Abstract
The cell surface protein TMPRSS2 (transmembrane protease serine 2) is an androgen-responsive serine protease important for prostate cancer progression and therefore an attractive therapeutic target. Besides its role in tumor biology, TMPRSS2 is also a key player in cellular entry by the SARS-CoV viruses. The COVID-19 pandemic caused by the coronavirus SARS-CoV-2 has resulted in huge losses in socio-economy, culture, and human lives for which safe and effective cures are highly demanded. The main protease (Mpro/3CLpro) of SARS-CoV-2 is a critical enzyme for viral propagation in host cells and, like TMPRSS2, has been exploited for treatment of the infectious disease. Numerous natural compounds abundant in common fruits have been suggested with anti-coronavirus infection in the previous outbreaks of SARS-CoV. Here we show that screening of these compounds identified tannic acid a potent inhibitor of both SARS-CoV-2 Mpro and TMPRSS2. Molecular analysis demonstrated that tannic acid formed a thermodynamically stable complex with the two proteins at a KD of 1.1 μM for Mpro and 1.77 μM for TMPRSS2. Tannic acid inhibited the activities of the two proteases with an IC50 of 13.4 μM for Mpro and 2.31 μM for TMPRSS2. Mpro protein. Consistently, functional assays using the virus particles pseudotyped (Vpp) of SARS-CoV2-S demonstrated that tannic acid suppressed viral entry into cells. Thus, our results demonstrate that tannic acid has high potential of developing anti-COVID-19 therapeutics as a potent dual inhibitor of two independent enzymes essential for SARS-CoV-2 infection.
Keywords: Tannic acid, COVID-19, SARS-CoV-2, main protease, TMPRSS2
Introduction
The cell surface protease transmembrane protease serine 2 (TMPRSS2) is an androgen-responsive serine protease important for prostate cancer progression [1-3]. Increased expression of TMPRSS2 frequently occurs in prostate cancer in response to androgen stimulation, and somatic fusion of TMPRSS2 with the ERG gene has been found to foster prostate cancer development [4]. The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been a serious global pandemic, resulting in enormous socio-economic losses. In spite of the intensive efforts invested, safe, effective, and cost-efficient drugs for COVID-19 are yet beyond the horizon. Importantly, cellular entry of SARS-CoV-2 entails the viral surface spike protein (S protein) which is activated by proteolytic processing the S protein to produce the S1 protein by TMPRSS2 [5]. The S1 protein is then recognized by and binds to the cell surface receptor ACE2 (angiotensin converting enzyme 2), a process triggers the fusion of the virus and cell membranes for virus entry [6]. Subsequent virus maturation of the virions in host cells is through a highly coordinated proteolytic cascade of processing the viral precursor polyprotein by the main protease (Mpro/3CLpro) [7-10]. The SARS-CoV-2 Mpro belongs to the chymotrypsin-like protease family and cleaves its substrate polypeptides at a well-defined cleavage sequence. Inhibition of Mpro blocks the process of viral polyproteins thus abrogates viral replication in host cells [11], making it an attractive therapeutic target of COVID-19. Numerous natural compounds have been suggested with anti-CoV activity during the previous epidemic outbreaks of SARS and MERS [12-17]. Interestingly, many of the natural compounds showing promising in suppressing viral infection are abundant in popular fruits, particularly berries, citrus, grape, and apples (see below). We therefore set out to determine the potency of these natural compounds in fruits for their inhibitory activities of SARS-CoV-2. Our results support tannic acid as a natural compound with potent anti-SARS-CoV-2 activity of targeting both SARS-CoV-2 Mpro and the cellular protease TMPRSS2.
Material and methods
Sample preparations
The SARS-CoV-2 Mpro cDNA and its fluorescent protein substrate were constructed in expression plasmids and purified as described previously [18]. Briefly, the bacterial clones harboring the IPTG-inducible expression vectors of the recombinant proteins were cultured in LB medium at 37°C until the culture reached an OD600 between 0.6-0.8. Expression of the recombinant proteins was induced with 0.5 mM IPTG or 0.2% L-rhamnose and further incubated at 20°C for 18 hr. The cells were pelleted by centrifugation and the pellets were suspended in buffer A containing 50 mM Tris pH 8.0, 0.5 M NaCl, 10% glycerol, 1 mM TCEP, 1 mM PMSF and lysed by sonication. Following centrifugation at 28,000 g, 4°C for 30 min, the supernatant was collected and separated through a Ni-column (GE Healthcare). The column was then washed by the sonication buffer supplemented by 10 mM imidazole, followed by elution with a 20-200 mM imidazole gradient in buffer A. Mpro recombinant protein was released by cleaving the N-terminal SUMO fusion tag with the TEV protease. Mpro and its substrate protein were then further purified by gel-filtration.
FRET-based enzyme activity assay of Mpro
Fluorescence-labeled protein substrate containing the cleavage site of Mpro was used for the FRET-based enzyme activity assay established in our previous study [18]. Briefly, SARS-CoV-2 Mpro protein (1 μM) was pre-incubated with different bioactive compounds (50 μM) for 30 min at room temperature in the assay buffer (20 mM Tris-HCl pH 7.8, 20 mM NaCl). The reaction was started by addition of 20 μM of the fluorescent protein substrate. The fluorescence signals of the cleaved products in the presence of different concentrations of tannic acid were monitored for emission at 474 nm by an excitation at 434 nm using a Synergy™ H1 hybrid multi-mode microplate reader (BioTek Instruments, Inc.). Initial velocity (V0) was calculated in the first 15 min of the reaction by linear regression. The IC50 was determined using two-fold serial dilutions and analyzed by the GraphPad Prism software. All experiments were performed in triplicates. The cleavage products derived from the fluorescent protein substrates by SARS-CoV-2 Mpro were confirmed by separation in 12% SDS-PAGE gels and visualized by Coomassie Blue staining.
Measurement of human TMPRSS2 activity by a FRET-based enzymatic assay
Purified MBP-tagged TMPRSS2 (residues 256-492, UniProt accession: O15393) was pre-incubated in the presence or absence of tannic acid (60 μM) in the assay buffer (25 mM Tris, 150 mM NaCl) for 30 min at room temperature. After adding 20 μM of the fluorescent protein substrate the reaction was monitored using a Synergy™ H1 hybrid multi-mode microplate reader with excitation at 506 nm and emission at 536 nm. DMSO was used as the negative control.
Surface plasmon resonance (SPR) analysis
Binding kinetics of tannic acid to SARS-CoV-2 Mpro was assessed using a Biacore T200 (Cytiva) at 25°C with a CM5 sensor chip. The purified SARS-CoV-2 Mpro was dialyzed into PBS buffer pH 7.4 at 4°C for overnight before applying to the chip. The surface of the CM5 chip was activated in EDC/NHS for 420 s. The final immobilization level of SARS-CoV-2 Mpro was estimated to be about 12000 response units. Tannic acid was dissolved in PBS buffer and flowed through the chip surface at a flow rate of 30 μl/min. The association and dissociation time for affinity analysis was set for 60 and 120 sec, respectively. Five dilutions of tannic acid from 25 to 1.56 μM were assessed to determine the dissociation constant (KD) using the BIA evaluation program.
Molecular docking
The SARS CoV-2 Mpro structure was retrieved from Protein Data Bank (PDB code 6W63). The structures of tannic acid and Mpro were prepared using the BIOVIA Discovery Studio software (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2020, San Diego: Dassault Systèmes, 2016). To investigate how tannic acid interacts with SARS CoV-2 Mpro, LibDock was employed to generate and score the docking poses.
Vpp pseudoviral particle infection assay
The SARS-CoV2-S pseudoviral particles were purchased from the RNAi core of the Academia Sinica, Taiwan. The human embryonic kidney cell line 293T stably expressing recombinant human ACE2 (293/hACE2) were maintained in Dulbecco’s MEM containing 10% fetal bovine serum, 100 unit penicillin, and 100 μg streptomycin. The 293/hACE2 cells were seeded into 96-well plates and pre-treated with different doses of tannic acid for 1 h, then inoculated with 100 μl of normal media containing the pseudovirions (MOI = 0.28). After overnight incubation, cells were fed with fresh media. At about 40 h post-inoculation, cells were lysed with 100 μl medium containing 50% Steady-Glo (Promega) at room temperature for 5 min. The transduction efficiency was measured by quantification of the luciferase activity using an ELISA reader. The cytotoxicity of tannic acid to 293T/hACE2 cells was assessed by using the Cell Counting Kit-8 kit (ab228554, Abcam) following the manufacture’s protocol. Briefly, 5×104 cells per well in 96 well plates were seeded in 100 μl of media and cultured for about 24 h, followed by treatment with different doses of tannic acid in 100 μl DMEM/high glucose medium containing 5% FBS for 24 h. CCK-8 solution (10 μl) was added and incubated for 3 h. The OD values (460 nm) were measured to represent cell proliferation ability. All experiments were performed in triplicates, and repeated three times.
The African green monkey kidney cells Vero E6 were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1x GlutaMAX, and 1% penicillin/streptomycin. Cells were seeded into 96-well plates and pre-treated with different doses of tannic acid for 1 hr, then inoculated with 50 ml pseudovirions (MOI = 0.28). After incubation for 24 h, cell viability was confirmed by the Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories). Each sample was then mixed with an equal volume of luciferase substrate of the Bright-Glo Luciferase Assay System (Promega), and luminescence was measured immediately by the GloMax Navigator System (Promega). Viability-normalized relative light unit (RLU) was set as 100% for the control group, and the relative infection efficiencies of the treated groups were calculated. The cytotoxicity of tannic acid to Vero E6 was assessed by seeding 2×104 cells per well in 24 well plates in 500 μl media and cultured for about 24 h, followed by treatment with different doses of tannic acid in 500 μl DMEM/high glucose medium containing 2% FBS for 48 h. The media was then removed and viable cells were detected with MTT assay which was read by absorbance at 570 nm by an ELISA reader. All experiments were performed in triplicates, and repeated three times.
Results
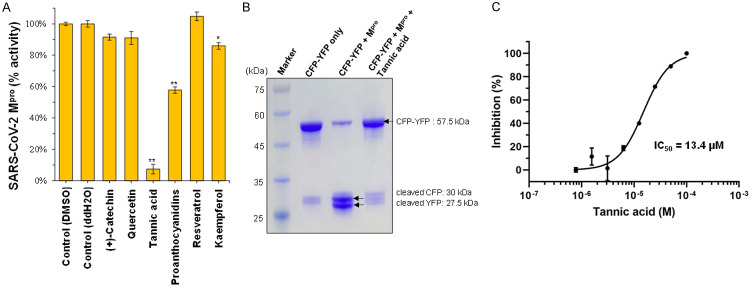
Six previously identified bioactive natural compounds that were effective against CoV, namely catechin [12], kaempferol [15], quercetin [15], proanthocyanidins [16], resveratrol [14], and tannic acid [12], were assessed for their ability to inhibit the enzymatic activity of SARS-CoV-2 Mpro using a fluorescence resonance energy transfer (FRET)-based assay to measure the enzymatic activity of Mpro [19,20] (Table 1). Among the six compounds tested, only tannic acid showed up with significant activity of inhibiting up to 90% of the enzymatic activity of SARS-CoV-2 Mpro at a concentration of 50 μM (Figure 1A; data not shown). The cleavage was verified with SDS polyacrylamide gel electrophoresis (SDA-PAGE) to visualize the produced peptides (Figure 1B). A dose-response analysis estimated a half-maximal inhibitory concentration (IC50) of tannic acid at 1.3×10-5 M in SARS-CoV-2 Mpro inhibition (Figure 1C). These results raise the question of whether tannic acid directly interacts with SARS-CoV-2 Mpro to inhibit its activity. To determine the dose-dependent affinity of tannic acid to SARS-CoV-2 Mpro, 5 dilutions of tannic acid from 25 μM to 1.56 μM were tested. The dissociation constant (KD) for tannic acid binding to SARS-CoV-2 Mpro was determined to be 1.1×10-6 M from the association and dissociation curves of the sensorgrams, using the Biacore evaluation program (Figure 2A). To further understand the mechanism of Mpro inhibition by tannic acid, the interaction between them was modeled by docking analysis. The crystal structure of SARS-CoV-2 Mpro in its ligand-free state has been resolved in previous studies [11,18,21]. Each SARS-CoV-2 Mpro molecule is composed of three domains. Domain I (residues 10 to 100) and II (residues 101 to 182) have a chymotrypsin-like, two-β-barrel fold commonly found in the structure of Mpro of other human and animal coronaviruses, such as PEDV, MERS-CoV, and SARS-CoV [9,22,23]. Domain III (residues 200 to 304) of SARS-CoV-2 Mpro includes five α-helices that form a globular structure. Given that the two viruses share 96.08% sequence identity, it is not surprising that the structure of the SARS-CoV-2 Mpro is highly similar to that of the SARS-CoV Mpro [21]. The domains I and II occupy the substrate-binding pockets and are evolutionarily highly conserved. In the helical domain III, on the other hand, the surface loops of CoV Mpros harbor regions of intrinsic variation [11]. The structure of tannic acid is characterized by a central glucose molecule esterified at the five hydroxyl groups with one or more gallic molecules [24]. Docking analysis showed that tannic acid occupies the pocket defined by binding with the key residue Cys145 via the pi-sulfur and hydrogen bond with 4.69 Å and 2.62 Å, respectively, as well as Asn142 and Met165 via the hydrogen bond and pi-alkyl interactions (Figure 2B).
Table 1.
List of dietary sources of fruit bioactive compounds shown to be active in suppression SARS-CoV infection
| Source fruits | Proposed targets | Ref. | |
|---|---|---|---|
| (+)-Catechin | apples, blueberries, gooseberries, grape seeds, kiwi, strawberries | 3CLpro | [12] |
| Quercetin | apples, citrus fruits, grapes, red raspberry, nectarine | PLpro | [15] |
| Tannic acid | berries, grapes, persimmons, pomegranate | 3CLpro | [12] |
| Proanthocyanidins | blueberries, cranberries, black currant, plums, grapes | infection | [16] |
| Resveratrol | blueberries, bilberries, cranberries, grapes | apoptosis | [14] |
| Kaempferol | apples, grapes, tomatoes, peaches, blackberries, raspberries | PLpro | [15] |
3CLpro, 3-chemotrypsin-like protease; PLpro, papain-like protease.
Figure 1.
Tannic acid is a promising direct inhibitor of Mpro. A. Screening of previously identified bioactive compounds in fruits that were effective in inhibiting Mpro of SARS-CoV-2. 50 μM bioactive compounds were pre-incubated with 1 μM of SARS-CoV-2 Mpro for 30 min at room temperature. The fluorescent substrates (20 μM) were then added to initiate the reaction. The first 15 min of the reaction was used to calculate initial velocity (V0) and then normalized to control. Tannic acid was dissolved in water, other compounds were dissolved in DMSO. Both water and DMSO were used as negative control as indicated. Data are shown as mean ± SEM from experiments performed in triplicate. Statistical significance was calculated using Student t-test, *P < 0.05, **P < 0.01. B. Inhibition of peptide cleavage activity of SARS-CoV-2 Mpro by tannic acid. M, marker. Lane 1, the fluorescent substrate (CFP-TSAVLQSGFRKM-YFP, 57.5 kDa) only. Lane 2, SARS-CoV-2 Mpro was incubated with the fluorescent substrate at 30°C for 1 hour. Two separate bands correspond to CFP-TSAVLQ (30 kDa) and SGFRKM-YFP (27.5 kDa) were detected. Lane 3, SARS-CoV-2 Mpro was incubated with the fluorescent substrate plus 50 μM tannic acid at 30°C for 1 hour. The fluorescent substrate remains uncleaved. C. Dose-effect course of tannic acid in inhibiting SARS-CoV-2 Mpro. The IC50 was calculated by plotting the initial velocity against different concentrations of tannic acid as shown by a dose-response curve generated with the Prism 8 software.
Figure 2.
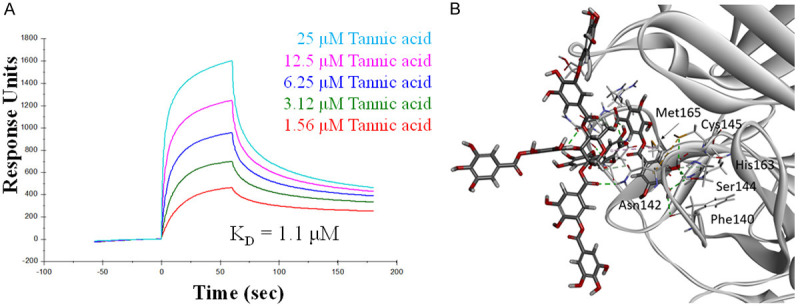
Association of tannic acid with SARS-CoV-2 Mpro. A. Binding kinetics of tannic acid to SARS-CoV-2 Mpro. Surface plasmon resonance analysis is performed using a BiacoreT200 with sensor chip CM5 at 25°C. Tannic acid was dissolved in PBS buffer pH 7.4 and flowed through the chip surface at a flow rate of 30 µl/min. B. Molecular docking of tannic acid to SARS-CoV-2 Mpro. Tannic acid and the interacting side-chain residues are shown in sticks. Hydrogen bonds are colored in green. Pi-sulfur bond is colored in yellow. Pi-alkyl bond is colored in light pink.
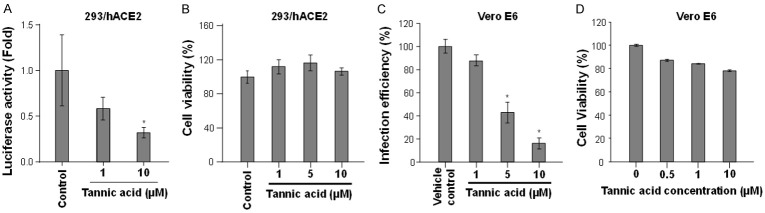
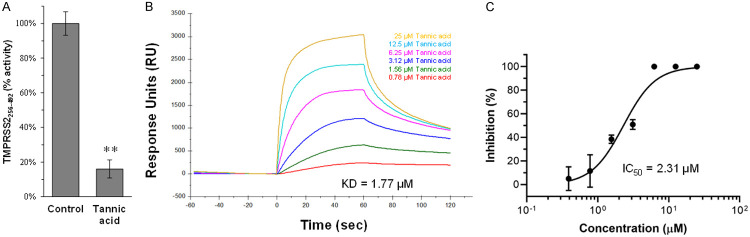
The potential activity of tannic acid in SARS-CoV-2 infection was assessed by a SARS-CoV-2 pseudovirus assay in which the human baby kidney 293T cells overexpressing the S protein receptor ACE2 were used as the target cells in a pseudoviral particle infection assay. Intriguingly, tannic acid treatment significantly inhibited the entry of SARS-CoV-2 pseudovirus in a dose-dependent manner in the absence of significant cytotoxicity (Figure 3A and 3B). Similar effect of entry inhibition by tannic acid was also observed in Vero E6 cells (Figure 3C and 3D). This result suggests that, besides the intracellular proteolytic enzyme Mpro, tannic acid can also target the mechanisms governing virus entry. Indeed, in vitro enzymatic assay of TMPRSS2 showed that treatment with tannic acid inhibited the protease activity of TMPRSS2 (Figure 4A). To further test whether tannic acid is a direct inhibitor of TMPRSS2, the dose-dependent affinity of tannic acid to human TMPRSS2 was determined by measuring 5 dilutions of tannic acid from 25 μM to 1.56 μM. Analysis of the association and dissociation curves of the sensorgrams showed a dissociation constant (KD) of 1.77×10-6 M for tannic acid binding to TMPRSS2 using the Biacore evaluation program (Figure 4B). Thus, these results together demonstrate that tannic acid is an inhibitor with dual inhibitory functions of blocking both viral and cellular serine proteases critical for viral infection.
Figure 3.
Tannic acid blocked cell entry of the SARS-CoV-2 pseudovirus. A. 293T/hACE2 cells were pre-incubated with the indicated doses of tannic acid and cell entry of the SARS-CoV2-S pseudoviral particles was assessed by a luciferase reporter assay. B. The cytotoxicity of 293/hACE2 cells treated with the indicated concentrations of tannic acid for 24 h as measured by cell viability. C. Vero E6 cells were pre-incubated with the indicated doses of tannic acid and cell entry of the SARS-CoV2-S pseudoviral particles was assessed by a luciferase reporter assay. D. The cytotoxicity of Vero E6 cells treated with the indicated concentrations of tannic acid for 48 h as measured by cell viability. The detail procedures of measuring viral entry and cytotoxicity are described in Materials and Methods. Bars, standard deviations; *, P < 0.05 as determined by Student’s t test.
Figure 4.
Tannic acid is an inhibitor of TMPRSS2. A. Recombinant TMPRSS2 was mock treated or treated with tannic acid (60 μM) and the enzymatic activity was determined. **, P < 0.01. B. Binding kinetics of tannic acid to TMPRSS2256-492. SPR analysis is performed using a BiacoreT200 with sensor chip CM5 at 25°C. Tannic acid was dissolved in PBS buffer pH 7.4 and flowed through the chip surface at a flow rate of 30 µl/min. Six dilutions of tannic acid from 25 to 0.78 μM were analyzed to determine the binding affinity to serine protease domain of TMPRSS2. The experiments were repeated. C. Tannic acid inhibits the protease activity of TMPRSS2256-492. IC50 (2.31 μM) of tannic acid was determined based on 7 dilutions from 25 to 0.39 μM and analyzed by GraphPad Prism software.
Discussion
Currently there are no clinically proven drugs specifically targeting SARS-CoV-2 except for remdesivir which has been approved by US FDA for emergency use. However, a randomized, double-blinded, placebo-controlled clinical studies have shown that treatment by remdesivir did not result in difference in time to clinical improvement (hazard ratio 1.23 [95% CI 0.87-1.75]) [25]. On the other hand, among the patients with symptom duration of 10 days or less, individuals received remdesivir did have a trend to achieve clinical improvement faster than those received placebo. The difference compared with a placebo, however, is not statistically significant (hazard ratio 1.52 [0.95-2.43]) [25]. Like remdesivir, the six compounds selected for this study were also based on the prior applications in coping with viral outbreaks, particularly for CoVs.
Tannic acid belongs to the tannin family. Tannins are the major components affecting the richness of texture of red wine. It is estimated that the enological concentrations of tannins in red wines range from 5~100 μM depending on the variety. Tannic acid is a water-soluble polyphenol frequently present in herbaceous and woody plants, legumes, sorghum, as well as the commonly consumed fruits such as raspberries, bananas, and persimmons [24]. The polyphenol compounds are well known for their redox activity as antioxidants and radical scavengers [26,27]. To this regards, a cohort of recent studies have demonstrated the activity of tannic acid in suppressing multiple biological functions in cancer cells from energy metabolism, cell proliferation, invasion, metastasis, to anti-inflammation, establishing tannic acid as a chemopreventing and chemosensitizing anti-cancer agent [28,29]. Further work is required to determine whether tannic acid inhibits cancer progression through targeting the TMPRSS2 membrane protease. Overall our results highlight the potential of mining natural sources as reservoirs of safe and effective remedies to curb the COVID-19 pandemic besides neoplastic diseases.
Acknowledgements
This research was supported by the start-up fund of the China Medical University, Taichung, Taiwan (to M.-C.H). Experiments and data analysis were performed in part through the use of the Medical Research Core Facilities Center, Office of Research & Development at China medical University.
Disclosure of conflict of interest
A provisional patent application has been submitted to the United States Patent and Trademark Office (63/106,938). Mien-Chie Hung, Yeh Chen, and Shao-Chun Wang are co-inventors.
References
- 1.Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 2.Ko CJ, Huang CC, Lin HY, Juan CP, Lan SW, Shyu HY, Wu SR, Hsiao PW, Huang HP, Shun CT, Lee MS. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75:2949–2960. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- 3.Lucas JM, Heinlein C, Kim T, Hernandez SA, Malik MS, True LD, Morrissey C, Corey E, Montgomery B, Mostaghel E, Clegg N, Coleman I, Brown CM, Schneider EL, Craik C, Simon JA, Bedalov A, Nelson PS. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Xia S, Wang Q, Xu W, Li W, Lu L, Jiang S. Broad-spectrum coronavirus fusion inhibitors to combat COVID-19 and other emerging coronavirus diseases. Int J Mol Sci. 2020;21:3843. doi: 10.3390/ijms21113843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 8.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 12.Chen CN, Lin CP, Huang KK, Chen WC, Hsieh HP, Liang PH, Hsu JT. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3’-digallate (TF3) Evid Based Complement Alternat Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, Zhang X, Hua SN, Yu J, Xiao PG, Li RS, Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SC, Ho CT, Chuo WH, Li S, Wang TT, Lin CC. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis. 2017;17:144. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JY, Yuk HJ, Ryu HW, Lim SH, Kim KS, Park KH, Ryu YB, Lee WS. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzyme Inhib Med Chem. 2017;32:504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang M, Jiang H, Suzuki Y, Li X, Xiao P, Tanaka T, Ling H, Yang B, Saitoh H, Zhang L, Qin C, Sugamura K, Hattori T. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang ST, Lai HC, Lin YC, Huang WT, Hung HH, Ou SC, Lin HJ, Hung MC. Principles and treatment strategies for the use of Chinese herbal medicine in patients at different stages of coronavirus infection. Am J Cancer Res. 2020;10:2010–2031. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YC, Yang WH, Yang CS, Hou MH, Tsai CL, Chou YZ, Hung MC, Chen Y. Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am J Cancer Res. 2020;10:2535–2545. [PMC free article] [PubMed] [Google Scholar]
- 19.Chuck CP, Chong LT, Chen C, Chow HF, Wan DC, Wong KB. Profiling of substrate specificity of SARS-CoV 3CL. PLoS One. 2010;5:e13197. doi: 10.1371/journal.pone.0013197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Yang WH, Huang LM, Wang YC, Yang CS, Liu YL, Hou MH, Tsai CL, Chou YZ, Huang BY, Hung CF, Hung YL, Chen JS, Chiang YP, Cho DY, Jeng LB, Tsai CH, Hung MC. Inhibition of severe acute respiratory syndrome coronavirus 2 main protease by tafenoquine in vitro. bioRxiv. 2020 2020.2008.2014.250258. [Google Scholar]
- 21.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galasiti Kankanamalage AC, Kim Y, Damalanka VC, Rathnayake AD, Fehr AR, Mehzabeen N, Battaile KP, Lovell S, Lushington GH, Perlman S, Chang KO, Groutas WC. Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element. Eur J Med Chem. 2018;150:334–346. doi: 10.1016/j.ejmech.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye G, Wang X, Tong X, Shi Y, Fu ZF, Peng G. Structural basis for inhibiting porcine epidemic diarrhea virus replication with the 3C-like protease inhibitor GC376. Viruses. 2020;12:240. doi: 10.3390/v12020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung KT, Wong TY, Wei CI, Huang YW, Lin Y. Tannins and human health: a review. Crit Rev Food Sci Nutr. 1998;38:421–464. doi: 10.1080/10408699891274273. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrade RG Jr, Dalvi LT, Silva JM Jr, Lopes GK, Alonso A, Hermes-Lima M. The antioxidant effect of tannic acid on the in vitro copper-mediated formation of free radicals. Arch Biochem Biophys. 2005;437:1–9. doi: 10.1016/j.abb.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Wu LT, Chu CC, Chung JG, Chen CH, Hsu LS, Liu JK, Chen SC. Effects of tannic acid and its related compounds on food mutagens or hydrogen peroxide-induced DNA strands breaks in human lymphocytes. Mutat Res. 2004;556:75–82. doi: 10.1016/j.mrfmmm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Nagesh PKB, Chowdhury P, Hatami E, Jain S, Dan N, Kashyap VK, Chauhan SC, Jaggi M, Yallapu MM. Tannic acid inhibits lipid metabolism and induce ROS in prostate cancer cells. Sci Rep. 2020;10:980. doi: 10.1038/s41598-020-57932-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baer-Dubowska W, Szaefer H, Majchrzak-Celińska A, Krajka-Kuźniak V. Tannic acid: specific form of tannins in cancer chemoprevention and therapy-old and new applications. Curr Pharmacol Rep. 2020;6:28–37. [Google Scholar]