Abstract
Hepatocellular carcinoma (HCC) is a worldwide malignancy with high morbidity and mortality. In this study, ubiquitin conjugating enzyme E2I (UBE2I), a small ubiquitin-like modifier E2 enzyme reportedly expressed in tumors, was examined for its potential effects in HCC. Bioinformatics analysis was performed based on HCCDB, TIMER, and Kaplan-Meier plotter databases to explore the clinical implications in HCC. An siRNA kit was used to downregulate UBE2I, and in vitro experiments-including migration, invasion and proliferation assays-were performed to examine UBE2I expression in HCC. Western blot (WB) was used to determine whether downregulated UBE2I expression influenced the prognosis of HCC via autophagy pathways. Finally, RNA-sequencing was performed to explore candidate molecular mechanisms underlying the effect of UBE2I. Bioinformatics analysis including stratification by alcohol ingestion and hepatitis status in HCC showed that highly expressed UBE2I was not only correlated with poor prognosis, but was also associated with immune infiltrates. In vitro experiments showed that high expression of UBE2I was associated with increased migration, invasion and proliferation of HCC cells. WB results indicated that downregulated expression of UBE2I was associated with higher levels of autophagy-related proteins including LC3A/B, Beclin-1 and ATG16L1. Moreover, RNA-sequencing results suggested that UBE2I was involved in hepatocarcinogenesis, non-alcohol fatty liver disease, steatohepatitis, liver fibrosis, inflammation, hepatoblastoma, tumor angiogenesis, type 2 mellitus diabetes, biliary tract disease and other diseases. We conclude that oncogene UBE2I is associated with poor prognosis of HCC via autophagy pathways and may be involved in hepatocarcinogenesis, tumor angiogenesis, non-alcohol fatty liver disease and inflammation.
Keywords: Hepatocellular carcinoma, UBE2I, autophagy pathway, prognosis, RNA-sequencing
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and ranks as the third leading cause of tumor-related deaths worldwide [1]. Due to delays in the early diagnosis of HCC, the disease has resulted in higher mortality worldwide [2,3]. Although several novel chemotherapeutic drugs have been introduced in clinical applications, surgery remains the most effective treatment for HCC patients [1]. However, surgical resection is limited in treating HCC due to the high incidence of recurrence, as well as intrahepatic and extra-hepatic metastasis [4-6], which means that the long-term survival of HCC patients remains poor. The highly sensitive biomarker alpha-fetoprotein has been used to predict the clinical survival of HCC patients after surgical resection, but the results have been unsatisfactory [7]. Therefore, it is critical to identify novel markers involved in the molecular mechanisms of HCC for early diagnosis and long-term surveillance.
In contrast to other post-translational modifications such as phosphorylation and acetylation, sumoylation adds a small ubiquitin-like modifier (SUMO) polypeptide to the ε-amino group of lysine residues [8-10]. In humans, there are four SUMO proteins, SUMO 1, 2, 3 and 4 [11]. SUMO 2 and 3 are highly homologous to each other and are similar to SUMO 1 [8,10]. Reminiscent of ubiquitination, sumoylation is catalyzed by a three-enzyme cascade, which is composed of a single heterodimeric E1 activating enzyme, a single E2 conjugating enzyme and multiple E3 ligases [9,10]. From yeast to humans, ubiquitin conjugating enzyme E2I (UBE2I), previously named ubiquitin conjugating enzyme 9 (UBC9), is the single SUMO E2 enzyme and thus provides a convenient intervention point for globally interrogating how sumoylation modulates many kinds of biological processes in these species [11]. In adipose tissue, UBE2I expression is positively associated with a marker of insulin resistance and corresponds with impaired browning of human white adipocytes, while the UBE2I/microRNA-30a axis regulates mitochondrial activity in human white adipocytes [12]. In lung cancer, Ping et al. demonstrated that upregulated UBE2I promotes cell invasion and metastasis of lung cancer cells, suggesting an important role in cancer progression [13]. In epithelial ovarian cancer, Li et al. showed that UBE2I promotes cell proliferation and therefore plays a pivotal role in this cancer through the PI3K/AKT pathway [14]. In glioma, Shengkui et al. found that UBE2I, also upregulated in glioma tissues, is negatively correlated with the prognosis of patients with glioma, indicating it may be a prognostic indicator of glioma [15]. Given the above research focused on UBE2I in tumors, we explored the potential role of UBE2I in HCC by bioinformatics analysis and functional experiments and identified candidate molecular mechanisms underlying the involvement of UBE2I in HCC progression.
Materials and methods
Analysis of differential expression and prognostic significance
The tumor immune estimation resource (TIMER; https://cistrome.shinyapps.io/timer/) database was used for differential analysis of UBE2I in humans and immune infiltrates in HCC [16,17]. In addition, the HCCDB (http://lifeome.net/database/hccdb/home.html) database was used to explore the prognostic significance of UBE2I in HCC and related co-expressed proteins [18].
Validation of prognostic significance and construction of a gene-gene interaction network using gene ontology (GO) terms and KEGG pathways of UBE2I-related co-expressed genes
For validation of differential expression and its prognostic significance, Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia2.cancer-pku.cn/#index) [19] and Kaplan-Meier plotter (https://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq) [20] databases were employed to examine prognostic significance to perform a stratified analysis by gender and race of UBE2I in HCC. Moreover, a gene-gene interaction network was visualized via geneMANIA plugin of Cytoscape version 3.8.0 [21,22]. To further explore potential molecular mechanisms underlying UBE2I involvement in HCC, UBE2I and its co-expressed related genes were used to construct an interaction network using the CluGO plugin of Cytoscape version 3.8.0 software [23]. Co-expressed genes were identified from the Cbioportal (https://www.cbioportal.org/) database [24,25].
Cell culture and cell transfection, RT-PCR and WB assays
To further determine the functions of UBE2I in HCC, HCCM and Huh7 cell lines were kindly provided by Prof. Guo-Dong Lu, National university of Singapore and were used for in vitro experiments [26]. Cells were cultured in high sugar Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, 8120113) with 10% fetal bovine serum (BI, 1903226), 1% of 100 U/mg penicillin and 100 μg/ml streptomycin (Solarbio, 20191203). Cells were cultured in 5% CO2 at 37°C. siRNA for UBE2I was purchased from the Ribobio company (siBDM1999A). Antibodies against UBE2I and GAPDH for western blot (WB) were purchased from Abcam (ab75854) and Aksomics (KC-5G5), respectively. Liposome lipo6000 was purchased from Beyotime (C0526).
Cell transfection was performed according to company instructions in five groups as follows: 5 μl of siRNA (1, 2, 3) +5 μl Lipo6000 transfection reagent, negative control (NC, 5 μl NC + 5 μl Lipo6000 transfection reagent), and mock group (5 μl DMEM + 5 μl Lipo6000 transfection reagent). Cells were treated with the siRNA mixtures for 6 h, then washed with sterile PBS and cultured in DMEM with 10% fetal bovine serum. The cells were extracted 48 h later using TRIzol reagent (Invitrogen, 15596026) to obtain total RNA for RT-PCR, or were lysed in RIPA reagent (Solarbio, R0010) after 72 h to obtain total protein for WB. Relative mRNA expression in the different groups was compared using the 2-ΔΔCT method. Primers for UBE2I and GAPDH were as follows: UBE2I Forward: AAAAATCCCGATGGCACGATG, Reverse: CTTCCCACGGAGTCCCTTTC; GAPDH Forward: GTCAGCCGCATCTTCTTT, Reverse: CGCCCAATACGACCAAAT. WB assays were performed to determine protein expression in the different groups.
Assays of cell invasion, migration and proliferation ability
Cell invasion ability was determined by Transwell assay. Matrix gels (8.7 mg/ml, 356234) was diluted according to instructions and was added to the upper chamber of Transwell plates (3422, Corning). A total of 1 × 105 cells was resuspended using DMEM and seeded in the upper chamber. The lower chamber received 800 μl medium containing FBS, penicillin and streptomycin. After 24 h, the Transwell chambers were firstly fixed in 800 μl of methanol and then stained in Crystal Violet Stain solution for 30 min. Then, cells were counted under the microscope before comparison.
Cell migration ability was determined using a wound healing assay. Cells were seeded in a six-well plate. Cell monolayers were scratched, and wound healing was assessed by measuring the gap distance at 0, 24 and 48 h. Cell proliferation assays were performed using MTT method (BB-4201, Bestbio). Cells transfected at 48 h were seeded into 96-well plates at 2000 cells/well. Then, cells were treated with 10 μl of MTT solution and incubated for 4 h. The cells were then extracted using 150 μl of MTT dissolving solution and mixed at low speed. Optical density was measured at 550 nm.
Determination of autophagy-related pathways of UBE2I
To further explore whether UBE2I utilizes an autophagy pathway, the Autophagy Antibody Sampler Kit (Cell Signaling Technology, #4445), including LC3A/B, ATG3 and Beclin-1 antibodies, was used for WB assay. Antibodies against these proteins were purchased from Cell Signaling Technology and diluted according to instructions. GAPDH was used as an internal control.
Further exploration of molecular mechanisms by RNA-sequencing analysis
We next explored molecular mechanisms by RNA-sequencing analysis using the DESeq2 method in HCCM and Huh7 cells in Novogene company [27]. First, differentially expressed genes (DEGs) were determined using the following criteria: |log2FoldChange| > 0, adjusted P ≤ 0.05. Then, enrichment analysis of GO terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Reactome pathways, disease ontology (DO) terms and DisGeNET was performed to further explore candidate molecular mechanisms underlying UBE2I involvement.
Statistical analysis
Graphpad 8.0 was used to construct box plots. Unpaired t-test was employed for comparisons for each two groups. Kaplan-Meier plots were constructed for survival analysis. Image J software was used for densitometric analysis. P ≤ 0.05 was considered statistically significant.
Results
Analysis of differential expression and clinical significance
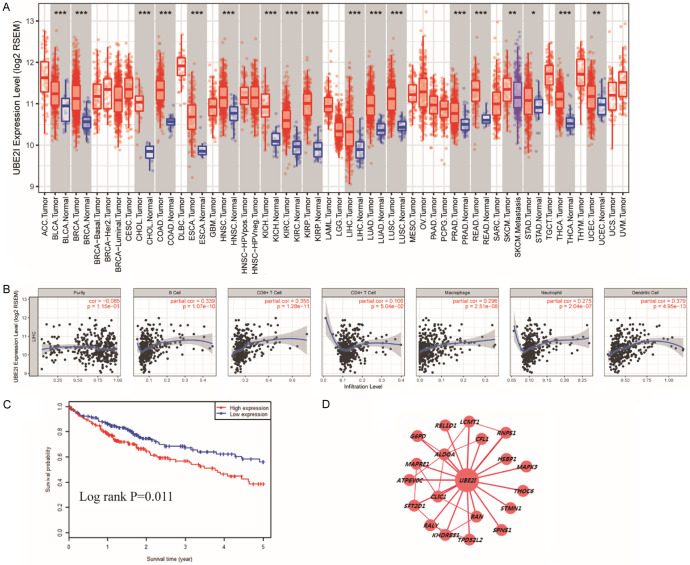
Analysis of differential expression showed that UBE2I was differentially expressed in several cancers, including HCC, bladder urothelial carcinoma, breast invasive carcinoma, cholangio carcinoma, colon adenocarcinoma, lung adenocarcinoma, rectum adenocarcinoma, kidney renal clear cell carcinoma, and others (all P < 0.001, Figure 1A). Immune infiltrate analysis indicated that UBE2I expression was positively correlated with B cell, CD8+ T cell, macrophage, neutrophil and dendritic cell infiltration levels (all P < 0.001, Figure 1B) but not with cell purity or CD4+ T cell infiltration. In addition, the HCCDB database showed that patients with high expression of UBE2I exhibited poor prognosis in HCC (Log-rank P = 0.011, Figure 1C). Co-expression network analysis indicated that UBE2I was associated with CFL1, RNPS1, HSBP1, MAPK3, THOC6, STMN1, CLIC1, SFT2D1, G6PD, LCMT1, ALDOA, and others (Figure 1D).
Figure 1.
Clinical analysis of UBE2I in HCC. A: Differential expression analysis of UBE2I in various cancers. B: Analysis of UBE2I expression and immune infiltrates. C: Survival analysis of UBE2I expression in HCC (low = 178, high = 178). D: Co-expression network of UBE2I and co-expression-related genes.
Validation of differential expression, prognostic significance and molecular mechanisms of related co-expressed-genes
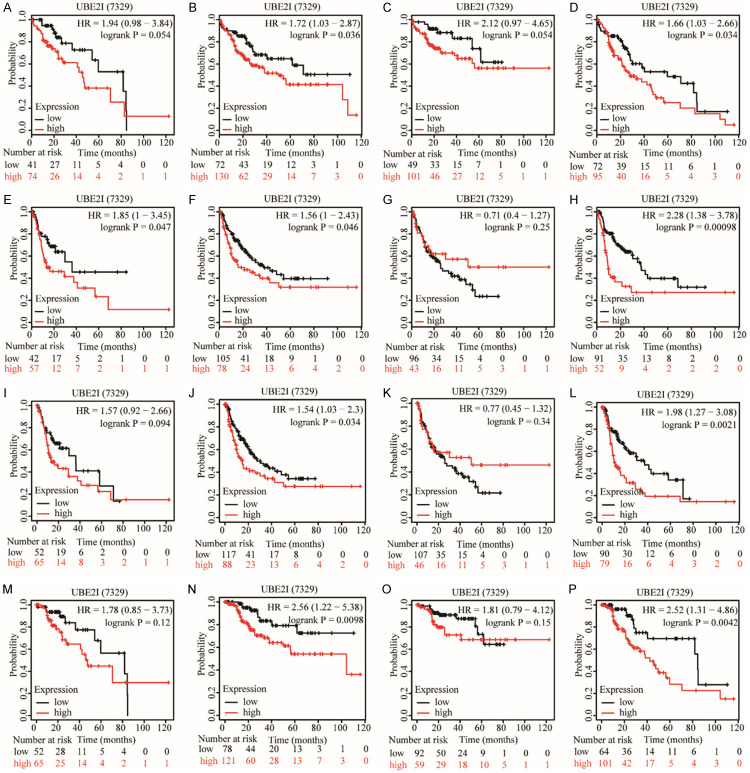
Differential expression of UBE2I was validated in the GEPIA database, which showed that UBE2I was differentially and highly expressed in HCC compared with non-tumorous tissue (P < 0.05, Figure 2A). A gene-gene interaction network was constructed which indicated that UBE2I had physical interactions with MITF, RANGAP1, RAD52, ETV6, and other proteins (Figure 2B). The prognostic significance of UBE2I was validated by the Kaplan-Meier plotter website, which showed that high expression of UBE2I was associated with poor OS, RFS, PFS, and DSS in HCC (Log-rank P = 0.016, 0.013, 0.011 and 0.015, respectively; Figure 2C-F). Then, stratified analysis according to HCC risk factors-alcohol consumption and hepatitis infection status-was performed for overall survival (OS), recurrence-free survival (RFS), progression-free survival (PFS) and disease-specific survival (DSS). In an alcohol-drinking population, high expression of UBE2I was associated with poor prognosis only for RFS (Log-rank P = 0.047, Figure 3E) not others (Figure 3A, 3I, 3M). However, in a non-drinking population, high expression of UBE2I was associated with poor prognosis for OS, RFS, PFS and DSS (all P ≤ 0.05, Figure 3B, 3F, 3J, 3N). In a hepatitis-infected population, UBE2I expression did not show any significant differences (Figure 3C, 3G, 3K, 3O). Nonetheless, high expression of UBE2I was associated with poor prognosis for OS, RFS, PFS and DSS (all P ≤ 0.05, Figure 3D, 3H, 3L, 3P).
Figure 2.
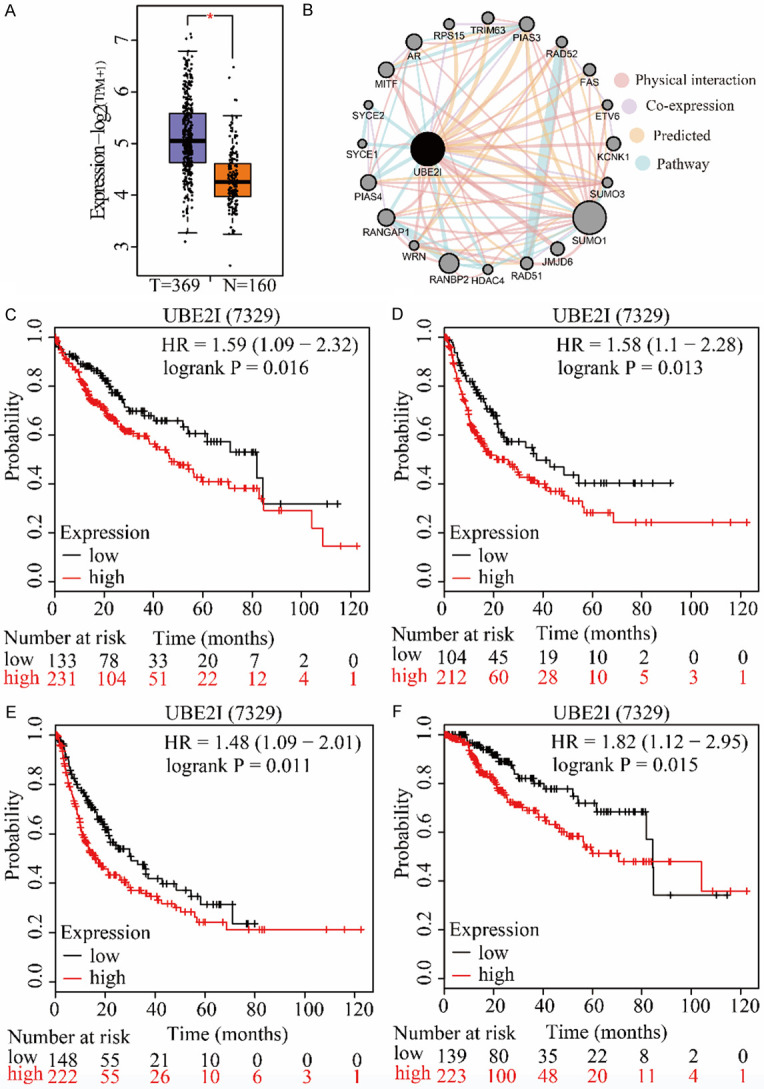
Clinical analysis and interaction network of UBE2I in HCC. A: Differential expression of UBE2I in HCC. B: Gene-gene interaction network of UBE2I and expression-related genes. C-F: Survival analysis of UBE2I expression affecting OS (low = 133, high = 231), RFS (low = 104, high = 212), PFS (low = 148, high = 222) and DSS (low = 139, high = 223) in HCC.
Figure 3.
Stratification analysis of UBE2I by alcohol ingestion and hepatitis status in HCC. A, E, I, M: Stratification analysis of UBE2I in a drinking population for OS (low = 41, high = 74), RFS (low = 42, high = 57), PFS (low = 52, high = 65) and DSS (low = 52, high = 65). B, F, J, N: Stratification analysis of UBE2I in a non-drinking population for OS (low = 72, high = 130), RFS (low = 105, high = 78), PFS (low = 117, high = 88) and DSS (low = 78, high = 121). C, G, K, O: Stratification analysis of UBE2I in a hepatitis patient population for OS (low = 49, high = 101), RFS (low = 96, high = 43), PFS (low = 107, high = 46) and DSS (low = 92, high = 59). D, H, L, P: Stratification analysis of UBE2I in a hepatitis-free population for OS (low = 72, high = 95), RFS (low = 91, high = 52), PFS (low = 90, high = 79) and DSS (low = 94, high = 101).
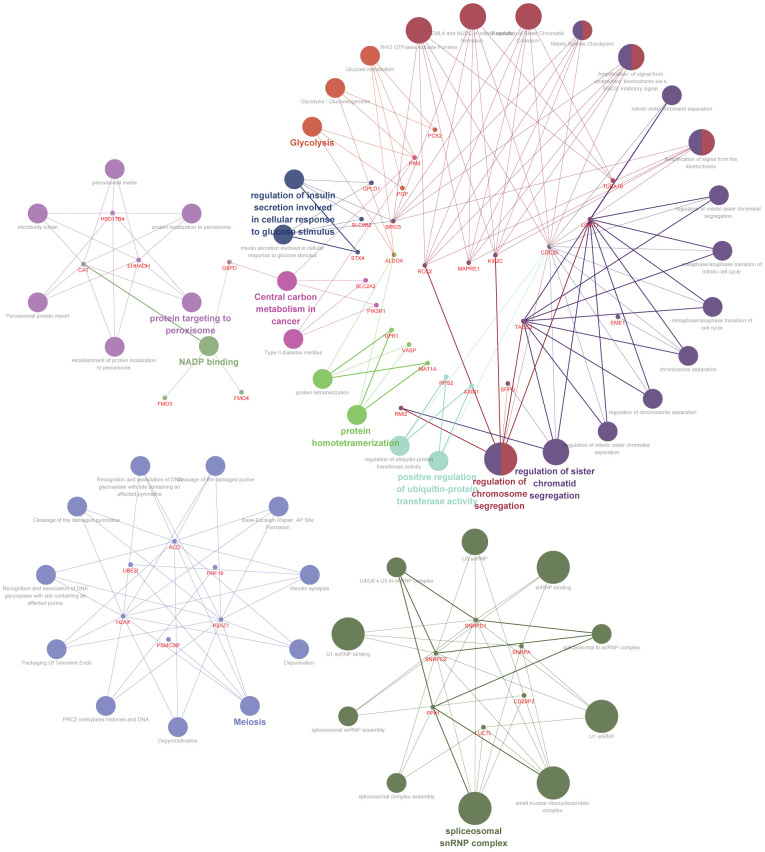
The top 100 co-expressed genes related to UBE2I were determined and are shown in Table S1. These results indicated that these genes were involved in protein targeting in peroxisome, glycolysis, positive regulation of ubiquitin-protein transferase activity, protein homotetramerization, regulation of chromosome segregation, regulation of chromatid segregation, central carbon metabolism in cancer, NADP binding and spliceosome snRNP complex, among others (Figure 4).
Figure 4.
Interaction network of candidate molecular mechanisms involving UBE2I and co-expression-related genes.
Downregulation of UBE2I expression by siRNA and assays of migration, invasion and proliferation
Downregulating expression of UBE2I was performed using an siRNA kit. Real-time PCR and WB consistently indicated that siRNA 2 (Si 2) was the most effective at suppressing UBE2I expression (Figure 5A-F). Therefore, siRNA 2 (Si 2) was used for further experiments.
Figure 5.
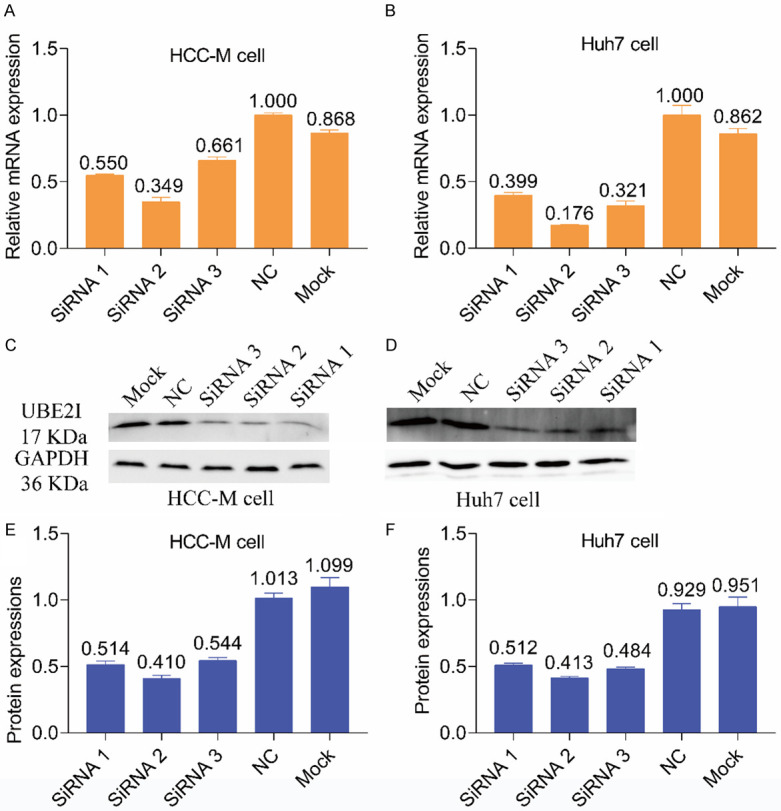
Results of real-time PCR and WB after silencing UBE2I. A, B: Results of real-time PCR after silencing UBE2I in HCCM and Huh7 cells. C, D: Results of WB after silencing UBE2I in HCCM and Huh7 cells. E, F: Histogram showing results of WB after silencing UBE2I in HCCM and Huh7 cells.
In invasion assays, downregulating UBE2I reduced cell movement through the transwell membrane compared with the NC group for both HCCM and Huh7 cells (both P < 0.001, Figure 6A-F), which indicated lower invasive capacity resulting from down-regulated expression of UBE2I. In migration assays, downregulating UBE2I resulted in reduced migration distance compared to the NC group for both HCCM and Huh7 cells at 24 h and 48 h (all P < 0.001, Figure 7A-D), which indicated lower migratory capacity produced by downregulated UBE2I expression. Proliferation assays showed that downregulation of UBE2I induced lower OD values compared with the NC group at 0 h, 24 h, 48 h and 72 h (all P < 0.05, Figure 8A, 8B), indicating lower proliferative ability with downregulated UBE2I expression. However, there was no significant difference between OD values in Huh7 cells at 24 h.
Figure 6.
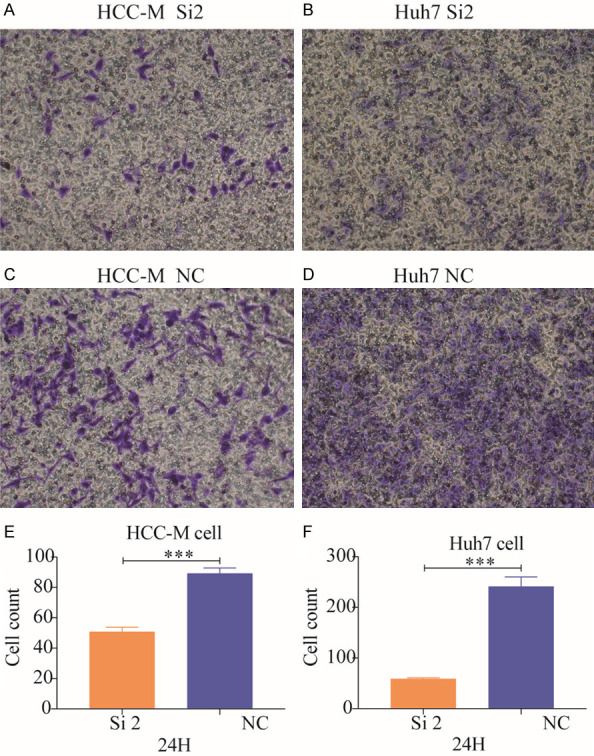
Results of cell invasion assays. A, B: Results of cell invasion assays in the siRNA-2 groups of HCCM and Huh7 cells. C, D: Results of cell invasion assays in the negative control groups for HCCM and Huh7 cells. E, F: Histograms comparing the two groups of HCCM and Huh7 cells.
Figure 7.
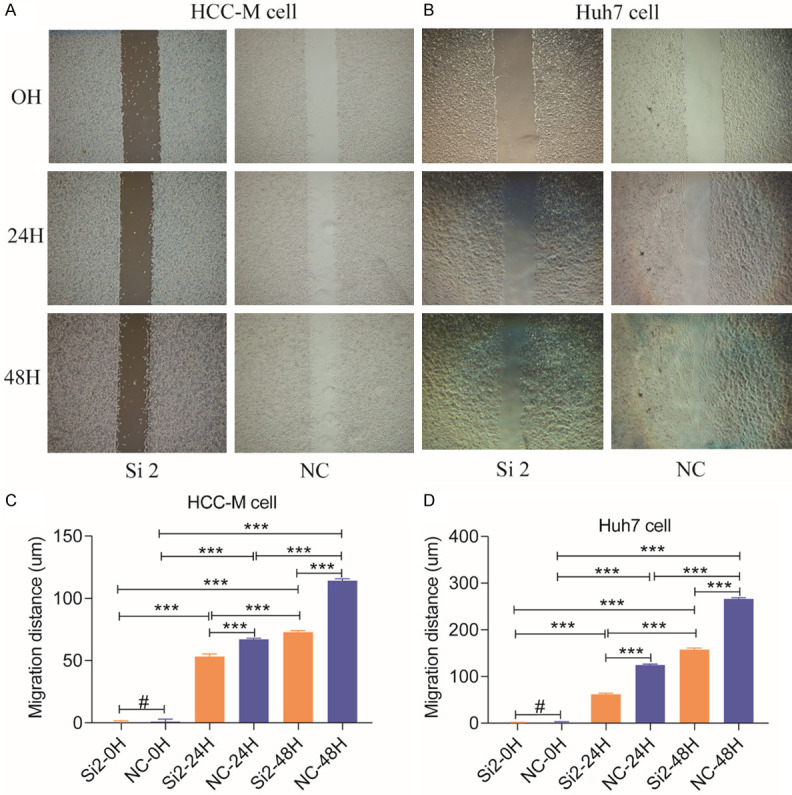
Results of cell migration assays. A, B: Results of cell invasion assays of HCCM and Huh7 cells. C, D: Histograms comparing the two groups of HCCM and Huh7 cells at 0, 24 and 48 h.
Figure 8.
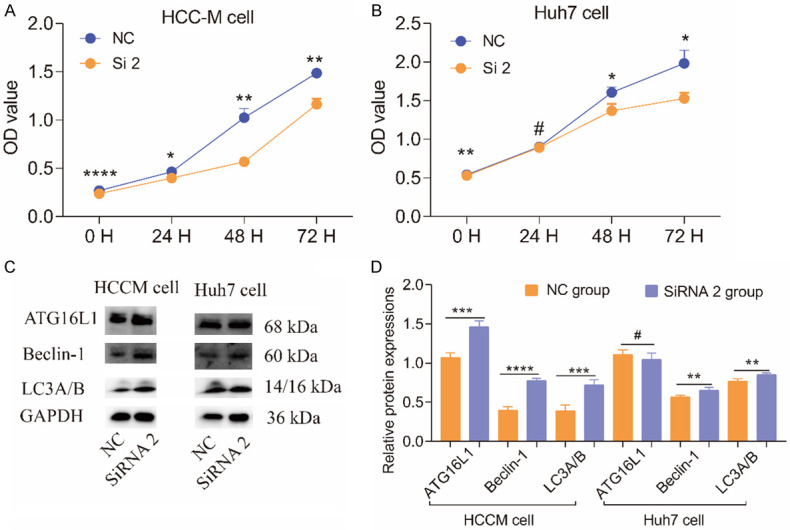
Results of cell proliferation assays and WB results of autophagy pathway-related proteins. A, B: Results of cell proliferation assays of HCCM and Huh7 cells at 0, 24, 48 and 72 h. C: WB results of autophagy pathway-related proteins. D: Histogram comparing autophagy pathway-related proteins in HCCM and Huh7 cells.
Involvement of autophagy-related pathways in UBE2I expression
To determine if UBE2I expression influences the prognosis of HCC via an autophagy pathway, we tested autophagy pathway-related proteins including LC3A/B, Beclin-1 and AGT16L1. WB results indicated that high expression of UBE2I was accompanied by low expression of LC3A/B, Beclin-1 and AGT16L1 in HCCM and Huh7 cells compared with downregulated UBE2I expression, especially in HCCM cells (all P < 0.05, Figure 8C, 8D). However, there was no significant difference between the two groups in terms of ATG16L1 expression in Huh7 cells.
Molecular mechanisms underlying UBE2I involvement in HCC by RNA-sequencing
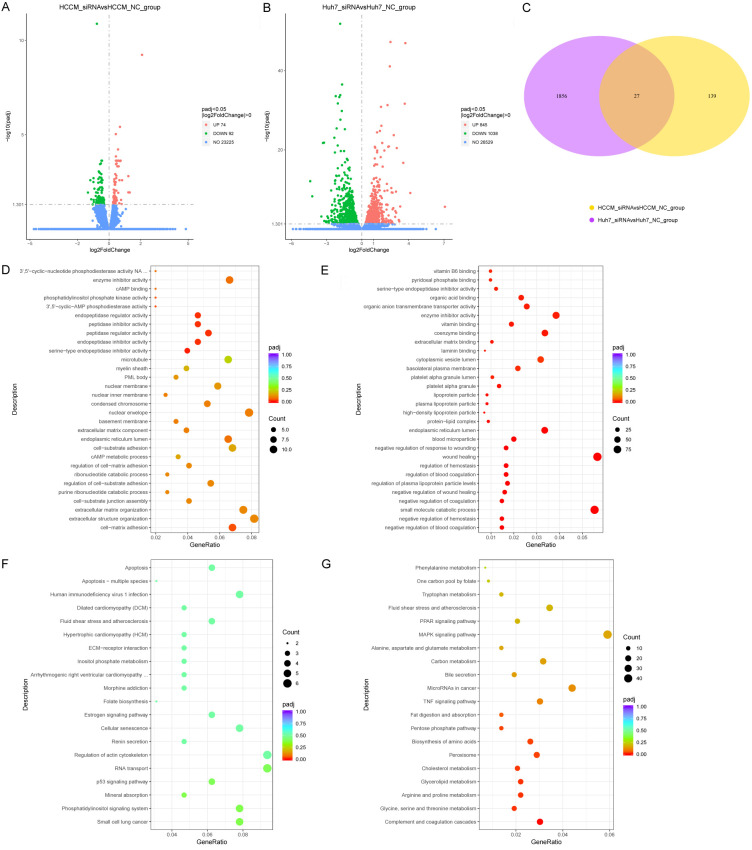
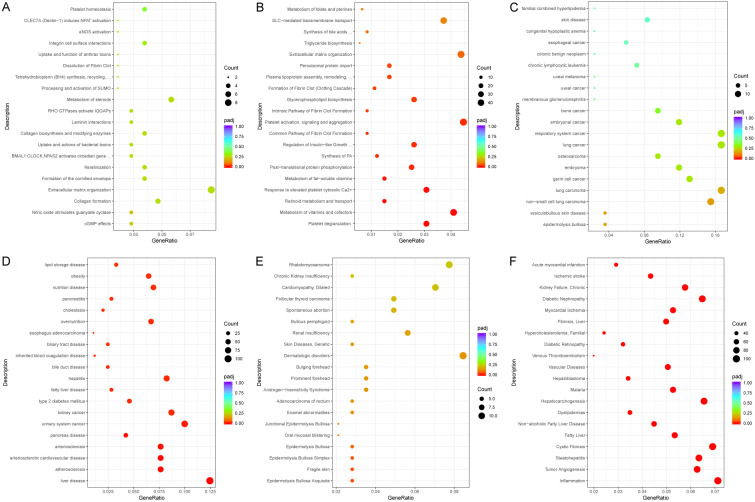
We further examined molecular mechanisms underlying UBE2I involvement in HCC by the RNA-sequencing method. A total of 166 DEGs, including 74 upregulated and 92 downregulated genes, were identified in HCCM cells. Meanwhile, 1883 DEGs, including 845 upregulated and 1038 downregulated genes, were identified in Huh7 cells (Figure 9A, 9B). A total of 27 DEGs intersected in the above two cell lines (Figure 9C). Details of DEGs in HCCM and Huh7 cells and intersecting DEGs are shown in Tables S2, S3, S4. Additionally, enrichment of GO terms and KEGG pathways showed the involvement of serine-type endopeptidase inhibitor activity, peptidase regulator activity, organic anion transmembrane transporter activity, extracellular matrix binding, laminin binding, wound healing, high-density lipoprotein particle, regulation of hemostasis, complement and coagulation cascades, among others (Figure 9D-G).
Figure 9.
Volcano plots, Venn diagram and dot plots based on DEGs in HCCM and Huh7 cells. A, B: Volcano plots in HCCM and Huh7 cells. C: Venn diagram of DEGs in HCCM and Huh7 cells. D, E: Dot plots showing enrichment of GO terms in HCCM and Huh7 cells. F, G: Dot plots showing enriched KEGG pathways in HCCM and Huh7 cells.
We then analyzed enrichment of reactome pathways, DO and DisGeNET. In the reactome pathway, we saw enrichment of perioximal protein import, plasma lipoprotein assembly, remodeling, formation of fibrin clot (clotting cascade), platelet degranulation, metabolism of vitamins and cofactors (Figure 10A, 10B). In DO, there was enrichment of liver disease, hepatitis, fatty liver disease, bile duct disease, biliary duct disease, obesity, lipid storage disease, type 2 mellitus diabetes and pancreas disease (Figure 10C, 10D). In DisGeNET, enrichment was seen for hepatocarcinogenesis, tumor angiogenesis, inflammation, steatohepatitis, fatty liver, non-alcoholic fatty liver disease, hepatoblastoma and liver fibrosis (Figure 10E, 10F).
Figure 10.
Dot plots of enrichment analysis results for the reactome pathway, disease ontology and DisGeNET. A, B: Dot plots of enrichment analysis results for reactome pathways in HCCM and Huh7 cells. C, D: Dot plots of enrichment analysis results for disease ontology in HCCM and Huh7 cells. E, F: Dot plots of enrichment analysis results for DisGeNET in HCCM and Huh7 cells.
Discussion
SUMO is a highly conserved protein family that is involved in protein modification after translation [28]. Although similar to ubiquitination in several aspects such as structure, conjugation process and attachment to targets, SUMOylation is different from ubiquitination in its biological consequences [28]. SUMOylation has been implicated in the modulation of transcriptional activity, protein activity, subcellular localization and protein-protein interactions [29,30]. The ubiquitination pathway employs many kinds of E2-conjugating enzymes for its metabolism [9]. In contrast, UBE2I is the only known E2-conjugating enzyme in the SUMO pathway and has been documented in yeast, invertebrates and vertebrates [31-33]. SUMOylation modulates a great number of cellular processes such as DNA replication and repair, chromosome integrity and segregation, signal transduction, cell cycle progression and nuclear transport, among others [34-37]. In agreement with previous reports, our study showed that UBE2I and its co-expression-related genes participated in protein targeting to peroxisome, NADP binding, central carbon metabolism in cancer, meiosis, protein homotetramerization, positive regulation of ubiquitin-protein transferase activity, regulation of chromosome segregation, regulation of sister chromatid segregation, glycolysis and regulation of insulin secretion involved in cellular response to glucose stimulus.
More recently, research has demonstrated that UBE2I can play an important role without relying on SUMOylation to regulate cell growth [38,39]. These studies suggested that changing UBE2I expression could influence cell growth and functions. For instance, Beck et al. suggested that overexpression of a UBE2I dominant-negative mutant was linked to increased drug sensitivity in breast cancer [40]. In yeast, defects in UBE2I expression induce increased sensitivity to genotoxic drugs [41]. In breast cancer, Mo et al. showed evidence that UBE2I promoted metastasis and invasion in a SUMOylation-independent manner via downregulation of a putative tumor suppressor microRNA-224 [42]. Su et al. suggested that high expression of UBE2I was correlated with poor response to chemotherapy and poor prognosis [43]. These research studies consistently indicate an oncogenic role for UBE2I expression in breast cancer.
In lung cancer research, He et al. demonstrated that high expression of UBE2I led to increased cell migration and invasion and suggested that it plays a pivotal role in lung cancer by promoting cell migration and invasion [43]. In osteosarcoma, Wu et al. revealed that downregulation of UBE2I suppressed tumorigenesis and enhanced chemosensitivity to HSV-TK/GCV by modulating connexin-43 SUMOylation [44]. Ran et al. found that UBE2I expression played a pivotal role in tumorigenesis and tumor progression of squamous cell carcinoma of the head and neck [45]. Shengkui et al. examined aberrant expression of microRNA-214 and UBE2I and found that high expression of UBE2I and low expression of microRNA-214 was associated with the poorest OS and was an independent prognostic predictor of glioma [15]. Their further stratification analysis demonstrated that high expression of UBE2I and low expression of microRNA-214 were obviously correlated with poor OS of glioma patients with high pathological grades [15]. The above studies thus suggested an oncogenic role for UBE2I expression in these malignancies. Parallel with them, we revealed an oncogenic role of UBE2I affecting HCC prognosis as assessed by OS, RFS, PFS and DSS. In addition, stratification analysis according to alcohol ingestion and hepatitis status suggested that high expression of UBE2I was an indicator of poor prognosis in HCC as well.
Autophagy is a catabolic mechanism and influences the growth of tumor cells. Autophagy maintains body development, aging and degeneration like a two-edged sword according to its changing activity [46]. Autophagy has been implicated as a disease-associated factor that is regulated in the liver cells of humans with liver diseases and accounts for the development and progression of various liver diseases such as hepatitis, steatosis, fibrosis, cirrhosis and HCC [47-50]. Autophagy protects liver cells from injury and cell death by eliminating injured organelles and proteins associated with liver diseases [51]. Beclin-1 is a key protein in the formation of autophagosomes [52]. LC3-I is transformed to LC3-II during autophagy and an increase in the LC3-II/LC3-I ratio indicates an enhancement of autophagy levels [53].
Jiang’s study focused on the role of matrine in HCC and showed that matrine could inhibit cell proliferation, migration and invasion and promote autophagy in HCC by regulating the circ_0027345/miR-345-5P/HOXD3 axis [54]. In agreement with this study, our research showed that silencing UBE2I expression led to decreased cell migration, invasion and proliferation and promoted protein expression of LC3A/B, Beclin-1 and ATG16L1 in the autophagy pathway in HCC cells. By inhibiting UBE2I expression to decrease the IC50 value of doxorubicin in HCC cells, Guo et al. showed that downregulating UBE2I could increase the sensitivity of HCC to doxorubicin [28]. They further demonstrated that downregulation of UBE2I has an apparent effect on the mitogen-activated protein kinase signaling pathway [28]. Recently, Lou et al. found that UBE2I promotes metastasis and was correlated with poor prognosis in HCC [55]. They suggested that downregulating UBE2I expression significantly inhibited the migration and invasion of HCC cells. Consistent with their results, our study found that knockdown of UBE2I decreased cell migration, invasion and proliferation. In addition, Lu et al. showed that loss of inhibition by has-miR-195-3P and dysregulation of UBE2I promoter methylation were associated with the overexpression of UBE2I in HCC. In contrast, we explored whether changing UBE2I expression influenced expression of autophagy pathway-related proteins and found that UBE2I knockdown was associated with high expression of autophagy pathway-related proteins, including LC3A/B, Beclin-1 and ATG16L1 in HCC cells.
Furthermore, we explored possible molecular mechanisms of UBE2I using the RNA-sequencing method. RNA-sequencing results indicated enrichment in GO terms and KEGG pathways involving serine-type endopeptidase inhibitor activity, peptidase regulator activity, organic anion transmembrane transporter activity, extracellular matrix binding, laminin binding, wound healing, high-density lipoprotein particle, regulation of hemostasis, complement and coagulation cascades. Enrichment analysis including the reactome pathway, DO, and DisGeNET was then performed. Results suggested involvement in hepatocarcinogenesis, tumor angiogenesis, inflammation, steatohepatitis, fatty liver, non-alcoholic fatty liver disease, hepatoblastoma, liver fibrosis, hepatitis, biliary duct disease, obesity, lipid storage disease, type 2 mellitus diabetes, pancreas disease, perioximal protein import, plasma lipoprotein assembly and remodeling. To date, a great deal of research has focused on non-alcohol fatty liver disease [56], liver fibrosis [57,58], hepatitis [59], steotohepatitis [60,61], obesity [62] and type 2 diabetes mellitus [63] and has implicated them in the occurrence and progression of HCC. Therefore, we have good reasons to believe that UBE2I may be involved in HCC through its relationship with the above diseases. However, this hypothesis needs further validation.
There were some limitations in the study that should be recognized. First, in vivo experiments need to be performed to explore the molecular mechanisms. Second, RNA-sequencing results need to be further validated in future studies. Third, associations between UBE2I expression and clinical pathological factors should be investigated. Then, relationship between overexpression of UBE2I and cell lines should further be attempted to validate. Moreover, RNA-sequencing results should be verified in the near future work.
Conclusion
We found that UBE2I was not only differentially expressed in HCC but also showed prognostic significance and association with immune infiltrates in HCC. In vitro experiments demonstrated that high expression of UBE2I was associated with increased cell migration, invasion and proliferation of HCC cells. WB indicated that downregulated expression of UBE2I was negatively correlated with higher expression of autophagy pathway proteins. Furthermore, we conducted RNA-sequencing after silencing UBE2I and found that it participated in the process of hepatocarcinogenesis, tumor angiogenesis, inflammation, steatohepatitis, non-alcoholic fatty liver disease, liver fibrosis, hepatitis, obesity, lipid storage disease, type 2 mellitus diabetes and others. Further validation of these results is warranted in future studies.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (No.: 81560535, 81802874), Natural Science Foundation of Guangxi Province of China (Grant No. 2018GXNSFBA138013, 2018GXNSFAA050119, 2020GXNSFAA159127), Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (GKE2018-01, GKE2019-11, GKEZZ202009), The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2018KY0110), Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (No. GXCDCKL201902), and 2018 Innovation Project of Guangxi Graduate Education (JGY2018037). As well as, the present study is also partly supported by Research Institute of Innovative Think-tank in Guangxi Medical University (The gene-environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention) and The central government in 2019 subsidized Guangxi’s major and difficult disease clinical collaboration construction project of Chinese and Western medicine. We would also acknowledge the supported by the Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education. This article does not contain any studies with human participants or animals performed by any of the authors.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen J, Zhang ZQ, Song J, Liu QM, Wang C, Huang Z, Chu L, Liang HF, Zhang BX, Chen XP. 18beta-glycyrrhetinic-acid-mediated unfolded protein response induces autophagy and apoptosis in hepatocellular carcinoma. Sci Rep. 2018;8:9365. doi: 10.1038/s41598-018-27142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J Hepatol. 2015;7:1964–1970. doi: 10.4254/wjh.v7.i15.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villanueva A, Llovet JM. Liver cancer in 2013: mutational landscape of HCC--the end of the beginning. Nat Rev Clin Oncol. 2014;11:73–74. doi: 10.1038/nrclinonc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levi Sandri GB, de Werra E, Mascianà G, Colasanti M, Santoro R, D’Andrea V, Ettorre GM. Laparoscopic and robotic approach for hepatocellular carcinoma-state of the art. Hepatobiliary Surg Nutr. 2016;5:478–484. doi: 10.21037/hbsn.2016.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyoda H, Kumada T, Tada T, Ito T, Maeda A, Kaneoka Y, Kagebayashi C, Satomura S. Changes in highly sensitive alpha-fetoprotein for the prediction of the outcome in patients with hepatocellular carcinoma after hepatectomy. Cancer Med. 2014;3:643–651. doi: 10.1002/cam4.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 11.Tahmasebi S, Ghorbani M, Savage P, Gocevski G, Yang XJ. The SUMO conjugating enzyme Ubc9 is required for inducing and maintaining stem cell pluripotency. Stem Cells. 2014;32:1012–1020. doi: 10.1002/stem.1600. [DOI] [PubMed] [Google Scholar]
- 12.Koh EH, Chen Y, Bader DA, Hamilton MP, He B, York B, Kajimura S, McGuire SE, Hartig SM. Mitochondrial activity in human white adipocytes is regulated by the ubiquitin carrier protein 9/microRNA-30a axis. J Biol Chem. 2016;291:24747–24755. doi: 10.1074/jbc.M116.749408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Niu H, Peng Y, Wang J, He P. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol Rep. 2013;29:1588–1594. doi: 10.3892/or.2013.2268. [DOI] [PubMed] [Google Scholar]
- 14.Dong M, Pang X, Xu Y, Wen F, Zhang Y. Ubiquitin-conjugating enzyme 9 promotes epithelial ovarian cancer cell proliferation in vitro. Int J Mol Sci. 2013;14:11061–11071. doi: 10.3390/ijms140611061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S, Jiao B, Geng S, Ma S, Liang Z, Lu S. Combined aberrant expression of microRNA-214 and UBC9 is an independent unfavorable prognostic factor for patients with gliomas. Med Oncol. 2014;31:767. doi: 10.1007/s12032-013-0767-5. [DOI] [PubMed] [Google Scholar]
- 16.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian Q, Wang S, Zhang G, Wang D, Luo G, Tang J, Chen L, Gu J. HCCDB: a database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics. 2018;16:269–275. doi: 10.1016/j.gpb.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (Oxford, England) 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu GD, Leung CH, Yan B, Tan CM, Low SY, Aung MO, Salto-Tellez M, Lim SG, Hooi SC. C/EBPalpha is up-regulated in a subset of hepatocellular carcinomas and plays a role in cell growth and proliferation. Gastroenterology. 2010;139:632–43. 643.e1–4. doi: 10.1053/j.gastro.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang S, Qiu J, Wu Z, Bai T, Guo W. Down-regulation of UBC9 increases the sensitivity of hepatocellular carcinoma to doxorubicin. Oncotarget. 2017;8:49783–49795. doi: 10.18632/oncotarget.17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 30.Zhou F, Xue Y, Lu H, Chen G, Yao X. A genome-wide analysis of sumoylation-related biological processes and functions in human nucleus. FEBS Lett. 2005;579:3369–3375. doi: 10.1016/j.febslet.2005.04.076. [DOI] [PubMed] [Google Scholar]
- 31.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 33.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 34.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 35.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 36.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li YJ, Stark JM, Chen DJ, Ann DK, Chen Y. Role of SUMO: SIM-mediated protein-protein interaction in non-homologous end joining. Oncogene. 2010;29:3509–3518. doi: 10.1038/onc.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Yang MC, Weissler JC, Yang YS. Modulation of PLAGL2 transactivation activity by Ubc9 co-activation not SUMOylation. Biochem Biophys Res Commun. 2008;374:570–575. doi: 10.1016/j.bbrc.2008.07.064. [DOI] [PubMed] [Google Scholar]
- 39.Kaul S, Blackford JA Jr, Cho S, Simons SS Jr. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J Biol Chem. 2002;277:12541–12549. doi: 10.1074/jbc.M112330200. [DOI] [PubMed] [Google Scholar]
- 40.Mo YY, Yu Y, Ee PL, Beck WT. Overexpression of a dominant-negative mutant Ubc9 is associated with increased sensitivity to anticancer drugs. Cancer Res. 2004;64:2793–2798. doi: 10.1158/0008-5472.can-03-2410. [DOI] [PubMed] [Google Scholar]
- 41.Jacquiau HR, van Waardenburg RC, Reid RJ, Woo MH, Guo H, Johnson ES, Bjornsti MA. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J Biol Chem. 2005;280:23566–23575. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
- 42.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SF, Gong C, Luo M, Yao HR, Zeng YJ, Su FX. Ubc9 expression predicts chemoresistance in breast cancer. Chin J Cancer. 2011;30:638–644. doi: 10.5732/cjc.011.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Yu K, Yang Z, Li Y, Ma X, Bian X, Liu F, Li L, Liu X, Wu W. Silencing Ubc9 expression suppresses osteosarcoma tumorigenesis and enhances chemosensitivity to HSV-TK/GCV by regulating connexin 43 SUMOylation. Int J Oncol. 2018;53:1323–1331. doi: 10.3892/ijo.2018.4448. [DOI] [PubMed] [Google Scholar]
- 45.Ronen O, Malone JP, Kay P, Bivens C, Hall K, Paruchuri LP, Mo YY, Robbins KT, Ran S. Expression of a novel marker, Ubc9, in squamous cell carcinoma of the head and neck. Head Neck. 2009;31:845–855. doi: 10.1002/hed.21048. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Choi ME. Autophagy in kidney health and disease. Antioxid Redox Signal. 2014;20:519–537. doi: 10.1089/ars.2013.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 48.Wang K. Autophagy and apoptosis in liver injury. Cell Cycle. 2015;14:1631–1642. doi: 10.1080/15384101.2015.1038685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czaja MJ. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Ke PY. Diverse functions of autophagy in liver physiology and liver diseases. Int J Mol Sci. 2019;20:300. doi: 10.3390/ijms20020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin S, Zhuang J, Zhu L, Jiang Z. Matrine inhibits cell growth, migration, invasion and promotes autophagy in hepatocellular carcinoma by regulation of circ_0027345/miR-345-5p/HOXD3 axis. Cancer Cell Int. 2020;20:246. doi: 10.1186/s12935-020-01293-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Gao S, Chen J, Lou W. UBE2I promotes metastasis and correlates with poor prognosis in hepatocellular carcinoma. Cancer Cell Int. 2020;20:234. doi: 10.1186/s12935-020-01311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Younossi Z, Stepanova M, Ong JP, Jacobson IM, Bugianesi E, Duseja A, Eguchi Y, Wong VW, Negro F, Yilmaz Y, Romero-Gomez M, George J, Ahmed A, Wong R, Younossi I, Ziayee M, Afendy A. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol. 2019;17:748–755. e743. doi: 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 57.Younossi Z, Henry L. Contribution of alcoholic and nonalcoholic fatty liver disease to the burden of liver-related morbidity and mortality. Gastroenterology. 2016;150:1778–1785. doi: 10.1053/j.gastro.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Kim D, Li AA, Perumpail BJ, Gadiparthi C, Kim W, Cholankeril G. Changing trends in etiology-based and ethnicity-based annual mortality rates of cirrhosis and hepatocellular carcinoma in the United States. Hepatology. 2019;69:1064–1074. doi: 10.1002/hep.30161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marengo A, Rosso C, Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 60.Younossi ZM, Stepanova M, Rafiq N, Makhlouf H, Younoszai Z, Agrawal R, Goodman Z. Pathologic criteria for nonalcoholic steatohepatitis: interprotocol agreement and ability to predict liver-related mortality. Hepatology. 2011;53:1874–1882. doi: 10.1002/hep.24268. [DOI] [PubMed] [Google Scholar]
- 61.Younossi ZM, Stepanova M, Rafiq N, Henry L, Loomba R, Makhlouf H, Goodman Z. Nonalcoholic steatofibrosis independently predicts mortality in nonalcoholic fatty liver disease. Hepatol Commun. 2017;1:421–428. doi: 10.1002/hep4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 63.Wainwright P, Scorletti E, Byrne C. Type 2 diabetes and hepatocellular carcinoma: risk factors and pathogenesis. Curr Diab Rep. 2017;17:20. doi: 10.1007/s11892-017-0851-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.