Abstract
FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) is one of the most common somatic mutations in acute myeloid leukemia (AML). However, the molecular structure characteristics and widely accepted prognostic factors for FLT3-ITD are still not well described. This study aimed to retrospectively examine 81 patients with FLT3-ITD-positive AML diagnosed and treated at the First Affiliated Hospital of Zhejiang University from December 2013 to March 2018 using the next-generation sequencing 185-gene platform. High variant allele frequency (VAF) [> 0.48, P = 0.0089 for overall survival (OS), P = 0.13 for relapse-free survival (RFS)], multiple ITDs (> 1 ITDs, P = 0.011 for OS, P = 0.033 for RFS) and longer insertion length (> 69 bp, P = 0.14 for OS, P = 0.0078 for RFS) predicted poor survival. The study further proposed an easily applicable scoring model for OS using the Least Absolute Shrinkage and Selector Operation (LASSO) Cox regression model. Also, an independent cohort of 30 patients was used for external model validation. The mode was expressed as follows: 0.659 × FLT3-ITD VAF + 0.375 × FLT3-ITD number + 0.807 × Age + 0.688 × DNMT3A + 1.939 × U2AF1 (FLT3-ITD VAF > 0.48 scored 1; FLT3-ITD number scored 1 if carried 1 ITD, 2 if carried ≥ 2 ITDs; age > 44 years scored 1, the presence of DNMT3A or U2AF1 scored 1; 0 for other conditions). It categorized patients into low-risk (L-R, score < 1, n = 20) and high-risk (H-R, score ≥ 1, n = 61) groups based on the risk score with a significant difference in survival (3-year OS, P < 0.0001; 3-year RFS, P = 0.0005). A prognostic nomogram that integrated these five factors was developed with a concordance index calculation [OS: 0.68, 95% CI (0.64-0.72)].
Keywords: Acute myeloid leukemia, FLT3-ITD, LASSO cox regression, next-generation sequencing, prognosis
Introduction
Acute myeloid leukemia (AML) is a common hematologic malignancy with strong heterogeneity and different cytogenetic and molecular defects presenting with completely different clinical properties and therapeutic response [1-3]. The FMS-like tyrosine kinase 3 (FLT3) gene encodes a receptor tyrosine kinase involved in hematopoiesis. FLT3 internal tandem duplication (FLT3-ITD) has been identified in patients with AML since 1996. The frequency of FLT3-ITD in adult AML [non-acute promyelocytic leukemia (APL)] is approximately 20% and much higher in patients with normal karyotype [4,5]. Numerous studies have confirmed that the ITD mutations in AML are strongly related to shorter remission duration and poorer survival outcomes [6-8]. However, in clinical practice, about 25% of patients with FLT3-ITD-mutated AML could finally achieve favorable prognosis, suggesting that some factors might be involved in the clinical outcome of FLT3-ITD-postive patients. Widely accepted prognostic factors predicting the clinical outcome of newly diagnosed FLT3-ITD-postive AML are still lacking. Hence, the molecular characteristics of the FLT3-ITD need to be studied, and more potential prognostic markers need to be assessed urgently.
Besides the age of onset, white blood cell (WBC) count, and other clinical characteristics, heterogeneous molecular structures of FLT3-ITD may also be crucial in predicting prognosis. Previous studies have observed different prognostic roles of FLT3-ITD in the number, insertion length, and insertion regions of ITD [9-11]. However, the variant allele frequency (VAF) cutoff value for the Chinese population and the mechanism by which the molecular characteristics of ITD mutations impact the prognosis are still controversial. Meanwhile, FLT3-ITD co-mutations have important prognostic significance, which may even change the treatment decisions. In the opinion of the European Leukemia Net (ELN), NPM1-mutated AML with an FLT3-ITD allele ratio (AR) < 0.5 (low AR) has a favorable prognosis, and allogeneic-hematopoietic stem cell transplant (allo-HSCT) in the first complete remission (CR1) period is not strongly recommended [12]. Also, FLT3-ITD-positive patients with DNMT3A may suffer from poor outcomes [13,14]. More co-mutations with prognostic significance remain to be discovered in this specific subgroup.
FLT3-ITD sequencing was widely applied in the diagnosis and classification of patients with de novo AML. In recent years, next-generation sequencing (NGS) has been used in genomic and epigenetic research with the advantages of high throughput, high sensitivity, and high stability. However, still no uniform statement is available on how to precisely identify the risk stratification of patients with FLT3-ITD. The present study were conducted with 81 patients with FLT3-ITD-positive AML having intermediate-risk cytogenetics to explore the characteristics and assess more potential prognostic markers. In the meantime, an easily applicable scoring model was proposed for overall survival (OS) in patients with newly diagnosed FLT3-ITD-positive AML.
Methods
Subjects
This study included 81 patients diagnosed between December 2013 and March 2018 and treated at the First Affiliated Hospital of Zhejiang University (Figure 1). All patients were FLT3-ITD-postive with intermediate-risk cytogenetic. Most of them (97.5%, 79/81) had de novo AML, and 2.5% (2/81) had mixed-phenotype acute leukemia (MPAL). APL and secondary AML were excluded. Patients who died before induction or receiving nonstandard dose induction therapies were also excluded. Thirty patients from Changhai Hospital with the same enrollment criteria were used for external model validation. AML was diagnosed according to the 2016 revision World Health Organization classification of myeloid neoplasms and acute leukemia [15]. Cytogenetic risks were classified based on the National Comprehensive Cancer Network Guidelines Version 3.2018 [16]. Written consents were obtained from all patients following the Declaration of Helsinki, and the study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University.
Figure 1.
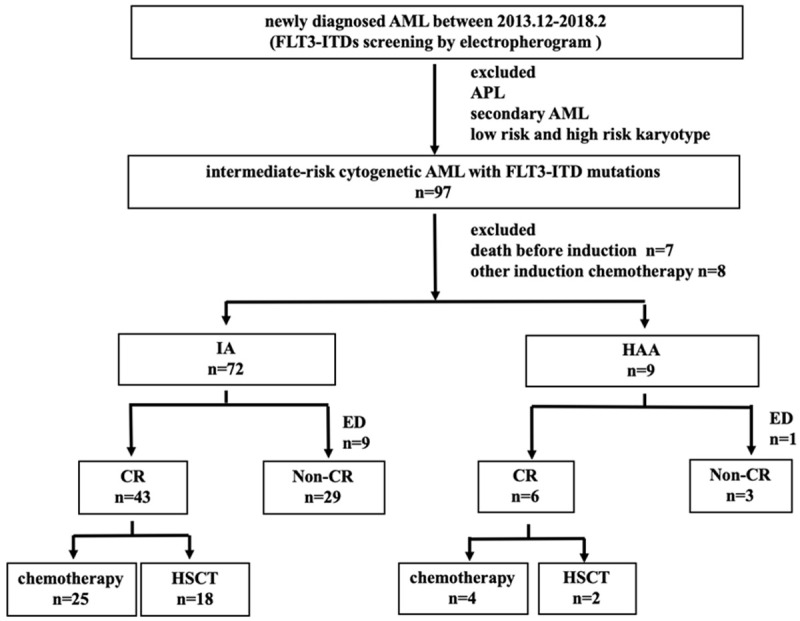
Patient enrollment and treatment response.
Treatments
Of the 81 patients, 88.9% (72/81) were treated with IA 3+7 regimen (idarubicin 10 mg/m2 for 3 days plus cytarabine (Ara-C) 100 mg/m2 for 7 days), and 11.1% (9/81) with HAA regimen (homoharringtonine 2 mg/m2, Ara-C 100 mg/m2 and aclarubicin 20 mg/d for 7 days). Patients who failed to achieve complete remission (CR) after first induction therapy were treated by re-induction (same regimen if partial remission (PR) after the first cycle and crossed regimen if no remission). Consolidation chemotherapy comprised at least two cycles of high-dose cytarabine (Ara-C 2.0 g/m2, every q12h for 3 days), and patients with a suitable donor underwent allogeneic-hematopoietic stem cell transplantation (allo-HSCT). FLT3-specific tyrosine kinase inhibitors-sorafenib was applied to 19 patients with refractory/recurrent disease.
NGS experiments design and methods
NGS was performed in all 81 patients using a panel of 185 genes, which covered all the mutation hotspots of acute leukemia, myelodysplastic syndromeand myeloproliferative neoplasms (Supplementary Table 1). The gene chip was provided by Acornmed Biotechnology Co., Ltd. Genomic DNAs were extracted from mononuclear cells isolated by Ficoll gradient centrifugation from bone marrow (BM) samples at primary diagnosis using a DNeasy 96 Blood & Tissue Kit (Qiagen, Hilden, Germany). Gene library amplification was performed using a KAPA Library Amplification Kit (KAPA, MA, USA). Roche NimbleGen kit (Roche, Basel, Switzerland) was used to capture the target region. The hybridized captured samples were sequenced using NovaSeq 6000 PE150.
Preprocessing of raw sequencing data and quality control statistics were performed using an indigenous QC program. Reads were aligned to the hg19 version of the human genome using a Burrows-Wheeler Alignment tool (BWA, version 0.7.12). Polymerase chain reaction (PCR) duplicates were marked using the MarkDuplicates tool in Picard. IndelRealigner and BaseRecalibrator on Genome Analysis Toolkit (GATK, version 3.8) were used to conduct realignment and recalibration of the BWA alignment results, respectively. Mutect2 was used for identifying paired-sample variant calling of SNV and INDEL. Candidate variations were obtained through background database filtering of normal samples. Pindel (version 0.2.4) [17] was used for detecting FLT3-ITD. FLT3-ITD quantitative analysis was performed by using in-house tools. The quantitative results were also compared with electropherogram to ensure the reliability of NGS. All the variants were annotated using ANNOVAR software [18].
Multiplexed libraries were sequenced using 150-bp paired-end runs on an Illumina Novaseq. The following criteria were used to filter raw variant results so as to ensure the quality of data: average effective sequencing depth on target per sample ≥ 1000 ×; allele mutation frequency ≥ 1% for single-nucleotide variation and insertion or deletion; all reads filtered with high mapping quality (≥ 30) and base quality (≥ 30); and the mutant reads supported by positive and negative strands. Further, synonymous variants, as well as single nucleotide polymorphisms (SNPs) reported in the 1000 Genomes Project database (October 2014 release) at a population frequency > 1% or the in-house SNP databases, were excluded.
Definition of clinical end points
CR was defined as less than 5% BM blasts and showing normal maturation of all cell lineages, no blast in the peripheral blood, absolute neutrophil count > 1.0 × 10E9/L, platelet count > 100 × 10E9/L, and no extramedullary leukemia. Early death referred to all causes of death within 30 days from the first day of induction chemotherapy. Relapse was defined as more than 5% blasts in the BM, reappearance of blasts in the peripheral blood, or extramedullary leukemia in patients with previously documented CR. OS was defined as the time from diagnosis to death or last follow-up. Relapse-free survival (RFS) was defined as the time from first CR to relapse, censoring at death in CR, or last follow-up. Patients were last followed up on February 12, 2019.
Statistical analysis
Data analysis was performed with SPSS (Version 25) and R (version 3.6.3). Differences in continuous variables were analyzed using the Mann-Whitney U test, and categorical variables were compared using the chi-square test or Fisher’s exact test. OS and RFS probabilities were estimated using the Kaplan-Meier survival analysis, and the differences in survival curves were compared using the log-rank test. Cox proportional hazards models were used to assess the clinical variables with survival. Factors for univariate analysis included: clinical characteristics [sex, age (44 years)], laboratory characteristics [white blood cells (median 60.4 × 109/L), hemoglobin (median 85 g/l), platelet count (median 46 × 109/L), lactate dehydrogenase level (median 886 U/L), BM blasts (median 78%)], molecular characteristics of FLT3-ITD [FLT3-ITD-VAF (0.48%), FLT3-ITD number (1 or more than 1), length (69 bp), location (JM/beta1-sheet/other domains)], and co-mutations detected in more than three patients. A statistical significance level of 0.1 in the univariate analysis was used to select variables in the risk score system. The least Absolute Shrinkage and Selector Operation (LASSO) Cox regression model was used for variable selection and predictive prognostic model construction. A two-tailed P < 0.05 indicated a statistically significant difference.
Results
Clinical characteristics of the FLT3-ITD-positive patients
The median age of the patients was 50 (13-70) years, 51.9% (42/81) were men. The FLT3-ITD mutation was seen in all French-American-British classification system (FAB) subgroups of AML except M7, and also seen in Mixed Phenotypic Acute Leukemia (MPAL). It was most common in M2 (40.7%, 33/81), followed by M5 (30.9%, 25/81). The presentation of ITD was related to younger age (76.5% patients were aged less than 60 years), high WBC count (median 60.4 × 10E9/L, 37.0% of 100 × 10E9/L) and high percentage of BM blasts (median 78.0%, range 20.5-97.0%). Among the 81 intermediate-risk cytogenetic patients, 90.1% (73/81) patients had normal karyotypes, and 7.4% (6/81) patients had +8 chromosomes. CR was achieved in 60.5% (49/81) patients, and 40.8% (20/49) patients relapsed after CR. No difference in the CR rate was found between the IA and HAA group (P = 1.000). Early deaths occurred in 12.3% (10/81) patients. The median follow-up duration was 9.5 (0.3-57.5) months for all patients and 22.6 (0.8-57.5) months for the surviving ones. The median OS was 11.4 (95% CI: 7.8-15.0) months, while the median RFS rate was not reached. The 3-year OS rate was 28.4% ± 6.0%, and the 3-year RFS rate was 57.8% ± 8.0%. Further, 40.8% (20/49) patients underwent HSCT in CR1, displaying significantly superior outcomes compared with those who did not undergo HSCT (OS P = 0.002, RFS P = 0.015). The details of the patients are summarized in Table 1.
Table 1.
Characteristics of 81 AML patients with FLT3-ITD mutations
| Variable | Number (%)/Median (range) |
|---|---|
| Sex | |
| Male | 42 (51.9) |
| Female | 39 (48.1) |
| Age, years, median (range) | 50 (13-70) |
| > 60 years | 19 (23.5) |
| WBC, 109/L | 60.4 (1.3-411.6) |
| ≥ 100 × 109/L | 30 (37.0) |
| Hb, g/L | 85 (38-141) |
| PLT, 109/L | 46 (7-241) |
| LDH, U/L | 668 (164-2515) |
| > 245 | 70 (86.4) |
| Bone marrow blasts, % | 78.0 (20.5-97.0) |
| ECOG | |
| 0 | 21 (25.9) |
| 1 | 43 (53.1) |
| 2 | 17 (21.0) |
| FAB | |
| M0 | 2 (2.5) |
| M1 | 13 (16.0) |
| M2 | 33 (40.7) |
| M4 | 5 (6.2) |
| M5 | 25 (30.9) |
| M6 | 1 (1.2) |
| MPAL | 2 (2.5) |
| Karyotype | |
| Normal karyotype | 73 (90.1) |
| Others | 8 (9.9) |
| HSCT | 20 (24.7) |
| Sorafenib | 19 (23.5) |
WBC, white blood cell count; Hb, Hemoglobin; PLT, platelet count; LDH, lactate dehydrogenase; ECOG, Eastern Cooperative Oncology Group; FAB, morphology according to French-American-British classification; MPAL, mixed phenotype acute leukemia; HSCT, Hematopoietic stem cell transplantation.
Mutational landscape of FLT3-ITD-positive patients
Thirty-seven different mutant genes were detected in the 81 patients, and the details of mutations are presented in Figure 2. The top five common concomitant mutations were NPM1 (55.6%), DNMT3A (32.1%), WT1 (17.3%), IDH2 (16.0%), and RUNX1 (16.0%). The median number of co-mutant genes was 4 (0-10), but the number of mutations did not affect the OS and RFS (both P > 0.05). Also, the genes were categorized by function, revealing that methylation-related genes (including DNA methylation and histone methylation, 37.6%), chromatin modifying genes (11.3%), and transcription factor genes (11.3%) were the most common. The gene association analysis was performed for genes detected in more than three patients, showing interesting coexistence and mutual exclusion relationships (Supplementary Figure 1). For example, NPM1-DNMT3A and NPM1-IDH2 tended to appear together (P = 0.00000; P = 0.005, respectively), while NPM1-WT1 and NPM1-RUNX1 appeared to be mutually exclusive (P = 0.00008; P = 0.0014, respectively).
Figure 2.
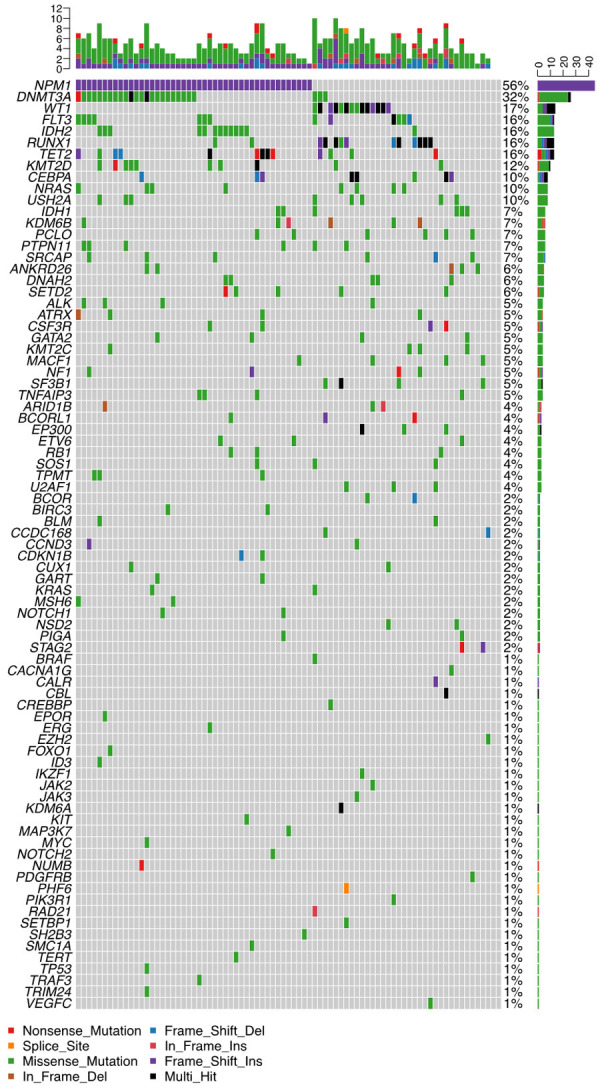
Mutational landscape in 81 patients with FLT3-ITD-positive AML. (Each row represents stated gene; each column represents a patient; the right side of the graph annotates the frequency and number of the gene; the upper histogram showed the number of gene mutations per patient; different colors below the graph represent different mutation patterns).
Molecular characteristics of the FLT3-ITD
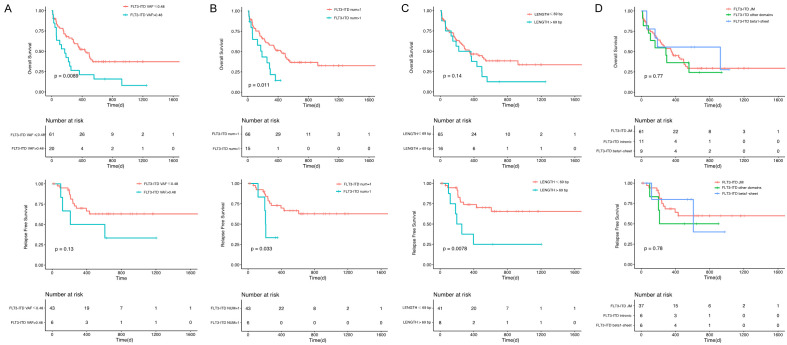
The molecular structure characteristics of the FLT3-ITD are summarized in Table 2. The OS and RFS according to different molecular characteristics of ITD are shown in Figure 3. For patients with multiple ITDs, ITD with the highest VAF was regarded as the predominant mutation; the analysis of the characteristics of these patients was based on the predominant ITDs. The cutoff for VAF 0.48 and length 69 bp were established by using the ROC curve for maximum OS and RFS difference, respectively.
Table 2.
Molecular characteristics of the FLT3-ITDs
| Variable | Number (%)/Median (range) |
|---|---|
| FLT3-ITD VAF | |
| median | 0.46 |
| range | 0.05-0.83 |
| Insertion length (bp) | |
| median | 51 |
| range | 18-207 |
| Insertion number | |
| 1 | 66 (81.5) |
| 2 | 13 (16.0) |
| 3 | 2 (2.5) |
| Insertion region | |
| JM-B | 2 (2.5) |
| JM-S | 6 (7.4) |
| JM-Z | 43 (53.1) |
| HR | 10 (12.3) |
| Beta1-sheet of TKD1 | 9 (11.1) |
| Other domains | 11 (13.6) |
FLT3-ITD, fms-like tyrosine kinase internal tandem duplication; VAF, variant allele frequency (VAF was calculated as mutant/total reads based on the sum of all ITD clones); JM-B, juxtamembrane domain-binding motif; JM-S, juxtamembrane domain-switch motif; JM-Z, juxtamembrane domain-zipper motif; HR, hinge region; TKD1, tyrosine kinase domains 1.
Figure 3.
OS and RFS in the entire patient group according to different molecular characteristics of FLT3-ITD. A. Mutation VAF; B. Mutation number; C. Mutation length; D. Mutation insertion site.
FLT3-ITD mutation VAF: VAF was calculated as mutant/total reads based on the sum of all ITD clones. The patients’ median VAF was 0.46 (0.05-0.8). According to the NCCN guidelines for AML, the VAF cutoff value of FLT3-ITD mutation between different stages of prognosis was 0.33. However, in the present study, 0.33 was not confirmed as the suitable cutoff value with or without considering the NPM1 mutation status (OS and RFS P > 0.05). Instead, patients with VAF > 0.48 had worse OS (P = 0.0089 for OS, P = 0.13 for RFS).
FLT3-ITD mutation number: Among 81 patients, 81.5% (66/81) carried 1 ITD, 16.0% (13/81) carried 2 ITDs, and only 2.5% (2/81) carried 3 ITDs. The number of ITDs was significantly related to prognosis. Patients with multiple ITDs were found to have worse clinical outcomes (P = 0.011 for OS, P = 0.033 for RFS).
FLT3-ITD mutation length: The median length of the insertion fragments was 51 (18-207) bp. Patients were grouped according to ITD size less or more than 69 bp, the longer the insertion (> 69 bp) the worse the RFS of the patient (P = 0.0078), but not the OS (P = 0.14).
FLT3-ITD mutation insertion region: ITD integration sites were categorized according to different functional regions of FLT3. In most patients (75.3%, 61/81), the ITD fragments were inserted in the juxtamembrane domain (JMD), containing JMD-binding motif (2.5%, 2/81), JMD switch motif (7.4%, 6/81), JMD-zipper motif (53.1%, 43/81), and hinge region (12.3%, 10/81). The other fragments were localized in the beta1-sheet of tyrosine kinase domain 1 (TKD1) (11.1%, 9/81) and other domains (13.6%, 11/81). No significantly prognostic differences were observed in JMD and other regions (P = 0.77 for OS, P = 0.78 for RFS).
Prognostic scoring model and nomogram construction
Prognostic factors with P < 0.1 in the univariate analysis were used to build the risk score system; the results of univariate analysis are listed in Supplementary Table 2. Five variables were incorporated in our scoring model using LASSO. A risk scoring model was developed incorporating the weighted coefficients of these variables: 0.659 × FLT3-ITD VAF + 0.375 × FLT3-ITD number + 0.807 × Age + 0.688 × DNMT3A + 1.939 × U2AF1 (FLT3-ITD VAF > 0.48 scored 1; FLT3-ITD number scored 1 if carried 1 ITD, 2 if carried ≥ 2 ITDs; age > 44 years scored 1, the presence of DNMT3A or U2AF1 scored 1; 0 for other conditions). Patients with FLT3-ITD-positive AML were categorized into two subgroups based on the risk score: low-risk (L-R, score < 1, n = 20) and high risk (H-R, score ≥ 1, n = 61) groups. The 3-year OS for the L-R and H-R groups was 73.8% ± 10.2% and 12.8% ± 5.5%, respectively (P < 0.0001), while the 3-year RFS was 92.3% ± 7.4% and 35.5% ± 10.5%, respectively (P = 0.0005) (Supplementary Figure 2A, 2B). Similar results were obtained in the validation cohort of 30 patients (L-R, n = 10 and H-R, n = 20; 3-year OS 80.0% ± 12.6% vs. 25.4% ± 10.3%, P = 0.01) (Supplementary Figure 2C). A comparison of the basic characteristics of patients in the experimental cohort and the validation cohorts is shown in Table 3.
Table 3.
Comparison of basic characteristics of patients in the experimental cohort and the verification cohort
| Factor | Experimental cohort | Verification cohort | P value |
|---|---|---|---|
| Number, n | 81 | 30 | |
| Age, years | 50 | 43 | 0.19 |
| Sex | 0.64 | ||
| Male | 42 (52%) | 14 (47%) | |
| Female | 39 (48%) | 16 (53%) | |
| BM blasts, % | 78 | 68.5 | 0.69 |
| WBC, 109/L | 60.4 | 17.37 | 0.004 |
| HB, g/L | 85 | 87 | 0.70 |
| PLT, 109/L | 46 | 68 | 0.014 |
| HSCT | 20 (25%) | 11 (37%) | 0.15 |
A prognostic nomogram that integrated all the five significantly independent factors from the LASSO Cox regression model was constructed (Figure 4). The predictive accuracy for OS in the experimental cohort as evaluated using the C-index was 0.68 (95% CI, 0.64-0.72). The calibration curves for predicting patient OS after 3 years showed a good correlation between the nomogram-predicted and actually observed values (Supplementary Figure 3A, 3B). The nomogram was externally confirmed in the validation cohort. The C-index of the validation cohort was 0.72 (95% CI, 0.60-0.84) (Supplementary Figure 3C, 3D).
Figure 4.
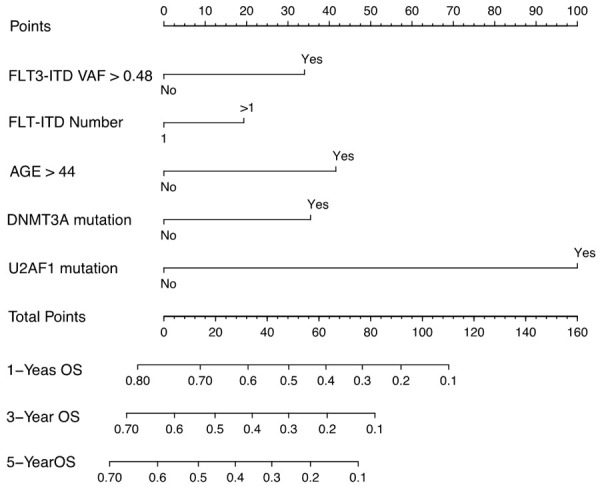
Nomogram for patients with newly diagnosed FLT3-ITD-positive AML. An individual’s value is located on each variable axis, and a line is drawn upward to determine the points received for each variable (corresponding points for each variable: FLT3-ITD VAF > 0.48, 31; FLT3-ITD number > 1, 20; age > 44 years, 41; the present of DNMT3A, 32; the present of U2AF1, 100). The sum of these points is located on the total point axis, and a line is drawn downward to the survival axes to determine the likelihood of 1, 3, or 5-year overall survival.
Discussion
This study analyzed the gene distribution of patients with FLT3-ITD having intermediate-risk cytogenetic using the NGS platform. It concluded that Chinese patients with FLT3-ITD had many coexisting gene mutations, and the interaction was very complicated. Meanwhile, the study explored the structural characteristics of FLT3-ITD and found that VAF > 0.48, length of the insertion fragments > 69 bp, and the number of ITDs > 1 had significant effects on prognosis. Since multiple factors could affect the prognosis, the present study was novel in proposing a scoring model combining clinical features, structural features, and gene mutations of FLT3-ITD-positive patients to predict outcomes. The databases from this study might help in further investigating the risk stratification and prognostic prediction among patients with intermediate-risk cytogenetic AML.
This study analyzed the gene expression of the 81 patients with de novo AML by target sequencing and discovered that most of the co-mutations clustered in methylation-related genes, chromatin-modifying genes and transcription factor genes. NPM1, DNMT3A, and WT1 mutations were the most common, while mutations such as TP53, JAK, and RAS were rare. The distribution was similar to other reported genomic and epigenetic landscapes of de novo AML in adult patients; however, NPM1 and DNMT3A mutations were more frequently identified in the present study [19,20].
Several studies showed controversial prognostic effects of the molecular trait of ITD. Some previously published studies have shown that VAF (ranging from 0.3 to 0.6) was the key prognostic factor [11,21-23]. The present study proved that VAF 0.33 (AR 0.5) was not the prognostic cutoff value for Chinese patients, but VAF 0.48 might be. This was probably due to the heterogeneity of the patients, difference in treatment approaches, and the effect of co-mutations. Similarly, Kim et al. proved that ITD length greater than 70 bp was significantly associated with inferior OS and EFS [21]. Chen et al. suggested that patients with longer ITDs (> 60 bp) had a worse OS and RFS [24]. On the contrary, some studies had the opposite conclusion; they found better survival in the long-ITD (> 70 bp) subgroup. In terms of the ITD mutation number, some studies supported the view that multiple ITDs had a poor prognosis, but some others found no prognostic difference in the number change [25]. Like most studies, the present study did not find a prognostic impact at different ITD locations. However, Kayser et al. believed that the insertion of FLT3-ITD in the tyrosine kinase domain-1 was associated with resistance to chemotherapy and poor outcomes [26], and Liu et al. suggested that the ITD integration site in the hinge region or beta1-sheet region was an unfavorable prognostic factor [11]. It is believed that the relationship between the molecular structure characteristics of ITD and prognosis will be further clarified with the application of NGS and more detailed results.
Less information is available to date for stratifying the risks and predicting the prognosis of patients with FLT3-ITD. In this study, a novel scoring model was established to distinguish patients with a good prognosis from a poor prognosis. Five variables were incorporated in the scoring model using LASSO, including FLT3-ITD VAF, FLT3-ITD number, age, DNMT3A mutation, and U2AF1 mutation, which were independent risk factors predicting the prognosis. The scoring model could accurately identify patients with a better prognosis. This new model might help doctors to interfere in advance for improving the prognosis and reducing the mortality of patients.
This study had some limitations. First, induction and consolidation regimens could not be completely unified due to the retrospective nature of the study. Second, the total number of patients in the experimental cohort was only 81, and the sample size was small, despite including a verification cohort. Hence, studies with a larger sample size are required to verify the new model further.
In this study, multi-gene sequencing and comprehensive prognostic analysis were performed for ITD-positive Chinese patients. These findings not only provided important information on the molecular structure characteristics of ITD but also revealed a novel scoring model that could stratify patients into two subgroups with distinct clinical outcomes.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC) (Grant No. 81800146) and Zhejiang Province Natural Science Foundation of China grants (LY19H080009).
Disclosure of conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting Information
References
- 1.Kuykendall A, Duployez N, Boissel N, Lancet JE, Welch JS. Acute myeloid leukemia: the good, the bad, and the ugly. Am Soc Clin Oncol Educ Book. 2018;38:555–573. doi: 10.1200/EDBK_199519. [DOI] [PubMed] [Google Scholar]
- 2.Estey EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol. 2018;93:1267–1291. doi: 10.1002/ajh.25214. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O’Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagunas-Rangel FA, Chavez-Valencia V. FLT3-ITD and its current role in acute myeloid leukaemia. Med Oncol. 2017;34:114. doi: 10.1007/s12032-017-0970-x. [DOI] [PubMed] [Google Scholar]
- 5.Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, Zang ZJ, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Ding LW, Alpermann T, Sun QY, Lin DC, Chien W, Madan V, Liu LZ, Tan KT, Sampath A, Venkatesan S, Inokuchi K, Wakita S, Yamaguchi H, Chng WJ, Kham SK, Yeoh AE, Sanada M, Schiller J, Kreuzer KA, Kornblau SM, Kantarjian HM, Haferlach T, Lill M, Kuo MC, Shih LY, Blau IW, Blau O, Yang H, Ogawa S, Koeffler HP. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126:2491–2501. doi: 10.1182/blood-2015-05-646240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao S, Wang C, Chen Y, Deng Y, Song L, Shi Y, Ling L, Ding B, He Z, Yu L. Prognosis and outcome of patients with acute myeloid leukemia based on FLT3-ITD mutation with or without additional abnormal cytogenetics. Oncol Lett. 2019;18:6766–6774. doi: 10.3892/ol.2019.11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu QC, Wang C, Bao XB, Yang J, Shen HJ, Ding ZX, Liu H, He J, Yao H, Chen SN, Li Z, Xue SL, Liu SB. The impact of FLT3 mutations on treatment response and survival in Chinese de novo AML patients. Hematology. 2018;23:131–138. doi: 10.1080/10245332.2017.1372248. [DOI] [PubMed] [Google Scholar]
- 9.Lyu M, Liao H, Shuai X, Jin Y, Su J, Zheng Q. The prognosis predictive value of FMS-like tyrosine kinase 3-internal tandem duplications mutant allelic ratio (FLT3-ITD MR) in patients with acute myeloid leukemia detected by GeneScan. Gene. 2020;726:144195. doi: 10.1016/j.gene.2019.144195. [DOI] [PubMed] [Google Scholar]
- 10.Liu SB, Dong HJ, Bao XB, Qiu QC, Li HZ, Shen HJ, Ding ZX, Wang C, Chu XL, Yu JQ, Tao T, Li Z, Tang XW, Chen SN, Wu DP, Li L, Xue SL. Impact of FLT3-ITD length on prognosis of acute myeloid leukemia. Haematologica. 2019;104:e9–e12. doi: 10.3324/haematol.2018.191809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu SB, Qiu QC, Bao XB, Ma X, Li HZ, Liu YJ, Chen SN, Song YH, Wu DP, Xue SL. Pattern and prognostic value of FLT3-ITD mutations in Chinese de novo adult acute myeloid leukemia. Cancer Sci. 2018;109:3981–3992. doi: 10.1111/cas.13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Lowenberg B, Bloomfield CD. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, Dunlap J, Paliga A, Traer E, Press R, Shen L, Fan G. DNMT3A co-mutation is required for FLT3-ITD as an adverse prognostic indicator in intermediate-risk cytogenetic group AML. Leuk Lymphoma. 2018;59:1938–1948. doi: 10.1080/10428194.2017.1397659. [DOI] [PubMed] [Google Scholar]
- 14.Tang S, Shen H, Mao X, Dai H, Zhu X, Xue S, Ding Z, Lu J, Wu D, Tang X. FLT3-ITD with DNMT3A R882 double mutation is a poor prognostic factor in Chinese patients with acute myeloid leukemia after chemotherapy or allogeneic hematopoietic stem cell transplantation. Int J Hematol. 2017;106:552–561. doi: 10.1007/s12185-017-2256-7. [DOI] [PubMed] [Google Scholar]
- 15.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 16.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: acute myeloid leukemia, V.3.2018. Nov 20, 2018. http://www.nccn.org/professionals/physician_gls/f_guidelines.asp.
- 17.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. Annovar: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br J Haematol. 2017;179:530–542. doi: 10.1111/bjh.14823. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Lee GD, Park J, Yoon JH, Kim HJ, Min WS, Kim M. Quantitative fragment analysis of FLT3-ITD efficiently identifying poor prognostic group with high mutant allele burden or long ITD length. Blood Cancer J. 2015;5:e336. doi: 10.1038/bcj.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blau O, Berenstein R, Sindram A, Blau IW. Molecular analysis of different FLT3-ITD mutations in acute myeloid leukemia. Leuk Lymphoma. 2013;54:145–152. doi: 10.3109/10428194.2012.704999. [DOI] [PubMed] [Google Scholar]
- 23.Sakaguchi M, Yamaguchi H, Najima Y, Usuki K, Ueki T, Oh I, Mori S, Kawata E, Uoshima N, Kobayashi Y, Kako S, Tajika K, Gomi S, Shono K, Kayamori K, Hagihara M, Kanda J, Uchiyama H, Kuroda J, Uchida N, Kubota Y, Kimura S, Kurosawa S, Nakajima N, Marumo A, Omori I, Fujiwara Y, Yui S, Wakita S, Arai K, Kitano T, Kakihana K, Kanda Y, Ohashi K, Fukuda T, Inokuchi K. Prognostic impact of low allelic ratio FLT3-ITD and NPM1 mutation in acute myeloid leukemia. Blood Adv. 2018;2:2744–2754. doi: 10.1182/bloodadvances.2018020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, Sun J, Yin C, Cheng J, Ni J, Jiang L, Wang Q, Yu G, Wei Y, Liu X, Sun J, Carter BZ, Jiang X. Impact of FLT3-ITD allele ratio and ITD length on therapeutic outcome in cytogenetically normal AML patients without NPM1 mutation. Bone Marrow Transplant. 2020;55:740–748. doi: 10.1038/s41409-019-0721-z. [DOI] [PubMed] [Google Scholar]
- 25.Azari-Yam A, Tavakkoly-Bazzaz J, Semnani Y, Davoudi-Dehaghani E, Ghodssi-Ghassemabadi R, Kianfar S, Saadat A, Masoudifard M, Yaghmaie M, Alimoghaddam K, Ghavamzadeh A, Zeinali S. FLT3 gene mutation profile and prognosis in adult acute myeloid leukemia. Clin Lab. 2016;62:2011–2017. doi: 10.7754/Clin.Lab.2016.160324. [DOI] [PubMed] [Google Scholar]
- 26.Kayser S, Schlenk RF, Londono MC, Breitenbuecher F, Wittke K, Du J, Groner S, Spath D, Krauter J, Ganser A, Dohner H, Fischer T, Dohner K. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood. 2009;114:2386–2392. doi: 10.1182/blood-2009-03-209999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.