubMed] [Google Scholar]
- Ben Maamar S, Hu J, Hartmann E M. Implications of indoor microbial ecology and evolution on antibiotic resistance. Journal of Exposure Science & Environmental Epidemiology. 2020;30(1):1–15. doi: 10.1038/s41370-019-0171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho C C C R, da Fonseca M M R. The remarkable Rhodococcus erythropolis. Applied Microbiology and Biotechnology. 2005;67(6):715–726. doi: 10.1007/s00253-005-1932-3. [DOI] [PubMed] [Google Scholar]
- Gilbert J A, Stephens B. Microbiology of the built environment. Nature Reviews. Microbiology. 2018;16(11):661–670. doi: 10.1038/s41579-018-0065-5. [DOI] [PubMed] [Google Scholar]
- Graham J E, Moore J E, Jiru X U, Moore J E, Goodall E A, Dooley J S, Hayes V E, Dartt D A, Downes C S, Moore T C. Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Investigative Ophthalmology & Visual Science. 2007;48(12):5616–5623. doi: 10.1167/iovs.07-0588. [DOI] [PubMed] [Google Scholar]
- Hayward A C, Fegan N, Fegan M, Stirling G R. Stenotrophomonas and Lysobacter: Ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. Journal of Applied Microbiology. 2010;108(3):756–770. doi: 10.1111/j.1365-2672.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- Hewitt K M, Gerba C P, Maxwell S L, Kelley S T. Office space bacterial abundance and diversity in three metropolitan areas. PLoS One. 2012;7(5):e37849. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai Y. Survival of bacteria under dry conditions; from a viewpoint of nosocomial infection. Journal of Hospital Infection. 1991;19(3):191–200. doi: 10.1016/0195-6701(91)90223-U. [DOI] [PubMed] [Google Scholar]
- Hoisington A, Maestre J P, Kinney K A, Siegel J A. Characterizing the bacterial communities in retail stores in the United States. Indoor Air. 2016;26(6):857–868. doi: 10.1111/ina.12273. [DOI] [PubMed] [Google Scholar]
- Kembel S W, Jones E, Kline J, Northcutt D, Stenson J, Womack A M, Bohannan B J, Brown G Z, Green J L. Architectural design influences the diversity and structure of the built environment microbiome. ISME Journal. 2012;6(8):1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel S W, Meadow J F, O’Connor T K, Mhuireach G, Northcutt D, Kline J, Moriyama M, Brown G Z, Bohannan B J M, Green J L. Architectural design drives the biogeography of indoor bacterial communities. PLoS One. 2014;9(1):e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepeis N E, Nelson W C, Ott W R, Robinson J P, Tsang A M, Switzer P, Behar J V, Hern S C, Engelmann W H. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. Journal of Exposure Science & Environmental Epidemiology. 2001;11(3):231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson E S, Zaneveld J, Mozer M C, Collman R G, Bushman F D, Knight R, Kelley S T. Bayesian community-wide culture-independent microbial source tracking. Nature Methods. 2011;8(9):761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krismer B, Weidenmaier C, Zipperer A, Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nature Reviews. Microbiology. 2017;15(11):675–687. doi: 10.1038/nrmicro.2017.104. [DOI] [PubMed] [Google Scholar]
- Lax S, Gilbert J A. Hospital-associated microbiota and implications for nosocomial infections. Trends in Molecular Medicine. 2015;21(7):427–432. doi: 10.1016/j.molmed.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Lax S, Nagler C R, Gilbert J A. Our interface with the built environment: immunity and the indoor microbiota. Trends in Immunology. 2015;36(3):121–123. doi: 10.1016/j.it.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith D P, Hampton-Marcell J, Owens S M, Handley K M, Scott N M, Gibbons S M, Larsen P, Shogan B D, Weiss S, Metcalf J L, Ursell L K, Vazquez-Baeza Y, Van Treuren W, Hasan N A, Gibson M K, Colwell R, Dantas G, Knight R, Gilbert J A. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends in Microbiology. 2009;17(10):458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Meadow J F, Altrichter A E, Kembel S W, Moriyama M, O’Connor T K, Womack A M, Brown G Z, Green J L, Bohannan B J. Bacterial communities on classroom surfaces vary with human contact. Microbiome. 2014;2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reasoner D J. Heterotrophic plate count methodology in the United States. International Journal of Food Microbiology. 2004;92(3):307–315. doi: 10.1016/j.ijfoodmicro.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Rodrigues C J C, de Carvalho C C C R. Rhodococcus erythropolis cells adapt their fatty acid composition during biofilm formation on metallic and non-metallic surfaces. FEMS Microbiology Ecology. 2015;91(12):fiv135. doi: 10.1093/femsec/fiv135. [DOI] [PubMed] [Google Scholar]
- Rose L, Jensen B, Peterson A, Banerjee S N, Arduino M J. Swab materials and Bacillus anthracis spore recovery from nonporous surfaces. Emerging Infectious Diseases. 2004;10(6):1023–1029. doi: 10.3201/eid1006.030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R P, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison M B, Berg G, Van Der Lelie D, Dow J M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nature Reviews. Microbiology. 2009;7(7):514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- Sawada H, Kuykendall L D, Young J M. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. Journal of General and Applied Microbiology. 2003;49(3):155–179. doi: 10.2323/jgam.49.155. [DOI] [PubMed] [Google Scholar]
- Sbihi H, Boutin R C, Cutler C, Suen M, Finlay B B, Turvey S E. Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy. 2019;74(11):2103–2115. doi: 10.1111/all.13812. [DOI] [PubMed] [Google Scholar]
- Schweizer C, Edwards R D, Bayer-Oglesby L, Gauderman W J, Ilacqua V, Juhani M, Jantunen H K, Lai M, Nieuwenhuijsen N, Künzli Indoor time-microenvironment-activity patterns in seven regions of Europe. Journal of Exposure Science & Environmental Epidemiology. 2007;17(2):170–181. doi: 10.1038/sj.jes.7500490. [DOI] [PubMed] [Google Scholar]
- Song L, Wang C, Wang Y. Optimized determination of airborne tetracycline resistance genes in laboratory atmosphere. Frontiers of Environmental Science and Engineering. 2020;14(6):95. doi: 10.1007/s11783-020-1274-5. [DOI] [Google Scholar]
- Towner K J. Acinetobacter: an old friend, but a new enemy. Journal of Hospital Infection. 2009;73(4):355–363. doi: 10.1016/j.jhin.2009.03.032. [DOI] [PubMed] [Google Scholar]
- Treu L, Kougias P G, de Diego-Díaz B, Campanaro S, Bassani I, Fernández-Rodríguez J, Angelidaki I. Two-year microbial adaptation during hydrogen-mediated biogas upgrading process in a serial reactor configuration. Bioresource Technology. 2018;264:140–147. doi: 10.1016/j.biortech.2018.05.070. [DOI] [PubMed] [Google Scholar]
- US Census Bureau . Classrooms More Racially and Ethnically Diverse. Suitland: US Census Bureau; 2018. [Google Scholar]
- Wyckoff K N, Chen S, Steinman A J, He Q. Impact of roadway stormwater runoff on microbial contamination in the receiving stream. Journal of Environmental Quality. 2017;46(5):1065–1071. doi: 10.2134/jeq2017.03.0116. [DOI] [PubMed] [Google Scholar]


