Abstract
Triple-negative breast cancer (TNBC) has high metastatic, drug-resistance, and recurrence rates, and is characterized by an angiogenic and fibrotic microenvironment that favors cancer malignancy. However, details of the mechanisms underlying malignancy are still largely unknown. Our mouse model indicated that knockdown of CDK6 inhibited lung metastasis significantly compared to parental cells. Immunohistochemical analyses revealed that the levels of collagen and the angiogenic marker matrix metalloproteinase (MMP)-2 were much lower in CDK6-deficient cells. To examine mechanisms in the CDK6-mediated phenotype of cancer cells, we studied its role in MMP-2 expression. CDK6 mediated the recruitment of transcription factors including c-Jun and Sp1 to the MMP2 promoter. Knockdown of CDK6 significantly suppressed the expression of MMP2 mRNA. Consistent with the in vitro data, the expression of CDK6 was positively correlated with the angiogenic and fibrotic tumor microenvironment in TNBC patient tissues as shown by MMP-2 and fibronectin staining, respectively. More importantly, after screening a small molecule library of 31 protein kinase inhibitors, we found that the Raf inhibitor sorafenib displayed the highest cytotoxicity in CDK6-depleted cells. These data indicate that CDK6 serves as an anti-microenvironment target and affects the drug response in retinoblastoma-proficient TNBC, suggesting that combining a CDK6 inhibitor and sorafenib leads to a synthetic effect that may represent a personalized therapeutic approach for patients with TNBC.
Keywords: CDK6, MMP2, c-Jun, tumor microenvironment, sorafenib
Introduction
Breast cancer is one of the most prevalent cancers in Taiwan with a rising incidence [1]. Its heterogeneity makes providing effective medical treatment difficult. Currently, the type of breast cancer and corresponding intervention strategies are determined using surface markers on cancer cells, including estrogen receptors, progesterone receptors, and human epidermal growth factor receptor 2. However, triple-negative breast cancer (TNBC), which lacks all three receptors, is the most aggressive and difficult subtype to treat because of its heterogeneity and the absence of any specific biomarker for targeted therapy [2]. Therefore, the development of specific and personalized treatments for TNBC is one of the most important issues in oncology research.
Cyclin-dependent kinases (CDKs) participate in controlling the cell cycle. CDK6 and CDK4 were the first of this family to be identified and were found to be activated upon mitogenic stimulation. After binding with cyclin D, CDK4/6 negatively regulates the retinoblastoma protein (Rb) and consequently releases the transcription factor E2F, promoting exit from the G1 to S phase [3-5]. A dramatic increase in CDK6 expression has been observed in several cancer types, including leukemia, glioblastoma, pancreatic cancer, and breast cancer [6-9]. However, breast cancer is a heterogeneous disease. Therefore, understanding the function of CDK6 in different breast cancer subtypes could lead to more potential strategies in personalized therapy.
Three selective CDK4/6 inhibitors have been approved by the United States Food and Drug Administration for treating estrogen receptor-positive breast cancer: palbociclib (PD0332991), ribociclib (LEE011), and abemaciclib (LY835219) [9-12]. Because these inhibitors achieve their antitumor effects by increasing Rb protein function, the expression of functional Rb has been proposed as a basis for treatment with them [13]. Notably, upregulation of cyclin D and CDK4/6 has been observed in TNBC. However, 20% of TNBC cases exhibit loss or mutation of Rb, which hinders the use of CDK4/6 inhibitors for treatment [14]. Nevertheless, available data suggest that high CDK4/6 and Rb-proficient TNBC may be still treatable with CDK4/6 inhibitors.
Combining CDK4/6 inhibitors with anti-estrogen therapies is beneficial in the treatment of estrogen receptor-positive breast cancer [15]. Other evidence indicates that the antitumor activity of trastuzumab against HER2-positive breast cancer can be enhanced when combined with CDK4/6 inhibitors [16,17]. Therefore, combinations of CDK4/6 inhibitors with other targeted therapies may improve efficacy and reduce drug resistance in TNBC [18]. In the current study, we revealed the interplay of CDK6 with its microenvironment in TNBC tissue and evaluated the therapeutic effect of CDK6 inhibition for TNBC. We also screened clinical and investigational drugs for candidates capable of producing therapeutic effects synergistically with CDK6 inhibition.
Materials and methods
Cell culture
The TNBC cell lines used in this study were 4T1, MDA-MB-231 and MDA-MB-468 (American Type Culture Collection (ATCC), Manassas, VA, USA). In addition, M10 mammary epithelial cells (Biosource Collection and Research Center, Hsinchu, Taiwan), SVEC4-10 lymphatic endothelial cells, and RMF-EG fibroblast cells (ATCC) were used. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM), DMEM/F12 medium, or M199 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Hyclone Laboratories, South Logan, UT, USA) and antibiotics at 37°C, equilibrated with 95% humidified air with 5% CO2.
Specimens
Tissue blocks from 37 patients with TNBC were selected from the Department of Pathology, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan. Institutional Review Board approval for using these tissues in this study was given by the Research Ethics Committee of the Kaohsiung Medical University Hospital (IRB:KMUHIRB-E(I)-20180321). Data were analyzed anonymously; therefore no additional informed consent was required. All methods were performed in accordance with the guidelines and regulations of the Kaohsiung Medical University Hospital.
Reagents
Antibodies against CDK6 (GTX103992), matrix metalloproteinase (MMP)-2 (GTX104577), MMP-9 (GTX100458), and fibronectin (GTX112794) were from GeneTex International (Irvine, CA, USA). β-Actin (#3700), Sp1 (#9389), c-Jun (#9165), antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Proliferating cell nuclear antigen (PCNA) (#ab29), lymphatic vessel endothelial hyaluronan receptor (LYVE-1) (#ab14917), and CD31 (#ab28364) antibodies were from Abcam (Cambridge, MA, USA). T-5224 was from Selleckchem (Houston, Texas, USA). Mithramycin A was purchased from Sigma-Aldrich (St. Louis, USA).
Western blotting
Protein extraction and immunoblotting were performed as described previously [19]. In brief, cells were lysed in protein lysis buffer containing a protease and phosphatase inhibitor cocktail (M-PERTM mammalian protein extraction buffer; Thermo Scientific, Rockford, IL, USA) on ice for 20 min and cellular debris was removed by centrifugation. Proteins (20-30 ug) were quantified, separated by 8-12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and immunoblotted using the indicated antibodies for overnight, followed by incubation with horsedish peroxide (HRP)-conjugated secondary for 1 h at room temperature.
Gene knockdown
To knock down the expression of CDK6 in MDA-MB-231 and 4T-1 cells, short hairpin (sh)RNA against human CDK6 (sh-CDK6) (sequence: #1: CAGACAGAGAAACCAAACTAA; #2: CAGACAGAGAAACCAAACTAA, and #3: CGTGGAAGTTCAGATGTTGAT), shRNA against luciferase (sequence: GCGGTTGCCAAGAGGTTCCAT), control mouse CDK6 shRNA (#1: CGGTTGCATC TTTGCAGAAAT; #2: CTTGACCACTTACTTGGATAA; #3: GCTCAACCCATCG AGAAGTTT), and control Luciferase shRNA (sequence: CTTCGAAATGTCCGTTCGGTT) were purchased from the National RNAi Core Facility (Academia Sinica, Taipei City, Taiwan). The transfection protocols for CDK6 and luciferase followed the manufacture’s guidelines.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA extraction and qRT-PCR protocols were performed as described previously [19]. Briefly, total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Synthesized cDNA was used as a template for PCR amplification with primers for human CDK6, hepatocyte growth factor (HGF), fibroblast-activating factor (FAF), interleukins (IL6 and IL8), transforming growth factor-β (TGFB1), platelet-derived growth factor (PDGF), vascular endothelial growth factor-A (VEGFA), MMP2, and MMP9. Quantitative RT-PCR was performed in a 20 μL reaction volume using the standard protocols provided with the LightCycler 480 II system (Roche, Basel, Switzerland). Expression of the abovementioned genes was determined as: ΔCT = CT (target gene)-CT (GAPDH); ΔΔCT = ΔCT (experimental group)-ΔCT (control group). The primers of CDK6 were (forward) 5’-CGTGGTCAGGTTGTTTGATG-3’ and (reverse) 5’-CGGGCTCTG GAACTTTATCC-3’; HGF were (forward) 5’-GAGGGACATAAGAAAAGAAGA-3’ and (reverse) 5’-GTGTGGTATCATGGAACTCCA-3’; IL-6 were (forward) 5’-GACAGCCACTCACCTCTTCA-3’ and (reverse) 5’-TGCAGGAAC TGGATCAGGAC-3’; IL-8 were (forward) 5’-CAGAGACAGCAGAGCACAC-3’ and (reverse) 5’-AGTTCTTTAGCACTCCTTGGC-3’; MMP-2 were (forward) 5’-TCTCCTGACATTGACCTTGGC-3’ and (reverse) 5’-CAAGGTGCTGG CTGAGTAGATC-3’; MMP-9 were (forward) 5’-TTGACAGCGACAAGAAGTGG-3’ and (reverse) 5’-GCCATTCACGTCGTCCTTAT-3’; PDGF were (forward) 5’-CAGCAGGCGTTGGAGATCAT-3’ and (reverse) 5’-GGAGTCGGCATG AATCGCTG-3’; TGF-β1 were (forward) 5’-GTCAATGTACAGCTGCCGCA-3’ and (reverse) 5’-CCCAGCATCTGCAAAGCTC-3’; VEGF-A were (forward) 5’-ACAGGTACAGGGATGAGGACAC-3’ and (reverse) 5’-AAGCAGGTGAG AGTAAGCGAAG-3’; VEGF-C were (forward) 5’-GCCAACCTCAACTCAAG GAC-3’ and (reverse) 5’-CCCACATCTGTAGACGGACA-3’; ANG-2 were (forward) 5’-TGGGATTTGGTAACCCTTCA-3’ and (reverse) 5’-CCTT GAGCGAATAGCCTGAG-3’; CXCL-5 were (forward) 5’-GCAAGGAGTTC ATCCCAAAA-3’ and (reverse) 5’-AAACTTTTCCATGCGTGCTC-3’; CXCL-6 were (forward) 5’-AGAGCTGCGTTGCACTTGTT-3’ and (reverse) 5’-GC AGTTTACCAATCGTTTTGGGG-3’; FAF were (forward) 5’-GAATGTT TCGGTCCTGTC-3’ and (reverse) 5’-CCATCCAGTTCTGCTTTC-3’; MMP2 promoter chip sequence for c-Jun binding were (forward) 5’-CTTCTCAAACTGTTCCCTGC-3’ and (reverse) 5’-GGAACGCCTGACTTCAGC-3’; MMP2 promoter chip sequence for Sp1 binding were (forward) 5’-GTCCTGGCAATCCCTTTGTA-3’ and (reverse) 5’-GGGGAAAAGAGGT GGAGAAA-3’.
Transwell co-culture wound healing assay
RMF-EG or SVEC4-10 cells were cultured in 6-well plates. A wound area was created using a 200 µL pipette tip on the confluent monolayer. The cells were then co-cultured with parental/sh-CDK6 MDA-MB-231 cells through a Transwell with 0.2 μm inserts on top. After co-culturing, images were captured at 0, 4, and 8 h. The experiments were performed in triplicate.
Tube formation assay
Matrigel Basement Membrane Matrix (BD Biosciences, Franklin Lakes, NJ, USA) was diluted with M199/DMEM and used to coat 24-well plates at 37°C for 1 h. Then, 5 × 104 SVEC4-10 cells were seeded onto the Matrigel. Conditioned medium from parental or sh-CDK6 MDA-MB-231 cells was added into the Transwells and incubated in the same well with SVEC4-10 cells. The tube-forming ability of SVEC4-10 cells was measured at 0.5, 1, and 2 h with or without parental or sh-CDK6 MDA-MB-231 conditioned medium.
Mice
Five-week-old female BALB/c mice (22-25 g) were purchased from the National Laboratory Animal Center (Taipei City, Taiwan). Mice were housed in the Kaohsiung Medical University Animal Research Facility. All experimental methods and procedures were approved by the Center for Laboratory Animals Committee at Kaohsiung Medical University (No. 107097) and performed according to the care and use of laboratory animal guidelines. Animals were kept in an Association for the Assessment and Accreditation of Laboratory Animal Care International-approved animal facility in Kaohsiung Medical University. Mice were divided randomly into two groups and subjected to treatment. Control and CDK6 knockdown 4T-1 cells (5 × 104 cells/mouse) were injected into the left fourth thoracic mammary fat pad. For ribociclib treatment, mice were then randomized into 2 groups: control and ribociclib (50 mg/kg/day) was given once daily for 5 days a week via oral gavage for 2 weeks. Tumor volumes were measured every 3 days from the second week after injection and were calculated using the formula: V = (length) × (width)2 × 0.5. After 45 days, animals were sacrificed and the tumors were excised and used for immunohistochemistry (IHC). For individual inhibitors, mice were randomized into two groups containing four mice each and treated with 0.2 mg/kg/day of Mithramycin A (i.p.) and 3 mg/kg/day of T-5224 (Oral gavage) five times per week for 42 days. After 42 days, were sacrificed and the tumors were excised and used for immunohistochemistry (IHC).
Immunohistochemistry (IHC) staining
IHC was performed as described previously [20]. In brief, paraffin-embedded tissue samples were cut into 4-μm-thick sections, de-paraffinized, and rehydrated. Antigen retrieval was achieved by autoclaving the sections at 121°C for 10 min in a pH 6.0 retrieval solution (DAKO, Carpinteria, CA, USA). Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide (Sigma-Aldrich, St. Louis, MO, USA) for 10 min. The sections were then incubated with primary antibodies (CDK6, MMP-2, CD31, PCNA, and fibronectin) at room temperature for 1 h. The REALTM EnVisionTM Detection System (DAKO) was then applied for 1 h. Finally, the sections were incubated in 3’, 3-diaminobenzidine for 5 min, counterstained with Mayer’s hematoxylin, and mounted. Negative controls were prepared by replacing the primary antibodies with nonimmune serum. The trichrome stain kit was purchased from Abcam (#ab150686).
Scoring
CDK6, MMP-2, and fibronectin expression in patients was scored through signal intensity (0, 1+, 2+, and 3+) and proportions of positive cells (0 = ≤ 10%, 1 = 11-25%, 2 = 26-50%, 3 = > 50%) [20]. Staining indices were calculated as the product of the intensity of signal and the proportion of positive cells; scores ranged from 0 to 9. A score ≤ 4 indicated low expression of CDK6 and MMP-2; a score ≥ 6 indicated high expression of CDK6 and MMP-2. Fibronectin expression was scored 1 or 0 according to whether or not it was staining was observed in the extracellular matrix.
Statistical analyses
The expression of CDK6, fibronectin, and MMP-2 in TNBC tissues, as determined by IHC staining, was compared using the chi-square test. A p value < 0.05 was considered to indicate a statistically significant difference between groups. All statistical analyses were performed using SPSS 19.0 software (IBM, Chicago, IL, USA).
Results
Suppression of CDK6 affected tumor growth and lung metastasis in an animal model of TNBC
To determine whether CDK6 was a biomarker and therapeutic target of TNBC, we performed patient-based analyses of RNA using the UALCAN database [21]. Among breast cancer types, TNBC showed the highest level of CDK6 at the RNA level (Figure S1). Notably, the level of CDK6 protein in Rb-proficient MDA-MB-231 cells was higher than in Rb null MDA-MB-468 cells (Figure S2). This finding suggests that targeting CDK6 may serve as an anticancer strategy in Rb-proficient TNBC.
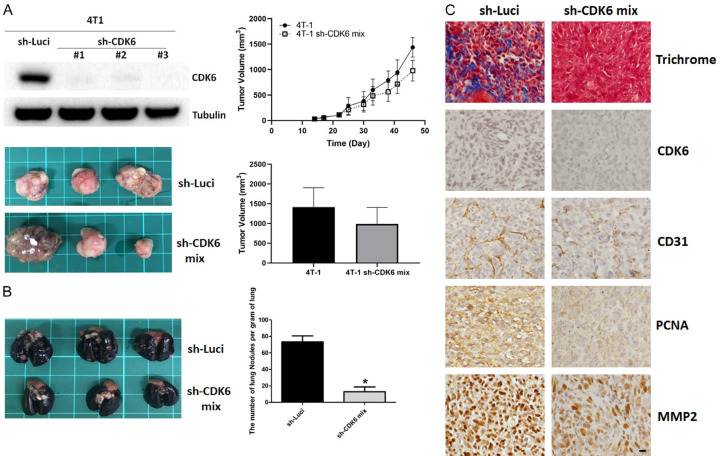
High metastasis and proliferation rates are characteristics of the TNBC phenotype. Murine 4T1 is a typical TNBC cell line that has been used as an allograft to induce the neoplastic process in immune-competent mice (e.g., BALB/c). To evaluate the effect of CDK6 on the growth and metastasis of TNBC cells in vivo, we established a murine 4T1 TNBC allogeneic model in BALB/c mice. Mock vector-and sh-CDK6-transfected cells were implanted orthotopically into the mammary glands of mice. The tumor growth of CDK6-depleted cells was not significantly changed relative to that of parental cells (Figure 1A). However, notably, we found that silencing CDK6 reduced lung metastasis significantly relative to control, mock-transfected tumor cells (Figure 1B). IHC revealed that levels of CD31 and MMP-2 were much lower in CDK6-deficient cells (Figure 1C). The level of PCNA was partially reduced in CDK6-depleted cancer tissue. More importantly, trichrome staining revealed decreased collagen in CDK6-depleted tumors (Figure 1C). These data suggest that continuous expression of CDK6 was required for the maintenance of a malignant phenotype. Furthermore, CDK6 silencing suppressed lung metastasis and modified the tumor microenvironment in vivo.
Figure 1.
Knockdown of CDK6 reduces lung metastasis and malignant phenotype in vivo. A. Left panel, levels of CDK6 in parental and sh-CDK6 transfected 4T1 cells, and representative images of breast cancer tissue with high or low expression of CDK6 in BALB/c mice. Right panel, quantitative analysis of tumor growth and size 45 days after tumor cell injection. Comparisons utilized the 2-tailed Student t test (*P < 0.05). B. Lungs were perfused with India ink after the mice were euthanized. Left panel, representative images of lung nodules. Right panel, numbers of visualized nodules per gram of lung tissue compared using the 2-tailed Student t-test (*P < 0.05). C. Representative immunohistochemical staining of CDK6, PCNA, CD31 and MMP-2, and collagen (Masson’s trichrome stain) in tumors of parental and CDK6-deficient breast cancer tissues. Original magnification: 40 ×, scale bar: 20 μm.
Knockdown of CDK6 in cancer cells abolishes cancer-induced fibroblast activation and migration
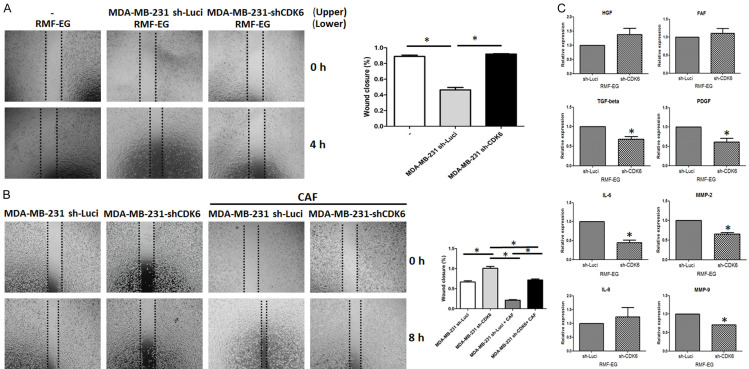
TNBC is characterized by higher grades of fibrosis than other types of breast cancer. Fibrotic tissue not only blocks the delivery of drugs into the tumor, but also provides a pro-metastatic microenvironment [22]. We found that CDK6 expression was positively correlated with fibrosis in tumor tissue (Figure 1C); therefore, we studied possible mechanisms that affected the activation of fibroblasts regulated by CDK6. We examined the migration ability of fibroblast cells by co-culturing with parental and CDK6-depleted breast cancer cells. As shown in Figure 2A, CDK6-deficient breast cancer cells did not increase the migration of fibroblasts, while parental breast cancer cells did (Figure 2A). Consistent with these findings, the migration of fibroblast was suppressed in CDK6-deficient 4T1 cells (Figure S3). On the other hand, cancer-associated fibroblasts did not promote the migration of CDK6-depleted cells (Figures 2B and S4).
Figure 2.
Knockdown of CDK6 impairs cancer-induced fibroblast activation and migration. A. For co-culture, 40,000 RMF-EG cells were seeded onto 24-well plates in 500 µL Dulbecco’s modified Eagle’s medium (DMEM). Additionally, 40,000 MDA-MB 231 or MDA-MB-231 sh-CDK6 pooling cells were seeded onto 0.2 µm Transwell inserts with 100 µL DMEM and co-cultured with RMF-EG cells for 48 h. Migration of RMF-EG cells into the scratch wound after 4 h was analyzed. Original magnification: 40 ×, scale bar: 500 μm. B. For the co-culture, 40,000 MDA-MB 231 or MDA-MB-231 sh-CDK6 pooling cells were seeded onto 24-well plates in 500 µL DMEM. Additionally, 40,000 cancer-associated fibroblasts were seeded onto 0.2 µm Transwell inserts in 100 µL DMEM and co-cultured with RMF-EG cells for 48 h. Migration of MDA-MB 231 and MDA-MB-231 sh-CDK6 pooling cells into the scratch wound after 8 h was analyzed. Original magnification: 40 ×, scale bar: 500 μm. C. The quantitative real-time polymerase chain reaction was used to investigate the mRNA expression of candidate genes in RMF-EG cells co-cultured with MDA-MB 231 or MDA-MB-231 sh-CDK6 pooling cells. Columns represent means of triplicate assays normalized to GAPDH expression (*P < 0.05).
Several proteins involved in fibroblast activation, including HGF, FAF, IL-6, IL-8, MMP-2, MMP-9, TGF-β, and PDGF, were examined. Among them, IL-6, MMP-2, MMP-9, TGF-β, and PDGF levels were significantly decreased in RMF-EG cells co-cultured with CDK6-deficient cells (Figure 2C). These results demonstrate that CDK6 plays an important role in the interplay between cancer cells and fibroblast activation.
Knockdown of CDK6 in cancer cells decreases cancer-induced tube formation and migration of endothelial cells
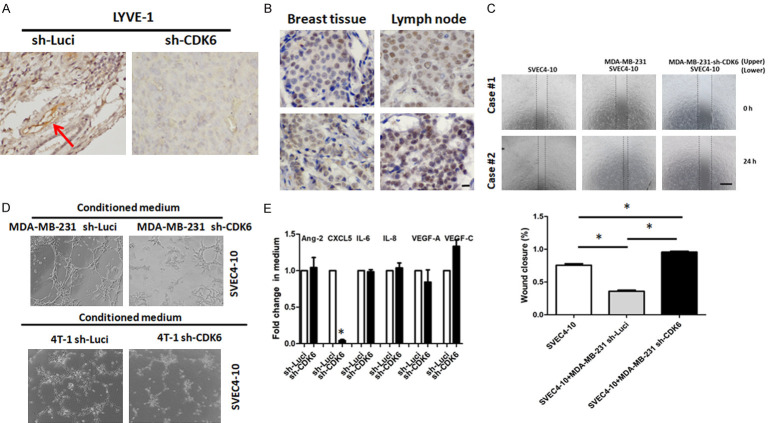
Lymphatic invasion is an aggressive biological phenotype. LYVE-1 is a specific marker for lymphatic blood vessels. As shown in Figure 3A, knockdown of CDK6 suppressed lymphangiogenesis. To validate the clinical significance of CDK6, we examined the correlation between CDK6 expression and lymphatic invasion by TNBC cells. Levels of CDK6 in lymph node tissue were higher than in primary tissue (Figure 3B). To address the mechanism by which CDK6 in TNBC cells induced lymphangiogenesis, lymphatic endothelial cells (SVEC4-10) were used. Co-culturing MDA-MB-231 cells with SVEC4-10 cells significantly increased the migration of SVEC4-10 cells. However, co-culturing SVEC4-10 cells with CDK6-knockdown MDA-MB-231 cells decreased their migration (Figure 3C).
Figure 3.
CDK6 in cancer cells mediates lymphangiogenesis. A. Levels of LYVE-1 in parental and sh-CDK6 transfected 4T1 cells, and representative images of breast cancer tissues with high or low expression of CDK6 in BALB/c mice. B. Representative immunohistochemical staining for CDK6 in primary tissue and lymph nodes of breast cancer patients. Original magnification: 40 ×, scale bar: 20 μm. C. For the co-culture, 40,000 SVEC4-10 cells were seeded onto 24-well plates in 500 µL Dulbecco’s modified Eagle’s medium (DMEM). Additionally, 40,000 MDA-MB 231 or MDA-MB-231 sh-CDK6 cells were seeded onto 0.2 µm Transwell inserts in 100 µL DMEM and co-cultured with SVEC4-10 cells for 48 h. Migration of SVEC4-10 cells into the scratch wound after 4 h was analyzed. Original magnification: 40 ×, scale bar: 500 μm. D. A total of 3 × 105 cells were seeded per 24 well with 1 mL of conditioned medium from either MDA-MB 231 or MDA-MB-231 sh-CDK6 pool or 4T1 or 4T1 sh-CDK6 pooling cells. Tube formation was assessed over the course of 4 h. E. Protein levels of candidate lymphangiogenesis-associated soluble factors were detected by enzyme-linked immunosorbent assay kits. Columns represent means of triplicate assays normalized to cell numbers (*P < 0.05).
These endothelial cells were also treated with conditioned medium from either control or CDK6-deficient cells, and their tube-forming abilities were then compared. We found that knockdown of CDK6 in MDA-MD-231 and 4T1 reduced cancer-induced tube formation (Figure 3D). Finally, we measured levels of lymphangiogenesis-associated factors including VEGF-A, VEGF-C, angiopoietin-2, CXCL5, CXCL6, IL-6, and IL-8. Only the protein levels of CXCL5 were reduced in CDK6-knockdown cells (Figure 3E). Taken together, these data indicate that endothelial cells with high CDK6 expression may be responsible for the pro-angiogenic abilities of cancer, while downregulating CDK6 levels in cancer cells can abolish this effect.
CDK6 upregulates MMP-2 transcription
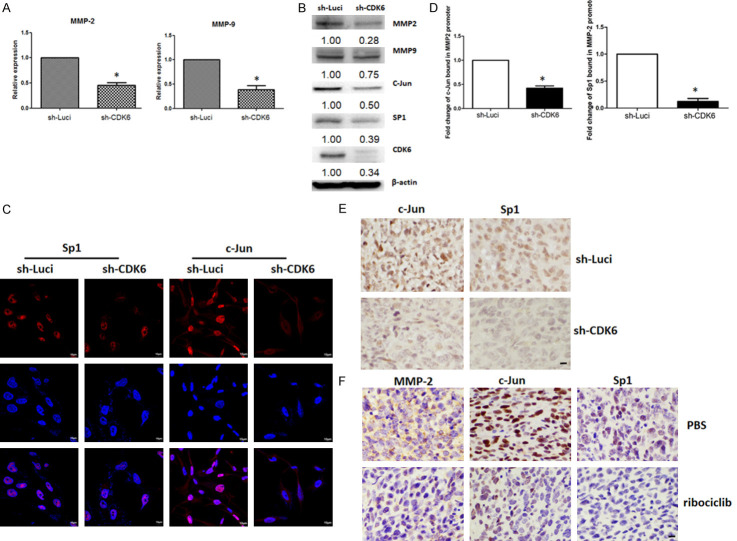
CDK6 is involved in kinase-dependent pathways that control cell-cycle progression. However, CDK6 is also a chromatin-bound cofactor that can directly control gene expression [23]. As CDK6 may be an important modulator of tumor-secreted factors that support angiogenesis and fibrosis, we screened for proangiogenic and profibrotic factors that may be regulated by CDK6 in cancer cells. MMPs not only play key roles in activating fibroblasts, but also promote tumor growth, metastasis, and angiogenesis [24,25]. Our data indicated that knockdown of CDK6 significantly reduced the mRNA expression levels of MMP2 and MMP9 (Figure 4A). Consistent with the mRNA levels, MMP-2 and MMP-9 protein levels also decreased in CDK6-knockdown cells (Figures 4B and S5).
Figure 4.
Knockdown of CDK6 suppresses the expression of MMP2 through transcriptional regulation. A. The quantitative real-time polymerase chain reaction (qRT-PCR) was used to investigate the expression of MMP2 and MMP9 mRNAs in MDA-MB 231 and MDA-MB-231 sh-CDK6 pooling cells. Columns represent means of triplicate assays normalized to GAPDH (*P < 0.05). B. Protein expression levels of CDK6, c-Jun, Sp1, MMP-2, and MMP-9 in MDA-MB 231 and MDA-MB-231 sh-CDK6 pooling cells were determined by western blotting. C. Immunohistochemistry confirmed nuclear translocation of c-Jun and Sp1 in MDA-MB 231 and MDA-MB-231 sh-CDK6 pooling cells. D. A chromatin immunoprecipitation-qRT-PCR analysis was performed to determine the status of c-Jun and Sp1 in the MMP-2 gene promoter. Experiments were performed in triplicate (*P < 0.05). E. Representative immunohistochemical staining for c-Jun and Sp1, in tumors with high and low CDK6 expression. Original magnification: 40 ×, scale bar: 10 µm. F. Representative immunohistochemical staining for c-Jun and Sp1, in tumors with Ribociclib. Original magnification: 40 ×, scale bar: 10 µm.
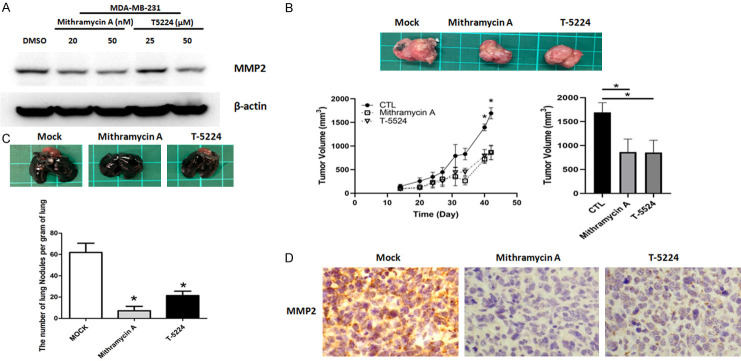
Sp1 and c-Jun are well-known regulators of MMP-2 promoter activity. As shown in Figure 4B, protein levels of c-Jun and Sp1 were decreased in CDK6-depleted cells. Notably, nuclear translocation of c-Jun and Sp1 was suppressed in CDK6-knockdown cells (Figures 4C and S6). Finally, the levels of c-Jun and Sp1 at the MMP-2 promoter were decreased in CDK6-deficient cells (Figure 4D). Consistent with these findings, IHC revealed that levels of c-Jun and Sp1 were much lower in CDK6-deficient cells and ribociclib treatment cells (Figure 4E and 4F). A gene-selective Sp1 inhibitor, Mithramycin A and c-Jun binding inhibitor, T-5224 also decreased the expression of MMP2 and tumor growth in vivo (Figure 5). Taken together, these results suggest that inhibiting CDK6 may have a therapeutic effect by preventing formation of a progressive tumor microenvironment through modulation of the c-Jun/Sp1-MMP-2 axis.
Figure 5.
Treatment of Mithramycin A and T-5224 decreases MMP2 expression lung metastasis and tumor growth in vivo. A. Protein expression levels of MMP-2 were shown in MDA-MB 231 cells treated with Mithramycin A and T-5224 by western blotting. B. Representative images of breast cancer tissue with the treatment of Mithramycin A and T-5224 in BALB/c mice. Lower panel, quantitative analysis of tumor growth and size 45 days after tumor cell injection. Comparisons utilized the 2-tailed Student t test (*P < 0.05). C. Lungs were perfused with India ink after the mice were euthanized. Upper panel, representative images of lung nodules. Lower panel, numbers of visualized nodules per gram of lung tissue compared using the 2-tailed Student t-test (*P < 0.05). D. Representative immunohistochemical staining of MMP-2 in tumors treated with Mithramycin A and T-5224 in BALB/c mice.
CDK6 is overexpressed in TNBC and positively correlated with angiogenesis and fibrosis within the tumor microenvironment
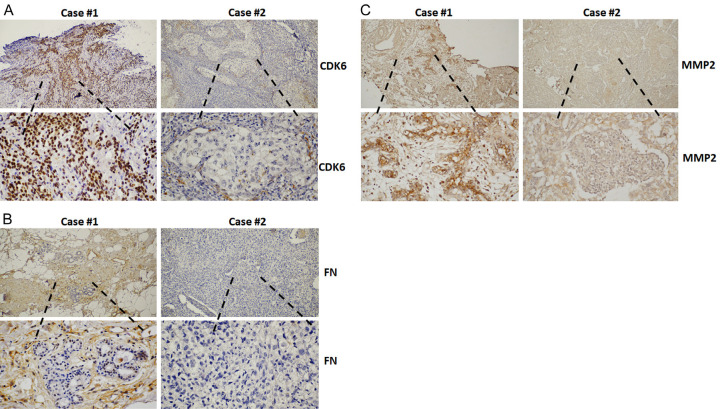
Angiogenesis and fibrosis in the tumor microenvironment are indicators of cancer progression and poor prognosis [26,27]. To improve our understanding of the role of CDK6 in tumor tissue, tumor tissues from 37 TNBC patients were subjected to immunodetection of CDK6, as well as MMP-2 and fibronectin, to reveal the extents of angiogenesis and fibrosis, respectively. The positive correlation of CDK6 with both MMP-2 and fibronectin is consistent with a critical role for CDK6 in tumor progression (Figure 6 and Table 1). This indicates that CDK6 may be a valuable therapeutic target in modulating the tumor microenvironment and reducing metastasis.
Figure 6.
CDK6 expression correlates positively with fibronectin and MMP-2 in breast cancer. Representative immunohistochemical images of CDK6 (A) and fibronectin (B) and MMP-2 (C) in 37 breast cancer tissue sections. Original magnification: 400×, scale bar: 20 µm. Associations between CDK6 and fibronectin or MMP-2 were determined by chi-square analyses, and the P values are shown.
Table 1.
Association of CDK6, Fibronectin expressions (ECM), and MMP-2 in TNBC tissues (n=37)
| Fibronectin (ECM) (%) | P-value | MMP-2 (%) | P-value | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Positive | negative | Positive | negative | ||||
| CDK6 | High | 11 (29.7) | 1 (2.7) | 10 (27.0) | 2 (5.4) | ||
| Low | 14 (37.8) | 11 (29.7) | P < 0.05 | 9 (24.3) | 16 (43.2) | P < 0.05 | |
CDK6 inhibition combined with sorafenib treatment induces synthetic lethality of TNBC cells
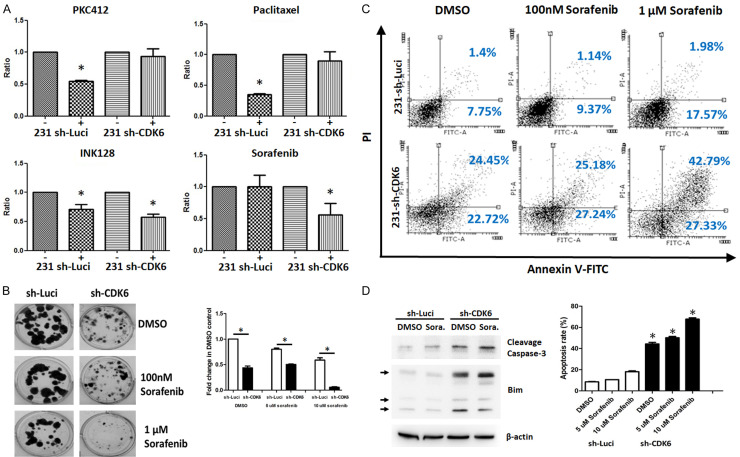
To test the ability of CDK6 inhibition to induce cancer cell death, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was utilized. In addition, we applied the concept of synthetic lethality, which involves combining effects on two different genes to further decrease cancer cell viability beyond that caused by either treatment alone [28]. We performed high-throughput analyses using clinical and pre-clinical drugs to screen for agents that could synergistically combine with a CDK6 inhibitor. The toxicities of 31 different protein kinase inhibitors were measured on both MDA-MB-231 and sh-CDK6 MDA-MB-231 cells. Among the tested combinations, sorafenib, a RAF/MEK/ERK pathway inhibitor, had the most significant effect on inducing cell death in CDK6-knockdown cells (Figure 7A). Consistent with these findings, knockdown of CDK6 led to enhanced cytotoxicity upon sorafenib treatment exposure, as compared to that in parental MDA-MB-231 cells through the analysis of colonies (Figure 7B). As shown in Figure 6C, the amount of cells in the early and late apoptotic phase was increased in CDK6-depleted cells (sh-Luci: 9.15%; sh-CDK6: 47.17%). More importantly, the treatment of sorafenib elevated the apoptosis in CDK6-depleted cells. These data indicated that CDK6 knockdown further enhances the apoptotic effect of sorafenib in MDA-MB-231 cells (Figure 7C). Considering the results of previous results, the regulatory mechanism of CDK6 may be assumed to affect the pro-survival proteins and pro-apoptotic proteins. Further, we found lower Bim and higher cleavage caspase-3 in CDK6-dpleted cells exposed with sorafenib treatment (Figure 7D). This result suggests that sorafenib could work with a CDK6 inhibitor to induce synthetic lethality in high CDK6 level/Rb-proficient TNBC cells.
Figure 7.
Knockdown of CDK6 enhances the cytotoxicity of sorafenib in breast cancer cells. A. MDA-MB 231 or MDA-MB-231 sh-CDK6 cells were treated with different compounds for 72 h. Cell viability was determined by the MTT assay (*P < 0.05). B. MDA-MB 231 or MDA-MB-231 sh-CDK6 cells were treated with sorafenib (0.1 or 1 μM) for 72 h, followed by culturing in fresh medium for 14 days. C. The percentage of apoptotic cells treated with indicated sorafenib for 72 h was detected via dual staining with annexin V-FITC and PI in MDA-MB 231 and MDA-MB-231 sh-CDK6 cells, followed by analysis of flow cytometry. The experiments were repeated three times. Between-group comparisons were performed using one-way ANOVA (*, P < 0.05). D. Protein expression levels of cleavage caspase-3, Bim and actin in MDA-MB 231 and MDA-MB-231 sh-CDK6 cells treated with vehicle and sorafenib for 72 h by western blotting.
Discussion
Uncontrolled cell cycle progression and proliferation are hallmarks of most types of cancer. Dysregulation of CDK6 expression and its activity are often associated with the development of cancers including lymphoma, leukemia, colorectal cancer, and estrogen receptor-positive breast cancer [29-31]. Notably, a previous report indicated that CDK6 was highly expressed in TNBC as compared to non-TNBC, and its overexpression correlated with a poor prognosis for TNBC [32]. However, the detailed mechanism still needed to be clarified. The present results agreed with those of previous studies showing that CDK6 expression correlated with an angiogenic and fibrotic tumor microenvironment. This suggests a positive correlation between CDK6 and a malignant tumor microenvironment.
The tumor microenvironment is an important regulator of cancer progression, a biomarker for prognosis, and a therapeutic target [33]. The microenvironment is composed of extracellular matrix, adipocytes, immune cells, endothelial cells, and fibroblasts [34]. Cancer cells interact with other cells within the microenvironment via soluble factors or intercellular receptor-ligand interactions. Different subtypes of breast cancer can create distinct microenvironments that favor malignant progression [35]. Among cells within the microenvironment, fibroblasts are a critical component for remodeling the environment to support cancer and participate in almost every step of cancer progression [36]. Fibroblasts interact with the tumor microenvironment through several factors that regulate inflammation and epithelial-to-mesenchymal events [33]. TGF-β, released by fibroblasts, affects TNBC tumor fibrosis, growth, and metastasis. These effects can be reversed by treatment with TGF-β antagonists and pirfenidone [37]. Fibroblast-regulated platelet-derived growth factor receptor (PDGFR)α, PDGFRβ, N2, and fibroblast activation protein α are highly expressed in TNBC and correlate with tumor invasiveness [38]. We found lower levels of IL-6, MMP-2, MMP-9, TGF-β, and PDGF in RMF-EG cells co-cultured with CDK6-deficient cells than in RMF-EG cells co-cultured with parental cells (Figure 2C). These results demonstrate that the level of CDK6 in cancer cells functions as a key modulator of fibroblast activation.
CDK6 has two major functions; first, its kinase-dependent function involves phosphorylating Rb and consequently releasing the transcription factor E2F. This promotes exit from the G1 phase, thereby increasing cell cycle progression [3-5]. Second, recent studies have also identified a novel kinase-independent function of CDK6 as a transcriptional regulator [39]. CDK6 potentially binds to transcription factors, such as NF-κB, STAT3, AP-1, SPI1, ETS, and PAX4, to function as a transcription cofactor [23,39,40]. CDK6 interacts with AP-1 and c-Jun to regulate the expression of VEGFA [39]. These findings indicate that CDK6 plays a dual role in carcinogenesis [40].
VEGF is increased aberrantly in TNBC tumors. It not only stimulates angiogenesis directly to provide vital nutrients for the tumor, but also boosts tumor-cell adhesion and migration via the cancer-endothelial cell interaction [41,42]. Revealing the regulatory mechanism of the tumor-microenvironmental interaction that supports cancer progression will certainly help in the development of novel therapeutic strategies for TNBC. Herein, our findings identify CDK6 as a potential target for cancer therapy that inhibits recruitment of c-Jun and Sp1 to the MMP-2 promoter to block lung metastasis.
To date, three CDK4/6 inhibitors, palbociclib, ribociclib, and abemaciclib, have been used in clinical cancer treatment. They exert their antitumor effects by targeting the cell-cycle progression function of CDKs in Rb-proficient tumor cells [18]. Clinical trials with these inhibitors have been performed in several cancer types, including breast cancer, leukemia, and soft tissue sarcomas [18]. In addition to inducing cell-cycle exit, CDK4/6 inhibitors may have other anticancer effects. For example, CDK4/6 inhibition promotes glycolytic and oxidative metabolism, increasing the number of mitochondria and reactive oxygen species accumulation via an Rb-dependent mechanism [43]. CDK4/6 inhibitor treatment also has been proposed to increase PD-L1 expression by blocking its degradation, as well as by increasing the number of PD-L1-expressing T cells and their infiltration [44,45]. Notably, CDK4-deficient supporting cells, but not cancer itself, also diminish the aggressive progression of oligodendroglioma [46]. Our data suggest a role for CDKs in developing the tumor microenvironment. Our findings also show that CDK6 plays an important role in the interaction between tumor and supporting cells, suggesting that inhibition of CDK6 may target the tumor microenvironment, resulting in a therapeutic effect.
An increasing number of early-phase clinical trials have examined the therapeutic effects of drug combinations [18]. According to the synthetic lethality theory, such combined treatments can minimize drug resistance and maximize antitumor effects [28]. In some types of cancer, resistance can be overcome by including a CDK4/6 inhibitor with signaling pathway-targeted inhibitor treatments, such as MEK and phosphoinositide 3-kinase/Akt inhibitors [47,48]. However, some have also reported an antagonistic effect of combined treatments used simultaneously due to interference between the treatments [48]. These findings demonstrate the necessity of designing an appropriate drug delivery schedule that depends on cancer type and biomarkers. After screening currently available signaling pathway-targeted drugs, we found that combined inhibition of CDK6 and Raf achieved the best cytotoxic effect in TNBC cells. Our results suggest that CDK6 inhibition could be a personalized treatment that works synergistically with chemotherapy drugs to improve overall therapeutic efficacy. However, further studies need to be conducted to determine the optimal sequence, timing, and dosages of combined CDK6/Raf inhibitor treatments for improving the therapeutic effect.
Conclusion
Summary, we provide evidence that CDK6 plays a significant role in the interaction between cancer and its microenvironment. CDK6 levels correlated positively with an increased angiogenic and fibrotic microenvironment in TNBC cells. This suggests that CDK6 can serve as both a biomarker and therapeutic target. CDK6 was involved in the cancer-microenvironmental interaction via the regulation of secreted factors that activated endothelial cells and fibroblasts. We also demonstrated that the Raf inhibitor, sorafenib, exerted a synergistic effect with CDK6 inhibition to induce synthetic lethality in TNBC cells. Overall, these results indicate that combining CDK6 inhibition with a Raf inhibitor could be a personalized treatment for the Rb-positive subpopulation of patients with TNBC.
Acknowledgements
We acknowledge the support from the following grants: (1) KMUH-DK-109001, KMUH107-7M12 and KMUH108-8R36 from the Kaohsiung Medical University Hospital; (2) MOHW109-TDU-B-212-010001 from the Ministry of Health and Welfare, Taiwan; (3) KMU-DK108011, KMU-DK-109003-1, KMUH-DK-109003-2 and KMUH-DK-109003-3 from KMU-KMUH Co-Project of Key Research; (4) 109-2314-B-037-019, 109-2314-B-037-132- and 109-2314-B-037-036-MY3 from the Ministry of Science and Technology, Taiwan; (5) 108CM-KMU-06 from Kaohsiung Medical University; (6) KMU-TC108A03-6 and KMU-TC109B05 from Kaohsiung Medical University Research Center Grant. The authors also thank the Center for Research Resources and Development in Kaohsiung Medical University for the assistance in flow cytometry analysis.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Liu FC, Lin HT, Kuo CF, See LC, Chiou MJ, Yu HP. Epidemiology and survival outcome of breast cancer in a nationwide study. Oncotarget. 2017;8:16939–16950. doi: 10.18632/oncotarget.15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, McCann A. The fate of chemoresistance in triple negative breast cancer (TNBC) BBA Clin. 2015;3:257–75. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner N, Reis-Filho J, Russell A, Springall R, Ryder K, Steele D, Savage K, Gillett C, Schmitt F, Ashworth A. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 4.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 5.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien HC, Lin CW, Huang PH, Chang ML, Hsu SM. Expression of cyclin-dependent kinase 6 (cdk6) and frequent loss of CD44 in nasal-nasopharyngeal NK/T-cell lymphomas: comparison with CD56-negative peripheral T-cell lymphomas. Lab Invest. 2000;80:893–900. doi: 10.1038/labinvest.3780093. [DOI] [PubMed] [Google Scholar]
- 7.Wiedemeyer WR, Dunn IF, Quayle SN, Zhang J, Chheda MG, Dunn GP, Zhuang L, Rosenbluh J, Chen S, Xiao Y, Shapiro GI, Hahn WC, Chin L. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci U S A. 2010;107:11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA Australian Pancreatic Cancer Genome Initiative. Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RS, Crown JP, Ettl J, Schmidt M, Bondarenko IM, Lang I, Pinter T, Boer K, Patel R, Randolph S, Kim ST, Huang X, Schnell P, Nadanaciva S, Bartlett CH, Slamon DJ. Efficacy and safety of palbociclib in combination with letrozole as first-line treatment of ER-positive, HER2-negative, advanced breast cancer: expanded analyses of subgroups from the randomized pivotal trial PALOMA-1/TRIO-18. Breast Cancer Res. 2016;18:67. doi: 10.1186/s13058-016-0721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkiewicz AK, Ertel A, McFalls J, Valsecchi ME, Schwartz G, Knudsen ES. RB-pathway disruption is associated with improved response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2012;18:5110–5122. doi: 10.1158/1078-0432.CCR-12-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton E, Infante JR. Targeting CDK4/6 in patients with cancer. Cancer Treat Rev. 2016;45:129–138. doi: 10.1016/j.ctrv.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Brough R, Gulati A, Haider S, Kumar R, Campbell J, Knudsen E, Pettitt SJ, Ryan CJ, Lord CJ. Identification of highly penetrant Rb-related synthetic lethal interactions in triple negative breast cancer. Oncogene. 2018;37:5701–5718. doi: 10.1038/s41388-018-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkiewicz AK, Cox D, Knudsen ES. CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes Cancer. 2014;5:261–272. doi: 10.18632/genesandcancer.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien N, Conklin D, Beckmann R, Luo T, Chau K, Thomas J, Mc Nulty A, Marchal C, Kalous O, von Euw E, Hurvitz S, Mockbee C, Slamon DJ. Preclinical activity of abemaciclib alone or in combination with antimitotic and targeted therapies in breast cancer. Mol Cancer Ther. 2018;17:897–907. doi: 10.1158/1535-7163.MCT-17-0290. [DOI] [PubMed] [Google Scholar]
- 18.Klein ME, Kovatcheva M, Davis LE, Tap WD, Koff A. CDK4/6 inhibitors: the mechanism of action may not be as simple as once thought. Cancer Cell. 2018;34:9–20. doi: 10.1016/j.ccell.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo CW, Wu CC, Ch’ang HJ. Radiation sensitization of tumor cells induced by shear stress: the roles of integrins and FAK. Biochim Biophys Acta. 2014;1843:2129–2137. doi: 10.1016/j.bbamcr.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Pan MR, Hou MF, Ou-Yang F, Wu CC, Chang SJ, Hung WC, Yip HK, Luo CW. FAK is required for tumor metastasis-related fluid microenvironment in triple-negative breast cancer. J Clin Med. 2019;8:38. doi: 10.3390/jcm8010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox TR, Erler JT. Fibrosis, cancer and the premetastatic niche. Breast Cancer Management. 2014;3:453–455. [Google Scholar]
- 23.Handschick K, Beuerlein K, Jurida L, Bartkuhn M, Müller H, Soelch J, Weber A, Dittrich-Breiholz O, Schneider H, Scharfe M, Jarek M, Stellzig J, Schmitz ML, Kracht M. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expression. Mol Cell. 2014;53:193–208. doi: 10.1016/j.molcel.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Bates AL, Pickup MW, Hallett MA, Dozier EA, Thomas S, Fingleton B. Stromal matrix metalloproteinase 2 regulates collagen expression and promotes the outgrowth of experimental metastases. J Pathol. 2015;235:773–783. doi: 10.1002/path.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer CF, Kronsteiner N, Marton E, Kubista M, Cullen KJ, Hirtenlehner K, Seifert M, Kubista E. MMP-2 and MMP-9 expression in breast cancer-derived human fibroblasts is differentially regulated by stromal-epithelial interactions. Breast Cancer Res Treat. 2002;72:69–77. doi: 10.1023/a:1014918512569. [DOI] [PubMed] [Google Scholar]
- 26.Longatto Filho A, Lopes JM, Schmitt FC. Angiogenesis and breast cancer. J Oncol. 2010;2010:576384–576391. doi: 10.1155/2010/576384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandler C, Liu T, Buckanovich R, Coffman LG. The double edge sword of fibrosis in cancer. Transl Res. 2019;209:55–67. doi: 10.1016/j.trsl.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat Rev Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 29.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 30.Tadesse S, Yu M, Kumarasiri M, Le BT, Wang S. Targeting CDK6 in cancer: State of the art and new insights. Cell Cycle. 2015;14:3220–3230. doi: 10.1080/15384101.2015.1084445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu Y, Xu J, Sun T. Cyclin-dependent kinases 4/6 inhibitors in breast cancer: current status, resistance, and combination strategies. J Cancer. 2019;10:5504–5517. doi: 10.7150/jca.32628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu YH, Yao J, Chan LC, Wu TJ, Hsu JL, Fang YF, Wei Y, Wu Y, Huang WC, Liu CL, Chang YC, Wang MY, Li CW, Shen J, Chen MK, Sahin AA, Sood A, Mills GB, Yu D, Hortobagyi GN, Hung MC. Definition of PKC-α, CDK6, and MET as therapeutic targets in triple-negative breast cancer. Cancer Res. 2014;74:4822–4835. doi: 10.1158/0008-5472.CAN-14-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu T, Di G. Role of tumor microenvironment in triple-negative breast cancer and its prognostic significance. Chin J Cancer Res. 2017;29:237–252. doi: 10.21147/j.issn.1000-9604.2017.03.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Place AE, Huh SJ, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13:227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casbas-Hernandez P, Sun X, Roman-Perez E, D’Arcy M, Sandhu R, Hishida A, McNaughton KK, Yang XR, Makowski L, Sherman ME, Figueroa JD, Troester MA. Tumor intrinsic subtype is reflected in cancer-adjacent tissue. Cancer Epidemiol Biomarkers Prev. 2015;24:406–414. doi: 10.1158/1055-9965.EPI-14-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao Y, Keller ET, Garfield DH, Shen K, Wang J, Reviews M. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takai K, Le A, Weaver VM, Werb Z. Targeting the cancer-associated fibroblasts as a treatment in triple-negative breast cancer. Oncotarget. 2016;7:82889–82901. doi: 10.18632/oncotarget.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SY, Kim HM, Koo JS. Differential expression of cancer-associated fibroblast-related proteins according to molecular subtype and stromal histology in breast cancer. Breast Cancer Res. 2015;149:727–741. doi: 10.1007/s10549-015-3291-9. [DOI] [PubMed] [Google Scholar]
- 39.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schäfer M, Fajmann S, Schlederer M, Schiefer A. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell. 2013;24:167–181. doi: 10.1016/j.ccr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tigan A, Bellutti F, Kollmann K, Tebb G, Sexl V. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene. 2016;35:3083–3091. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 41.Bender RJ, Mac Gabhann F. Expression of VEGF and semaphorin genes define subgroups of triple negative breast cancer. PLoS One. 2013;8:e61788. doi: 10.1371/journal.pone.0061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TH, Avraham HK, Jiang S, Avraham S. Vascular endothelial growth factor modulates the transendothelial migration of MDA-MB-231 breast cancer cells through regulation of brain microvascular endothelial cell permeability. J Biol Chem. 2003;278:5277–5284. doi: 10.1074/jbc.M210063200. [DOI] [PubMed] [Google Scholar]
- 43.Franco J, Balaji U, Freinkman E, Witkiewicz AK, Knudsen ES. Metabolic reprogramming of pancreatic cancer mediated by CDK4/6 inhibition elicits unique vulnerabilities. Cell Rep. 2016;14:979–990. doi: 10.1016/j.celrep.2015.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, Chhabra S, Huang W, Liu H, Aref AR, Ivanova E, Paweletz CP, Bowden M, Zhou CW, Herter-Sprie GS, Sorrentino JA, Bisi JE, Lizotte PH, Merlino AA, Quinn MM, Bufe LE, Yang A, Zhang Y, Zhang H, Gao P, Chen T, Cavanaugh ME, Rode AJ, Haines E, Roberts PJ, Strum JC, Richards WG, Lorch JH, Parangi S, Gunda V, Boland GM, Bueno R, Palakurthi S, Freeman GJ, Ritz J, Haining WN, Sharpless NE, Arthanari H, Shapiro GI, Barbie DA, Gray NS, Wong KK. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 2018;8:216–233. doi: 10.1158/2159-8290.CD-17-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciznadija D, Liu Y, Pyonteck SM, Holland EC, Koff A. Cyclin D1 and cdk4 mediate development of neurologically destructive oligodendroglioma. Cancer Res. 2011;71:6174–6183. doi: 10.1158/0008-5472.CAN-11-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cretella D, Fumarola C, Bonelli M, Alfieri R, La Monica S, Digiacomo G, Cavazzoni A, Galetti M, Generali D, Petronini PG. Pre-treatment with the CDK4/6 inhibitor palbociclib improves the efficacy of paclitaxel in TNBC cells. Sci Rep. 2019;9:13014. doi: 10.1038/s41598-019-49484-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.