Abstract
Abnormal circular RNA (circRNA) expression correlates with human traits such as many kinds of cancers. Though circRNAs have links to cancer, they have less characterization in metastatic castration-resistant prostate cancer (PCa), which is main reason for PCa mortality. Therefore, high-throughput sequencing was used for selected circRNA profiles. The result showed that circ-TRPS1 was upregulated significantly in high-grade PCa tissues or cell lines. High circ-TRPS1 expression correlated to aggressive PCa phenotypes. Knockdown of circ-TRPS1 suppressed PCa proliferation and metastasis through targeting miR-124-3p/EZH2 axis-mediated stemness in PCa, which was validated by luciferase reporter assays. EZH2 overexpression or miR-124-3p inhibition reversed the inhibition of circ-TRPS1 silencing in PCa cell migration and proliferation by recovering stemness. In summary, data demonstrated that circ-TRPS1 suppressed PCa progression through functioning similar to a miR-124-3p sponge to enhance EZH2 expression and cancer stem-like cell differentiation. Thus, circ-TRPS1 might be a candidate target for PCa treatment.
Keywords: Prostatic cancer, circ-TRPS1, miR-124-3p, EZH2, cancer stem-like cell
Introduction
Prostate cancer (PCa) is amongst the most popular carcinomas in male. Organ-confined PCa is efficiently treated through radical prostatectomy [1]. For advanced PCa, androgen deprivation therapy is regarded as first-line treatment [2]. If hormone resistance occurs, advanced PCa is usually severe because of PCa metastasis [3]. Therefore, studying molecular mechanisms and characterizing new targets for the development of therapeutics for patients with PCa is indispensable.
Studies find that noncoding RNAs (ncRNAs), including circular RNAs (circRNAs) and microRNAs (miRNAs) function in cancer progression [4]. CircRNAs arise from backsplicing of a 3’ splice donor to an upstream 5’ splice acceptor. CircRNAs are transcripts that are closed covalently and are resistant to exoribonuclease degradation, which are usually more stable than linear transcripts [5]. Aberrant circRNA expression has been discovered in many kinds of cancers such as breast [6], colorectal [7], gastric [8], and lung [9]. For prostate cancer, studies show that circLMTK2 acts as tumor suppressor in PCa through regulating microRNA-183 expression [10]. Silencing circular RNA circZNF609 restrains growth, invasion and migration through upregulating microRNA-186-5p in Pca [11]. Studies also find that circular RNA 17 [4], circAMOTL1L [12], circ-102004 [13], and circSMARCA5 [14] also function in PCa progression. Although some circRNA involvement is reported in PCa, investigations into biological mechanism and regulatory functions regarding circRNAs in PCa remain largely unclear. In this manner, the regulatory mechanisms regarding circRNAs in cancer need to be elucidated.
This investigation studied circRNA expression profiles in different grades of PCa tissues. We identified that circ-TRPS1 was increased significantly in high grade-PCa tissues and was related closely to PCa prognosis. We also discovered that circ-TRPS1 might act as an antioncogene sponge of miR-124-3p to enhance EZH2 expression and promote cancer stem-like cell-mediated PCa progression. As a result, circ-TRPS1 increases could function as biomarkers for prognosis and prediction, and a candidate PCa therapeutic target.
Materials and methods
Tissue samples
We collected 80 fresh PCa tissues after getting patient consent that informed in Renji Hospital Affiliated Shanghai Jiaotong University (Shanghai, China). Cancer tissues removed surgically from patients with PCa were frozen immediately in liquid nitrogen, which were stored at -80°C. We evaluated clinicopathological features of PCa following the revised International System for Staging Prostatic Cancer. The Ethics Committee in Renji Hospital Affiliated Shanghai Jiaotong University (Shanghai, China) approved this study.
Strand-specific high-throughput RNA-Seq library construction
We extracted total RNA from high-grade PCa (H-PCa) and low-grade PCa (L-PCa) tissues with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). We used about total RNA of 3 μg from each sample with VAHTS Total RNA-seq (H/M/R) Library Prep kits from Illumina (Vazyme Biotech Co., Ltd, Nanjing, China) to remove ribosomal RNA, and retain RNA classes including noncoding RNAs and mRNAs. We treated RNA through 40 U RNase R (Epicenter) at 37°C for three hours, followed by TRIzol purification. We prepared RNA-seq libraries via KAPA Stranded RNA-Seq Library Prep kits (Roche, Basel, Switzerland) and used them for deep sequencing (Illumina HiSeq 4000 at Aksomics, Inc., Shanghai, China).
Animals
We utilized BALB/c nude mice at 4 weeks old weighing 15~20 g (SLARC, Shanghai, China). Ethics Committee at Renji Hospital Affiliated Shanghai Jiaotong University (Shanghai, China) approved the animal experiments.
Cell culture and transfection
We cultured PC3, LnCaP, and DU145 cells. We maintained them in RPMI 1640 medium (Gibco, Grand Island, NY, USA) with penicillin of 100 units/ml and streptomycin of 100 μg/ml. We maintained RWPE-1 cells in K-SMF medium (Gibco, Grand Island, NY, USA) supplemented with epidermal growth factor of 5 ng/mL and bovine pituitary extract of 50 μg/mL. We incubated cultures in humidified environment with CO2 of 5% and 37°C.
We transfected small interfering RNAs for circ-TRPS1 (si-circ-TRPS1, 5’-TGCAGGACATCAATTCTTCAAGG-3’), miR-124-3p mimics (5’-UAAGGCACGCGGUGAAUGCC-3’), miR-124-3p inhibitors (5’-CGUGUUCACAGCGGACCUUGAU-3’), small interfering RNA for EZH2 (siEZH2, 5’-GAGGGAAAGUGUAUGAUAATT-3’), and the EZH2 pCDNA3.1 overexpression vector, and their NCs into cultured PC3 or LnCaP cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to standard process. To verify the circ-TRPS1 effects using in vivo experiments, lentiviral stabilized circ-TRPS1 silenced (sh-circ-TRPS1) PC3 cells were constructed.
Quantitative real-time polymerase chain reaction (RT-qPCR)
We used TRIzol reagent (Invitrogen, Carlsbad, CA, USA) to isolate total RNA from cultured tissues and cells. We determined RNA concentration and purity through a Nano-Drop 2000 spectrophotometer (Thermo Scientific, USA). We used PrimeScript™ RT reagent Kit (Takara, Japan) to synthesize cDNA. We determined miR-124-3p expression through miRNA qRT-PCR Starter kits (Riobobio, USA) and U6 and miR-124-3p (Riobobio) primers. We used the SYBR® Green Real time PCR Master Mix (Toyobo, Japan) to measure circ-TRPS1 and EZH2 expression. We normalized miR-124-3p, circ-TRPS1 and EZH2 expression to U6 and GADPH. We calculated relative miR-124-3p, circ-TRPS1 and EZH2 expressions using the 2-ΔΔCt method. Primers for circ-TRPS1, miR-124-3p, and EZH2 were: circ-TRPS1, (Forward, 5’-CTAAATGGGACCTATG-3’; Reverse, 5’-CAATTGTCTTAGAAGACGCTC-3’); miR-124-3p, (Forward, 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGCATTCA-3’; Reverse, 5’-ACACTCCAGCTGGGTAAGGCACGCGGTGAATGCC-3’); EZH2 (Forward, 5’-GCCATTATGGACATCGCGGTGC-3’; Reverse, 5’-CTTCGCGCTCGCGTGCCG-3’); U6 (Forward, 5’-CTCGCTTCGGCAGCACA-3’; Reverse, 5’-AACGCTTCACGAATTTGCGT-3’); GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Forward, 5’-ACAACTTTGGTATCGTGGAAGG-3’; and Reverse, 5’-GCCATCACGCCACAGTTTC-3’).
Fluorescence in situ hybridization (FISH)
We obtained specific probes for circ-TRPS1 (Dig-5’-CTATGATGTGCAGATGTACAGATCAGCTC-3’-Dig) from Geneseed Biotech (Guangzhou, China). Cells were immunostained with Cy3-conjugated antidigoxin and Cy3-conjugated antibiotin antibodies (Jackson ImmunoResearch Inc., West Grove, PA, USA). We counterstained nuclei through 4,6-diamidino-2-phenylindole (DAPI). We visualized staining through Zeiss LSM 700 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Bioinformatic analysis
We identified circRNA/miRNA target genes with CircularRNA Interactome. We predicted interactive correlations between miR-124-3p and EZH2 through TargetScanHuman.
Tumor sphere formation assays
We harvested PC3 and LnCaP cells, and resuspended them as single cells in serum-free medium. After cell counting, we added 200 cells/well in 200 μL serum-free medium to 96-well plates, 10 wells per group, changing medium every 2 days. We obtained images of five randomly selected regions for each group of wells with a camera-equipped microplate reader (Leica, Wetzlar, Germany) and calculated sphere percentage as number of spheres/200.
Cell proliferation assays
We employed Cell Counting Kit-8 assays (CCK-8; Gibco, Logan, Utah, USA) to quantify cellular proliferation. We seeded transfected cells into plates with 96 wells with 5,000 cells/well in triplicate, and determined cell viability with a CCK-8 system at 0, 24, 48 and 72 h after seeding, following given procedures.
For colony formation assays, we seeded cells that transfected into plates with 6 wells at 2,000 cells/well and maintained them in 1640 containing FBS of 10% for ten days. We imaged and counted colonies after fixing and staining.
Transwell assays
For invasion assays, we placed Transwell assay inserts (Millipore, Billerica, MA, USA) into plates with 24 wells, coating membranes in upper chambers with Matrigel (BD Biosciences). We placed 500 μL 1640 containing 10% FBS into bottom chambers and 10,000 cells in 200 μL serum-free 1640 in upper chambers. After 1~2 days, we utilized methanol to fix cells on membranes and stained them with crystal violet before observing them via microscope (Leica).
Western blots
We extracted protein from tissues or cells with RIPA lysis buffer and performed western blots as previously described [15]. Primary antibodies were against OCT4, Nanog and GAPDH (Cell Signaling Technology, Beverly, MA, USA) and blots were stained following standard procedures. We visualized immunoreactivity with chemiluminescence detection kits (Western Blotting Substrate, Donghuan Biotech, China).
Dual-luciferase reporter assays
We inserted binding sites for circ-TRPS1, termed circ-TRPS1-WT and circ-TRPS1-Mut, and the EZH2 3’-UTR, termed EZH2-3’UTR-WT, and EZH2-3’UTR-Mut, into HindIII and KpnI sites regarding a pGL3 promoter vector (Realgene, Nanjing, China) for dual-luciferase reporter assays. We plated cells into plates with 24 wells, and transfected 80 ng plasmid, 50 nM miR-124-3p mimics, 5 ng Renilla luciferase vector pRL-SV40, and NC reagents into cells through Lipofectamine 2000 (Invitrogen). We collected and measured cells 2 d after transfection with Dual-Luciferase Assay kits (Promega, Madison, WI, USA), following standard instructions. We independently repeated all experiments three times.
Animal studies
For xenograft assays, we subcutaneously injected 1 × 106 modified (circ-TRPS1-silenced) or NC PC3 cells into right sides of male nude mice. We calculated tumor volumes (length * width2 * 0.5) at timepoints indicated and excised tumors 1 m after injection.
For metastasis analysis, we transfected 2 × 105 NC or circ-TRPS1-silenced PC3 cells with luciferase expression vectors, and injected cells intravenously into mouse tails. After 30 d, PC3 cell metastases were analyzed by bioluminescence imaging following an intravenous injection of luciferin (150 mg luciferin/kg body weight) into tails.
Immunohistochemistry
We fixed tumor tissue samples in formalin 10% and embedded them in paraffin. We stained sections (5-μm) with Ki67 to explore proliferation. We examined sections with an Axiophot light microscope and imaged them via digital camera.
Statistics analyses
We assessed differences among groups via paired/unpaired two-tailed t-tests using Pearson’s correlation tests to obtain associations among groups. We denoted data by mean ± SEM. P-values < 0.05 indicated significant. We performed all statistical analysis with GraphPad Prism (GraphPad Inc., San Diego, CA, USA).
Results
High Circ-TRPS1 expression predicted unfavorable prognoses for PCa
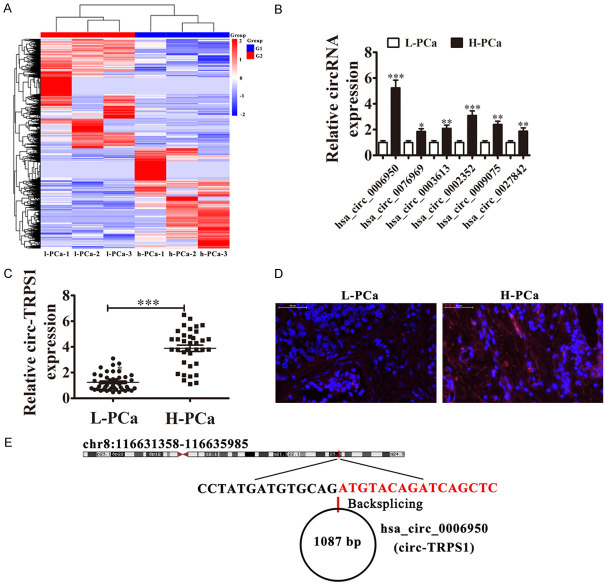
To determine correlations between circRNA expression and PCa progression, three PCa samples from H-PCa and three from L-PCa were used in circRNA high-throughput sequencing. The results showed a series differentially expressed circRNA in PCa tissues comparing (Figure 1A). RT-qPCR illustrated the six upregulated circRNA expressions in H-PCa and L-PCa tissues. The results showed that hsa_circ_0006950 (circ-TRPS1) had the highest expression in H-PCa tissues comparing with L-PCa tissues (Figure 1B). RT-qPCR also showed that circ-TRPS1 expression increased in H-PCa tissues compared to L-PCa patients (Figure 1C). FISH assays demonstrated that circ-TRPS1 expression increased in H-PCa tissues compared with L-PCa tissues. Circ-TRPS1 was localized predominantly to the cytoplasm (Figure 1D). To further understand the prognostic value of circ-TRPS1 expression, we analyzed correlations with patient characteristics. High circ-TRPS1 expression correlated with higher pathological T stage, lymph node metastasis, and distant metastasis (Table 1). Circ-TRPS1 is derived from circularizing exons from gene TRPS1, which are at chr8:116631358-116635985. TRPS1 is 4627 bp and its spliced mature circRNA is 1087 bp (Figure 1E). The findings indicated that circ-TRPS1 functioned in PCa progression.
Figure 1.
Analysis of circular RNA expression in human PCa tissues and cell lines. A. Heat map with expression of circRNA from high-grade PCa (H-PCa) and low-grade PCa (L-PCa) after high-throughput sequencing. B. RT-qPCR of expression of six upregulated circRNAs in H-PCa and L-PCa tissues. Data are means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 vs. L-PCa group. C. RT-qPCR for circ-TRPS1 expression in L-PCa (n = 44) and H-PCa (n = 36). Data are means ± SD; ***P < 0.001 vs. L-PCa group. D. Fluorescence in situ hybridization (FISH) of circ-TRPS1 (red) combined with nuclear DAPI staining (blue) in L-PCa and H-PCa tissues. E. Location of circ-TRPS1 on the human chromosome.
Table 1.
Correlation between circ-TRPS1 expression and clinicopathologic characteristics of PCa patients
| Clinicopathological Features | Overall (n = 80) | High (n = 36) | Low (n = 44) | p-Value |
|---|---|---|---|---|
| Clinical T Stage | ||||
| T1 and T2 | 59 | 22 | 37 | 0.006 |
| T3 and T4 | 21 | 14 | 7 | |
| Lymph Node Metastasis | ||||
| Yes | 16 | 12 | 4 | 0.01 |
| No | 64 | 24 | 40 | |
| Distant Metastasis | ||||
| Yes | 14 | 10 | 4 | 0.03 |
| No | 66 | 26 | 40 |
Downregulation of circ-TRPS1 suppressed PCa proliferation in vivo and in vitro
To identify circ-TRPS1 functions in PCa proliferation, we employed RT-qPCR to detect circ-TRPS1 expression. Circ-TRPS1 expression increased in PCa cell lines PC3, LnCaP, and DU145 compared with RWPE-1 (Figure 2A). PC3 and LnCaP cells had the highest circ-TRPS1 levels, so they were selected for further experiments. Circ-TRPS1 expression decreased significantly in both PC3 and LnCaP cells after transfection with an siRNA against circ-TRPS1 (Figure 2B). CCK8 (Figure 2C, 2D) and colony formation assays (Figure 2E, 2F) showed that circ-TRPS1 silencing decreased cell proliferation in PC3 and LnCaP cells. Xenograft results validated that circ-TRPS1 knockdown suppressed PC3 tumor growth (measured as volume and weight) comparing with an NC group (Figure 2G-I). Immunohistochemical Ki67 detection demonstrated that circ-TRPS1 silencing suppressed Ki67 expression in tumor tissues (Figure 2J, 2K), supporting that circ-TRPS1 knockdown suppressed tumor growth. Together, these data suggested that knocking down circ-TRPS1 suppressed the proliferation ability of PCa cells in vivo and in vitro.
Figure 2.
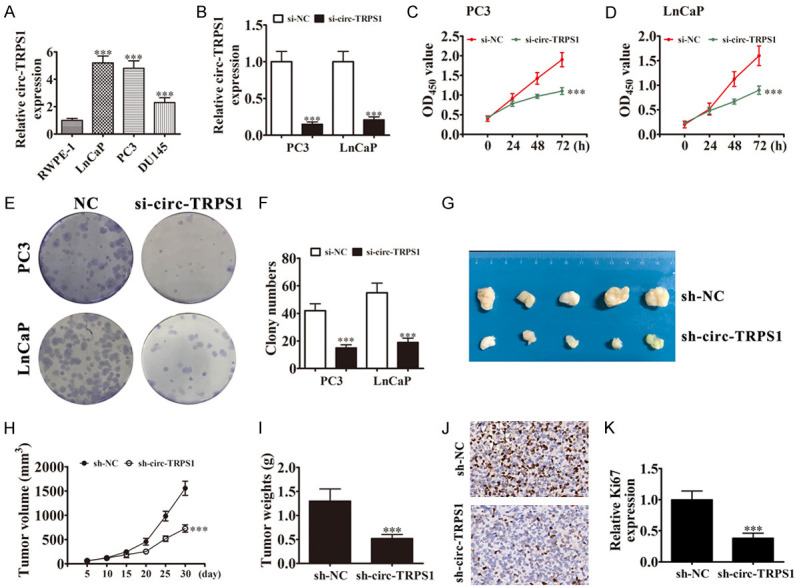
Downregulation of circ-TRPS1 suppressed PCa proliferation in vitro and in vivo. (A) Expression of circ-TRPS1 in noncancer and cancer cell lines analyzed by RT-qPCR. Data are means ± SD; ***P < 0.001 vs. RWPE-1 group. (B) RT-qPCR detected circ-TRPS1 in PC3 and LnCaP after transfecting with negative control (NC) or siRNA against circ-TRPS1 (si-circ-TRPS1). Data are means ± SD; ***P < 0.001 vs. si-NC group. (C and D) CCK8 detection showed proliferation of PC3 and LnCaP cells. Data are means ± SD; ***P < 0.001 vs. si-NC. (E and F) Colony formation assays showing proliferation of PC3 and LnCaP cells. Data are means ± SD; ***P < 0.001 vs. si-NC. (G) PC3 cells transfected with NC or sh-circ-TRPS1 and subcutaneously injected into nude mice. (H) Tumor growth curves, (I) tumor weight. Data are means ± SD; ***P < 0.001 vs. sh-NC. (J and K) Immunohistochemistry for the percentage of Ki67-positive cells. The relative Ki67 expression was calculated. Data are means ± SD; ***P < 0.001 vs. sh-NC.
circ-TRPS1 downregulation suppressed PCa invasion in vitro and in vivo
Transwell detection showed that circ-TRPS1 knockdown suppressed cell migration in PC3 and LnCaP cells (Figure 3A, 3B). The metastasis ability of PC3 cells also decreased after circ-TRPS1 silencing by decreased metastatic foci using live imaging analysis of metastasis model mice (Figure 3C, 3D). Together, these data suggested that knocking down circ-TRPS1 suppressed invasion by PCa cells in vivo and in vitro.
Figure 3.
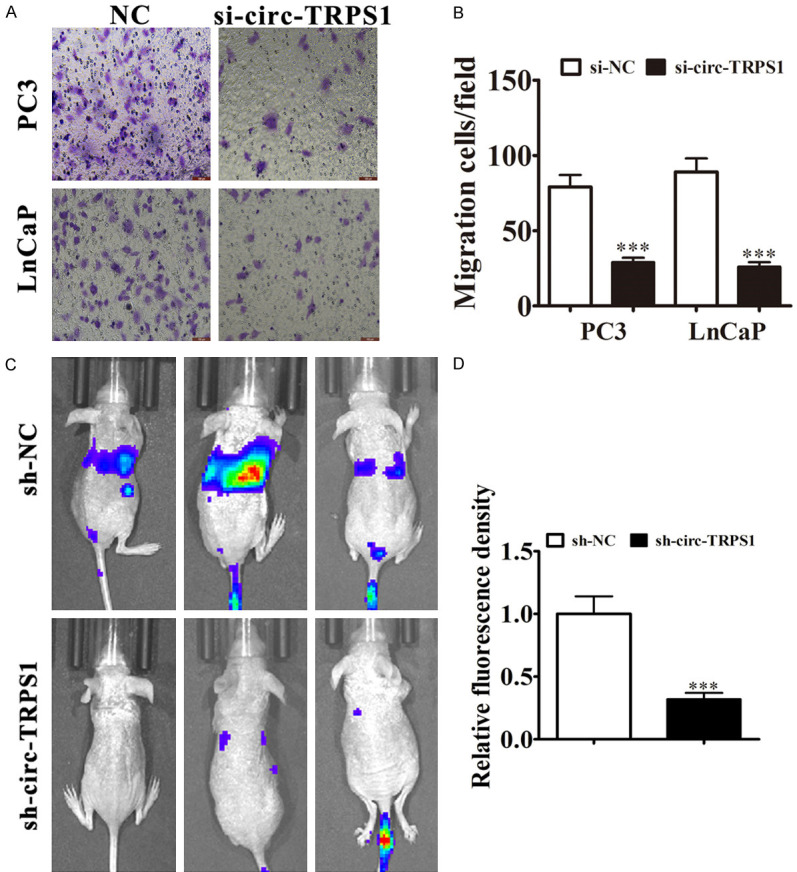
Downregulation of circ-TRPS1 decreased PCa invasion in vitro and in vivo. A, B. Cell migration assessed in PC3 and LnCaP cells using Transwell assays. Data are means ± SD; ***P < 0.001 vs. NC. C, D. Live imaging showing the effects of circ-TRPS1 on metastasis of PC3 cells 30 d after intravenous tail injection. The relative fluorescence density was calculate. Data are means ± SD; ***P < 0.001 vs. sh-NC.
miR-124-3p and EZH2 are downstream targets of circ-TRPS1 in PCa
Next, we investigated how circ-TRPS1 regulation affected PCa proliferation and invasion. Previous studies found that circRNA acts as a miRNA sponge. Given that circ-TRPS1 is mainly localized to the cytoplasm, we hypothesized that circ-TRPS1 might regulate tumor biological behavior by acting as a sponge to miRNAs. We employed Interactome to predict miRNAs. The data showed that miR-124-3p might have circ-TRPS1 acting as sponge (Figure 4A). To validate the association between circ-TRPS1 and miR-124-3p, we prepared wild-type (WT) or mutated (MUT) circ-TRPS1 sequences that included miR-548 binding sequences into a luciferase reporter vector (Figure 4A). We transfected this reporter vector into 293T cells with or not miR-124-3p mimic. Luciferase reporter analysis suggested that miR-124-3p inhibited luciferase activity in WT-transfected, while not MUT-transfected cells (Figure 4B), suggesting that miR-124-3p was circ-TRPS1 target. FISH assays demonstrated that both circ-TRPS1 and miR-124-3p were co-localization in the cytoplasm (Figure S1).
Figure 4.
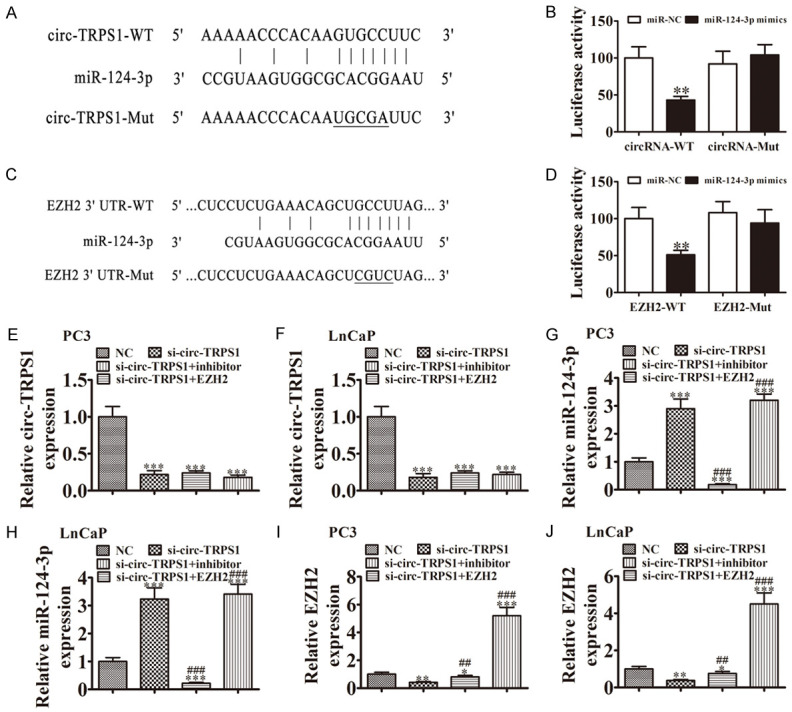
MiR-124-3p and EZH2 were downstream targets of circ-TRPS1. (A) Bioinformatics analysis predicting binding sites of miR-124-3p in circ-TRPS1. Mutant version of circ-TRPS1 is shown. (B) Relative luciferase activity determined 48 h after transfection of HEK293T cells with miR-124-3p mimic/NC or circ-TRPS1 wild-type/Mut. Data are means ± SD. **P < 0.01. (C) Prediction of miR-124-3p binding sites in the 3’-UTR of EZH2. Mutant version of 3’-UTR-EZH2 is shown. (D) Relative luciferase activity 48 h after transfection of HEK293T cells with miR-124-3p mimic/NC or 3’-UTR-EZH2 wild-type/Mut. Data are means ± SD. **P < 0.01. (E-J) RT-qPCR showing expression of (E and F) circ-TRPS1, (G and H) miR-124-3p, (I and J) EZH2 in PC3 and LnCaP cells. Data are means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC. ##P < 0.01, ###P < 0.001 vs. si-circ-TRPS1.
Bioinformatic analyses (TargetScanHuman) discovered that EZH2 was a miR-124-3p downstream target. To detect the association between miR-124-3p and EZH2, we constructed WT or MUT 3’UTR-EZH2 sequences that included miR-548 binding sequences into a luciferase reporter vector (Figure 4C). We transfected this reporter vector into 293T cells with or not miR-124-3p mimic. Luciferase reporter analysis demonstrated that miR-124-3p inhibited luciferase activity in WT-transfected but not MUT-transfected cells (Figure 4D), suggesting that EZH2 was a miR-124-3p target.
To identify the regulatory relationships among circ-TRPS1, miR-124-3p and EZH2, we transfected PC3 and LnCaP cells with si-circ-TRPS1, miR-124-3p inhibitor or EZH2 overexpression vector, either singly or in combination. RT-qPCR verified that circ-TRPS1 expression decreased significantly after transfection with siRNA against circ-TRPS1. MiR-124-3p downregulation or EZH2 overexpression did not reverse circ-TRPS1 expression in PC3 and LnCaP cells (Figure 4E, 4F). Circ-TRPS1 downregulation increased miR-124-3p expression in miR-124-3p cells. Treating cells with an miR-124-3p inhibitor suppressed miR-124-3p expression, but EZH2 overexpression did not reverse miR-124-3p expression (Figure 4G, 4H). We discovered that silencing circ-TRPS1 decreased EZH2 expression in LnCaP and PC3 cells, while treatment with an miR-124-3p inhibitor partially recovered EZH2 expression. Transfecting with an EZH2 overexpression vector increased EZH2 expression (Figure 4I, 4J). The results demonstrated that miR-124-3p and EZH22 were downstream targets of circ-TRPS1 and that EZH2 was a miR-124-3p downstream target.
Downregulating miR-124-3p or overexpressing EZH2 reversed inhibition by si-circ-TRPS1 of PCa proliferation and migration via cancer stem cell differentiation
Colony formation assays (Figure 5A-C) validated that silencing circ-TRPS1 suppressed PC3 and LnCaP cell proliferation. Further silencing miR-124-3p or overexpressing EZH2 rescued PC3 and LnCaP cell proliferation. Transwell assays to measure cell migration (Figure 5D-F) also found that silencing miR-124-3p or overexpressing EZH2 rescued migration of both PC3 and LnCaP cells.
Figure 5.
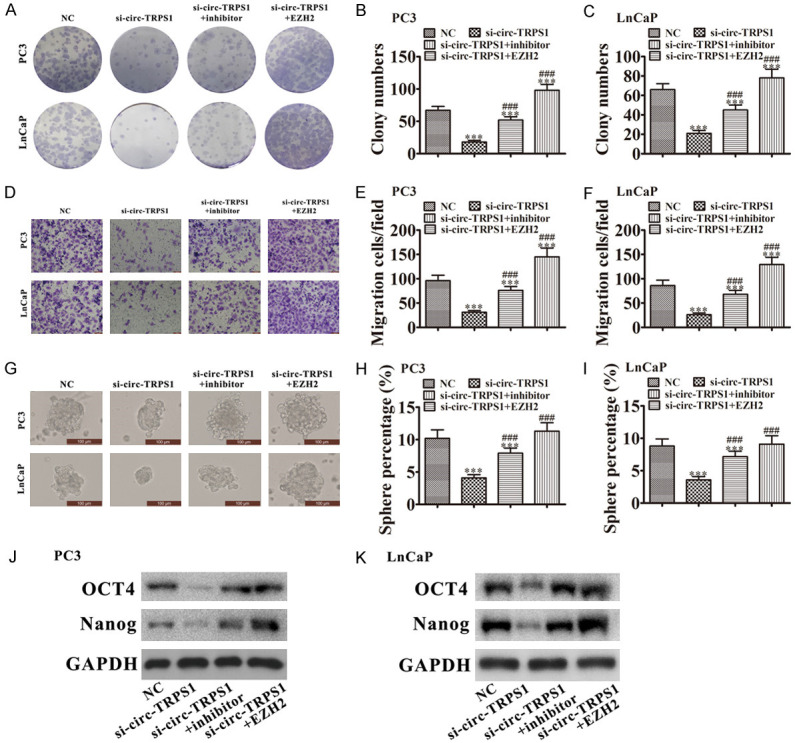
Downregulation of miR-124-3p or overexpression of EZH2 reversed the inhibitory effect of si-circ-TRPS1 on PCa proliferation and migration. A-C. Colony formation assays showing the proliferation of PC3 and LnCaP cells. Data are means ± SD; ***P < 0.001 vs. NC. ###P < 0.001 vs. si-circ-TRPS1. D-F. Cell migration was assessed in PC3 and LnCaP cells using Transwell assays. Data are means ± SD; ***P < 0.001 vs. NC. ###P < 0.001 vs si-circ-TRPS1. G-I. Images of tumor sphere formation assays in PC3 and LnCaP cells (200 cells/well); scale bar, 100 μm. Data are means ± SD; ***P < 0.001 vs. NC. ###P < 0.001 vs. si-circ-TRPS1. J, K. Western blots for the expression of stemness markers OCT4 and Nanog in PC3 and LnCaP cells.
We conducted tumor sphere formation assays. Downregulation of circ-TRPS1 reduced cancer stem cell differentiation by inhibiting tumor sphere formation. These data implied that downregulating miR-124-3p or overexpressing EZH2 enhanced the sphere percentage in LnCaP and PC3 cells (Figure 5G-I). Western blots showed that downregulating miR-124-3p or overexpressing EZH2 promoted expression of stemness markers OCT4 and Nanog in both PC3 and LnCaP cells even after circ-TRPS1 silencing (Figure 5J, 5K). These results suggested that circ-TRPS1 promotion enhanced PCa progression via regulating miR-124-3p/EZH2 axis-mediated stemness.
Overexpressing EZH2 restored cancer stem cell-mediated invasion and PCa cell proliferation after miR-124-3p overexpression
RT-qPCR illustrated that miR-124-3p expression increased significantly in PC3 and LnCaP cells after transfection with a miR-124-3p mimic. Upregulating EZH2 did not reverse miR-124-3p levels (Figure 6A, 6B). RT-qPCR demonstrated that miR-124-3p upregulation suppressed EZH2, but after transfecting with an EZH2 overexpression vector, expression of EZH2 significantly increased (Figure 6C, 6D). Colony formation (Figure 6E-G) assays validated that upregulating miR-124-3p suppressed PC3 and LnCaP cell proliferations. Overexpressing EZH2 rescued PC3 and LnCaP cell proliferation. Transwell assays to measure cell migration (Figure 6H-J) found that overexpressing EZH2 rescued migration of LnCaP and PC3 cells after upregulation of miR-124-3p.
Figure 6.
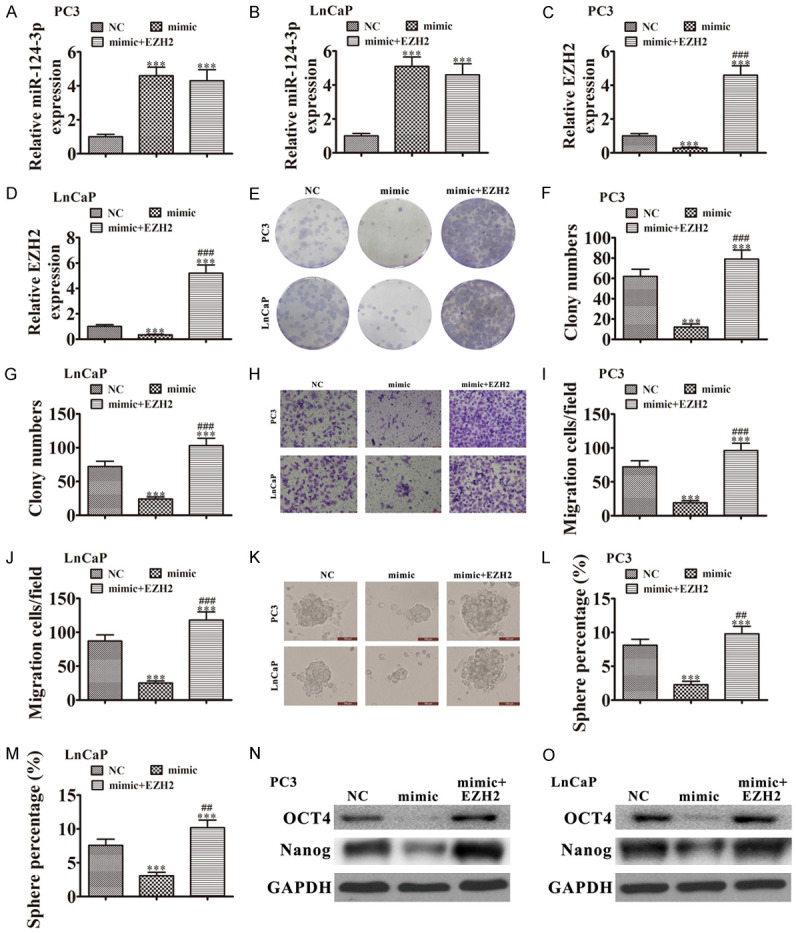
Overexpression of EZH2 reversed the inhibitory effect of miR-124-3p on PCa proliferation and migration. (A-D) RT-qPCR showing expression of (A and B) miR-124-3p and (C and D) EZH2 in PC3 and LnCaP cells. Data are means ± SD; ***P < 0.001 vs. NC. (E-G) Colony formation assays showing proliferation of PC3 and LnCaP cells. Data are means ± SD; ***P < 0.001 vs. NC. ###P < 0.001 vs. mimic. (H-J) Cell migration was assessed in PC3 and LnCaP cells using Transwell assays. Data are means ± SD; ***P < 0.001 vs. NC. ###P < 0.001 vs. mimic. (K-M) Images of tumor sphere formation assays in PC3 and LnCaP cells (200 cells/well); scale bar, 100 μm. Data are means ± SD; ***P < 0.001 vs. NC. ##P < 0.01, ###P < 0.001 vs. mimic. (N and O) western blots for expression of stemness markers OCT4 and Nanog in PC3 and LnCaP cells.
We conducted tumor sphere formation assays. Upregulating miR-124-3p decreased cancer stem cell differentiation by inhibiting tumor sphere formation. These results indicated that overexpressing EZH2 increased the sphere percentage in LnCaP and PC3 cells (Figure 6K-M). Western blots showed that overexpressing EZH2 increased expression of stemness markers OCT4 and Nanog expression in LnCaP and PC3 cells even after upregulating miR-124-3p (Figure 6N, 6O). The results suggested that miR-124-3p decreased PCa progression by regulating EZH2-mediated stemness.
Downregulation of miR-124-3p or overexpression of EZH2 restores the growth ability of PCa after silence circ-TRPS1 in in vivo
Xenograft results validated that circ-TRPS1 knockdown suppressed PC3 tumor growth (measured as volume and weight) comparing with an NC group. Downregulation of miR-124-3p or overexpression of EZH2 restores the growth ability of PCa after silence circ-TRPS1 (Figure 7A-C). Immunohistochemical Ki67 detection demonstrated that circ-TRPS1 silencing suppressed Ki67 expression in tumor tissues. Downregulation of miR-124-3p or overexpression of EZH2 restores Ki67 expression (Figure 7D, 7E), supporting that circ-TRPS1 knockdown suppressed tumor growth by regulation miR-124-3p/EZH2 axis.
Figure 7.
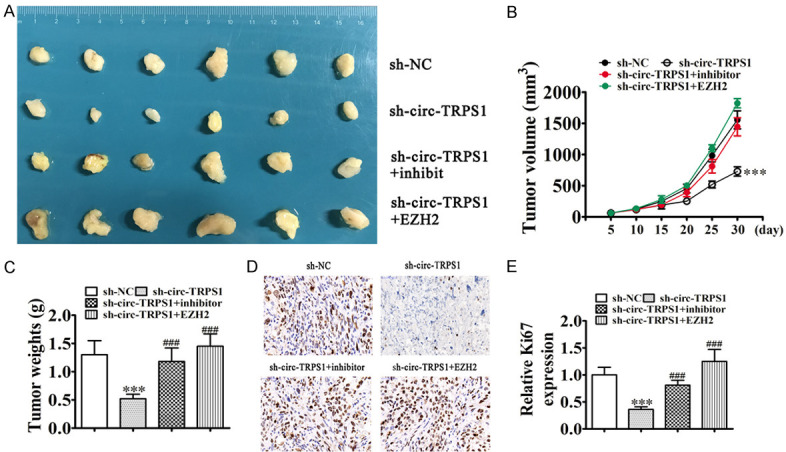
Downregulation of miR-124-3p or overexpression of EZH2 restores the growth ability of PCa after silence circ-TRPS1 in in vivo. (A) PC3 cells transfected with NC, sh-circ-TRPS1, miR-124-3p inhibitor or EZH2 overexpression vector single ro combine, then subcutaneously injected into nude mice. (B) Tumor growth curves, (C) tumor weight. Data are means ± SD; ***P < 0.001 vs. sh-NC. ###P < 0.001 vs. sh-circ-TRPS1. (D and E) Immunohistochemistry for the percentage of Ki-67-positive cells. The relative fluorescence density was calculated. Data are means ± SD; ***P < 0.001 vs. sh-NC. ###P < 0.001 vs. sh-circ-TRPS1.
Discussion
Increasingly, studies find that abnormal expression of circRNA is associated with human traits such as cancers [2]. Studies have found that silencing circZNF609 restrains growth, invasion and migration through upregulating microRNA-186-5p in Pca [11]. Upregulated circZMIZ1 enhances PCa cell proliferation and is a valuable plasma marker [16]. In this study, we investigated expression of circRNA using H-PCa and L-PCa tissues and high-throughput sequencing. The results showed that circ-TRPS1 significantly increased in H-PCa samples compared with L-PCa. High expression of circ-TRPS1 indicated poor progress, high pathological T stage, lymph node metastasis, and distant metastasis. This result suggested that circ-TRPS1 was involved in the progress of PCa. FISH validated that circ-TRPS1 was distributed mainly in the cytoplasm, similar to other circRNAs [17].
Further study found that circ-TRPS1 expression increased in PCa cell lines. Downregulation of circ-TRPS1 significantly decreased invasion and proliferation in in vitro and in vivo, suggesting that circ-TRPS1 had oncogenic characteristics. CircRNAs are reported to interact with miRNAs sponging or stabilizing them [18,19]. The CircInteractome web tool was developed to map RNA-binding protein- and miRNA-binding sites on human circRNA [20]. We used this website for analysis and found that circ-TRPS1 interacted with many miRNAs. The combination with miR-124-3p is the most conservative. Luciferase reporting assays confirmed that miR-124-3p interacted with circ-TRPS1. RT-qPCR showed that silencing circ-TRPS1 promoted miR-124-3p expression, but miR-124-3p overexpression did not reverse circ-TRPS1 lever after circ-TRPS1 silence. This result suggested that miR-124-3p was a circ-TRPS1 downstream target. Research shows that miR-124-3p expression inhibits cancers such like gastric [21] and bladder cancer [22], osteosarcoma [23], and glioma [24]. Previous investigations also validated that miR-124-3p expression is downregulated in Pca [25], suggesting that high expression of circ-TRPS1 promotes the progress of PCa by adsorption of miR-124-3p.
Bioinformatic and luciferase reporting experiments confirmed that EZH2 was a direct miR-124-3p target. circ-TRPS1 downregulation or miR-124-3p overexpression suppressed EZH2 expression. However, overexpression of EZH2 did not reverse circ-TRPS1 and miR-124-3p levels. In vitro experiments confirmed that miR-124-3p downregulation or EZH2 overexpression recovered proliferation and migration after circ-TRPS1 silencing. Overexpression of EZH2 reversed miR-124-3p inhibitory effects on PCa cell proliferation and migration, suggesting that downregulation of circ-TRPS1 suppressed PCa progress by acting as a sponge to miR-124-3p and enhancing EZH2 expression.
Enhancer of Zeste 2 (EZH2) is a gene that is highly conserved as well as expressed in multiple human cancers [26]. EZH2 is aberrantly expressed in glioma and has strong effects on PCa proliferation and metastasis [27-29]. Studies show that EZH2 functions in the maintenance of cancer stem-like cell (CSC) properties [30]. CSCs take part in tumor proliferation, recurrence, invasion, and chemoradioresistance, which makes a new concept of tumorigenicity and progression [31]. In this study, we found that circ-TRPS1 silencing suppressed CSC differentiation using tumor sphere formation assays and western blots. Overexpression of EZH2 recovered the CSC properties of PCa after circ-TRPS1 knockdown or miR-124-3p upregulation, suggesting that downregulation of circ-TRPS1 suppressed PCa progress via regulating miR-124-3p/EZH2 axis-mediated stemness.
Conclusions
In conclusion, our investigation revealed that circ-TRPS1 was increased in PCa patient samples and cell lines, and upregulation was correlated to aggressive PCa phenotypes. In addition, circ-TRPS1 silencing suppressed PCa metastasis and proliferation in vivo and in vitro. We also discovered that circ-TRPS1 downregulation suppressed PCa migration and proliferation through targeting the miR-124-3p/EZH2 axis. We identified circ-TRPS1 as a potential therapeutic target in PCa. Further studies regarding the circ-TRPS1/miR-124-3p/EZH2 axis could provide a basis to develop new potential PCa therapeutic strategies.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81572536, 81672850, 81772742, 81702840, 81702542), Science and Technology Commission of Shanghai Municipality (19ZR143100, 14140901700, 16411969800, 19411967400), Shanghai Shenkang Hospital Development Center (SHDC12015125, 16CR3049A), Shanghai Municipal Education Commission (15ZZ058), Shanghai Municipal Commission of Health and Family Planning (201640247), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152215), Shanghai Jiao Tong University (YG2016ZD08, YG2017MS47, YG2017MS52); Innovation Fund for Translational Research of Shanghai Jiao Tong University School of Medicine (15ZH4002), and Incubating Program for clinical Research and Innovation of Renji Hospital Shanghai Jiao Tong University School of Medicine (PYZY 16-008, PYXJS16-015). All authors agreed to publish this paper.
Disclosure of conflict of interest
None.
Abbreviations
- FISH
Fluorescence in situ hybridization
- EZH2
Enhancer of Zeste 2
- CSC
cancer stem-like cells
- circRNA
circular RNA
- RNA-seq
high-throughput RNA sequencing
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- CCK-8
Cell Counting Kit-8
- miRNAs
microRNA
- PCa
prostate cancer
Supporting Information
References
- 1.Wetherill YB, Hess-Wilson JK, Comstock CE, Shah SA, Buncher CR, Sallans L, Limbach PA, Schwemberger S, Babcock GF, Knudsen KE. Bisphenol A facilitates bypass of androgen ablation therapy in prostate cancer. Mol Cancer Ther. 2006;5:3181–3190. doi: 10.1158/1535-7163.MCT-06-0272. [DOI] [PubMed] [Google Scholar]
- 2.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto A, Inoue A, Yokoi S, Nozumi K, Miyazaki K, Hosoki S, Nagata M, Yamaguchi K. Evaluation of docetaxel plus estramustine in the treatment of patients with hormone-refractory prostate cancer. Int J Urol. 2009;16:687–691. doi: 10.1111/j.1442-2042.2009.02341.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, Niu Y, Wu D, Chang C. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019;10:37. doi: 10.1038/s41419-018-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Gong P, Yang Y, Yang C, Yang B, Ren L. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through Let-7a-5p/DUSP7 axis. Cancer Manag Res. 2020;12:2327–2337. doi: 10.2147/CMAR.S238513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Ren Y, Ma S, Wang S. Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Onco Targets Ther. 2020;13:1941–1951. doi: 10.2147/OTT.S240642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He YX, Ju H, Li N, Jiang YF, Zhao WJ, Song TT, Ren WH. Association between hsa_circ_0006156 expression and incidence of gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3030–3036. doi: 10.26355/eurrev_202003_20667. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Yu J, Huang Z, Fu B, Tao Y, Qi X, Mou Y, Hu Y, Wang Y, Cao Y, Jiang D, Xie J, Zhao J, Xiong W. Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. J Exp Clin Cancer Res. 2020;39:57. doi: 10.1186/s13046-020-01556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin C, Zhao W, Zhang Z, Liu W. CircLMTK2 acts as a tumor suppressor in prostate cancer via regulating the expression of microRNA-183. Life Sci. 2020;241:117097. doi: 10.1016/j.lfs.2019.117097. [DOI] [PubMed] [Google Scholar]
- 11.Jin C, Zhao W, Zhang Z, Liu W. Silencing circular RNA circZNF609 restrains growth, migration and invasion by up-regulating microRNA-186-5p in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47:3350–3358. doi: 10.1080/21691401.2019.1648281. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Qu CB, Zhang Y, Zhang WF, Wang DD, Gao CC, Ma L, Chen JS, Liu KL, Zheng B, Zhang XH, Zhang ML, Wang XL, Wen JK, Li W. Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway. Oncogene. 2019;38:2516–2532. doi: 10.1038/s41388-018-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X, He S, Li Z, Wang Y, Tang Y, Ye C. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol. 2019;122:1235–1243. doi: 10.1016/j.ijbiomac.2018.09.076. [DOI] [PubMed] [Google Scholar]
- 14.Kong Z, Wan X, Zhang Y, Zhang P, Zhang X, Qi X, Wu H, Huang J, Li Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493:1217–1223. doi: 10.1016/j.bbrc.2017.07.162. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, Xu P, Sun G, Xu J, Lv J, Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang H, Lv DJ, Song XL, Wang C, Yu YZ, Zhao SC. Upregulated circZMIZ1 promotes the proliferation of prostate cancer cells and is a valuable marker in plasma. Neoplasma. 2020;67:68–77. doi: 10.4149/neo_2019_190213N116. [DOI] [PubMed] [Google Scholar]
- 17.Yuan X, Yuan Y, He Z, Li D, Zeng B, Ni Q, Yang M, Yang D. The regulatory functions of circular RNAs in digestive system cancers. Cancers (Basel) 2020;12:770. doi: 10.3390/cancers12030770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Zhong H, Jiao L, Wen Y, Zhou Y, Zhou J, Lu X, Song X, Ying B. MiR-124-3p inhibits the migration and invasion of gastric cancer by targeting ITGB3. Pathol Res Pract. 2020;216:152762. doi: 10.1016/j.prp.2019.152762. [DOI] [PubMed] [Google Scholar]
- 22.Fu W, Wu X, Yang Z, Mi H. The effect of miR-124-3p on cell proliferation and apoptosis in bladder cancer by targeting EDNRB. Arch Med Sci. 2019;15:1154–1162. doi: 10.5114/aoms.2018.78743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Wu X, Pei Y, Wang W, Zheng K, Qiu E, Zhang X. PTHR1 may be involved in progression of osteosarcoma by regulating miR-124-3p-AR-Tgfb1i1, miR-27a-3p-PPARG-Abca1, and miR-103/590-3p-AXIN2 Axes. DNA Cell Biol. 2019;38:1323–1337. doi: 10.1089/dna.2019.4880. [DOI] [PubMed] [Google Scholar]
- 24.Deng D, Luo K, Liu H, Nie X, Xue L, Wang R, Xu Y, Cui J, Shao N, Zhi F. p62 acts as an oncogene and is targeted by miR-124-3p in glioma. Cancer Cell Int. 2019;19:280. doi: 10.1186/s12935-019-1004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentino A, Calarco A, Di Salle A, Finicelli M, Crispi S, Calogero RA, Riccardo F, Sciarra A, Gentilucci A, Galderisi U, Margarucci S, Peluso G. Deregulation of microRNAs mediated control of carnitine cycle in prostate cancer: molecular basis and pathophysiological consequences. Oncogene. 2017;36:6030–6040. doi: 10.1038/onc.2017.216. [DOI] [PubMed] [Google Scholar]
- 26.Orzan F, Pellegatta S, Poliani PL, Pisati F, Caldera V, Menghi F, Kapetis D, Marras C, Schiffer D, Finocchiaro G. Enhancer of Zeste 2 (EZH2) is up-regulated in malignant gliomas and in glioma stem-like cells. Neuropathol Appl Neurobiol. 2011;37:381–394. doi: 10.1111/j.1365-2990.2010.01132.x. [DOI] [PubMed] [Google Scholar]
- 27.Xu SG, Yu JJ, Shi Q, Niu Q, Guo Z, Guo BY, Zhou GC, Gu X, Wu YX. Conditionally replicative adenovirus carrying shRNA targeting EZH2 inhibits prostate cancer growth and invasion. Oncol Rep. 2019;42:273–282. doi: 10.3892/or.2019.7157. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, Xu W, Niu Y, Cheng L, Maity SN, Jiang R, Chang C. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571. doi: 10.1038/s41467-019-09784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai Y, Zhang Z, Cheng L, Wang R, Chen X, Kong Y, Feng F, Ahmad N, Li L, Liu X. Inhibition of enhancer of zeste homolog 2 (EZH2) overcomes enzalutamide resistance in castration-resistant prostate cancer. J Biol Chem. 2019;294:9911–9923. doi: 10.1074/jbc.RA119.008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorodetska I, Lukiyanchuk V, Peitzsch C, Kozeretska I, Dubrovska A. BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype. Int J Cancer. 2019;145:2974–2985. doi: 10.1002/ijc.32323. [DOI] [PubMed] [Google Scholar]
- 31.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.