Abstract
Infection with the human papillomaviruses (HPV) often involves the epigenetic modification of the host genome. Despite its prevalence among the population, host genome methylation in HPV-induced warts is not clearly understood. In this study, genome-wide methylation profiling was carried out on paired healthy skin and wart samples in order to investigate the effects that benign HPV infection has on gene methylation status. To overcome this gap in knowledge, paired wart (n = 12) and normal skin (n = 12) samples were obtained from Arab males in order to perform DNA extraction and subsequent genome-wide methylation profiling on the Infinium Methylation EPIC Bead Chip microarray. Analysis of differential methylation revealed a clear pattern of discrimination between the wart and normal skin samples. In warts, the most differentially methylated (DM) genes included long non-coding RNAs (AC005884, AL049646.2, AC126121.2, AP001790.1, and AC107959.3), microRNAs (MIR374B, MIR596, MIR1255B1, MIR26B, and MIR196A2),snoRNAs (SNORD114-22, SNORD70, and SNORD114-31), pseudogenes (AC069366.1, RNU4ATAC11P, AC120057.1, NANOGP3, AC106038.2, TPT1P2, SDC4P, PKMP3, and VN2R3P), and protein-coding genes (AREG, GJB2, C12orf71, AC020909.2, S100A8, ZBED2, FABP7, and CYSLTR1). In addition, pathway analysis revealed that, among the most differentially methylated genes, STAT5A, RARA, MEF2D, MAP3K8, and THRA were the common regulators. It can be observed that HPV-induced warts involve a clear and unique epigenetic alteration to the host genome.
Keywords: DNA methylation, Epigenetics, HPV, Warts
1. Introduction
The human papillomaviruses (HPV) are a group of double-stranded DNA viruses that exclusively infect the epithelia of mucosal or cutaneous surfaces (Mcmurray et al., 2001). HPV is an obligate intracellular parasite, entering the host cell, i.e. the basal keratinocyte, through a micro-abrasion in the epithelial layer and inducing its rapid growth and proliferation (Bacaj and Burch, 2018, Horvath et al., 2010). HPV transmission can occur via both sexual and non-sexual contact with infected individuals as well as through fomites (Houlihan et al., 2019). High-risk HPV types have been identified as a major causative agent in cervical cancer, while low-risk types have been associated with formation of skin lesions known as warts (Bosch et al., 2002, Senapati et al., 2016).
Warts are benign hyperproliferative tumors that are induced by low-risk HPV infection, with cases often resolving in a spontaneous manner (Jabłońska et al., 1997). Depending on the HPV type, warts can differ from one another based on their morphology, histology, anatomical localization, and potential to cause malignant transformation (Egawa et al., 2015). The most prevalent type of cutaneous wart is the common wart (Verruca vulgaris), which manifests as a discolored or skin-colored lesion with a hyperkeratotic surface (Mulhem and Pinelis, 2011). Common warts are associated with the low-risk HPV types 1–5, 27, 29, and 57, the latter of which are tightly linked to the differentiation cycle of basal keratinocytes (Al Aboud and Nigam, 2018, Bruggink et al., 2012, Graham, 2017, Mulhem and Pinelis, 2011). Due to the ephemeral nature of warts, epigenetic mechanisms such as DNA methylation are posited to be involved in their formation (Milavetz and Balakrishnan, 2015, von Knebel Doeberitz and Prigge, 2019).
DNA methylation, which involves the enzymatic addition of a methyl group to the fifth cytosine carbon, mostly occurs on CpG dinucleotides and plays an essential role in normal mammalian development (Hackett and Azim Surani, 2013, Smith and Meissner, 2013, Yong et al., 2016). Differentiated cells possess a stable and distinct pattern of methylation that helps regulate the transcription of their genes in a tissue-specific manner (Moore et al., 2013). Under normal conditions, the majority of the human genome can be found in a methylated state, but pockets of hypomethylation can be found within CpG islands, which are short and interspersed throughout the genome (Deaton and Bird, 2011). Modulated patterns of DNA methylation have been reported in skin pathogeneses, including autoimmune and malignant disorders (Mervis and McGee, 2020). Furthermore, aberrant methylation of the host cell genome is often induced by HPV infection and has been observed in a number of high-risk HPV-associated diseases (Dankai et al., 2019, Feng et al., 2018, Verlaat et al., 2018, von Knebel Doeberitz and Prigge, 2019).
Due to their carcinogenic potential, high-risk HPV infection has been the subject of a much greater amount of research compared to its low-risk counterpart. Our previous studies on warts focused on the methylation status of CpG islands, CpG sites, promoters, and tiling regions, with significant differential methylation being reported that significantly differed depending on the type of gene region (Al-Eitan et al., 2019a, Al-Eitan et al., 2020a, Al-Eitan et al., 2020b, Al-Eitan et al., 2020c). In this study, genome-wide methylation profiling was carried out on paired healthy skin and wart samples in order to investigate the effects that benign HPV infection has on gene methylation status.
2. Materials and methods
2.1. Ethics approval and consent to participate
Ethical approval was obtained from the Institutional Review Board (IRB) committee at Jordan University of Science and Technology (Ref. # 19/105/2017). All participants gave written informed consent before taking part in this study.
2.2. Sample collection
The same case cohort from previously published works was used in the current study to minimize the effects of inter-individual genetic variation (Al-Eitan et al., 2020a, Al-Eitan et al., 2020b, 2020d, 2020c, 2019a, 2019b). Participants (n = 12) were Arab males presenting with common warts (Verruca vulgaris) ranging between 18 and 27 years old in age and not reporting any comorbidities. Common warts were identified by a resident dermatologist based on typical clinical diagnosis (black thrombosed capillaries or pinpoint bleeding when pared and keratotic surfaces), and, after application of a local anesthetic, superficial shave biopsies of the warts (n = 12) and adjacent normal skin (n = 12) were obtained. The anatomic localization of the biopsies included the dorsal sides of the hand (n = 20) and foot (n = 2) as well as the upper part of the forehead (n = 2).
2.3. DNA extraction and methylation profiling
A QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) was employed in order to extract genomic DNA, which was later treated with RNase A. The quality of the extracted DNA was determined using agarose gel electrophoresisand the BioTekPowerWave XS2 Spectrophotometer (BioTek Instruments, Inc., Winuski, VT, USA). DNA samples that passed quality control were shipped on dry ice to the Australian Genome Research Facility (AGRF), where their quality and quantity were reassessed using the QuantiFluor® dsDNA System (Promega, Madison, WI, USA) and 0.8% agarose gel electrophoresis. After normalization to around 500 ng of DNA per 45 μL, the DNA samples were bisulfite converted using the Zymo EZ DNA Methylation kit (Zymo Research, Owen, CA, USA). Genome-wide methylation profiling was performed on the Infinium MethylationEPICBeadChip microarray (Illumina, San Diego, CA, USA), the latter of which analyzes the methylation patterns of over 850,000 CpG sites.
2.4. Data processing
The computational R package RnBeads was altered for the processing and analysis of the raw intensity data (IDAT files) produced by the methylation chip (Assenov et al., 2014). Following the RnBeads pipeline, all probes and samples were subject to quality control assurance, after which they were pre-processed, adjusted for batch effects, and normalized.
2.5. Differential methylation and statistical analysis
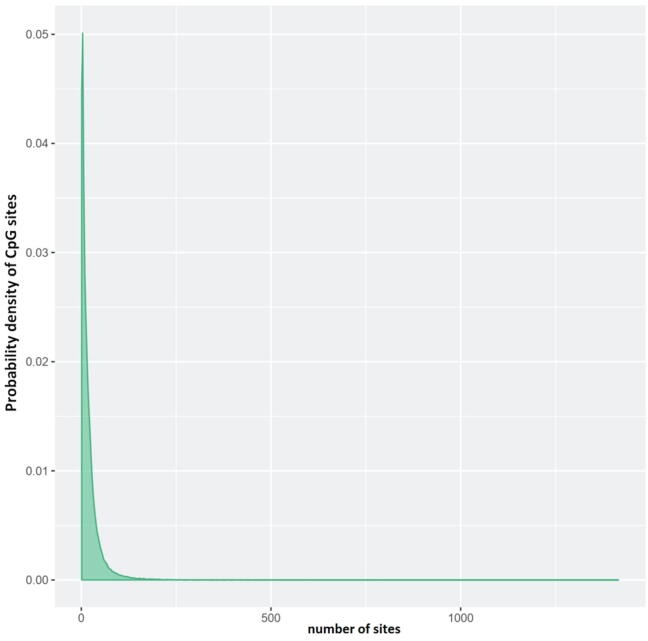
At the gene level, the mean of the mean β (mean.mean.β) values of all tested CpG sites in each gene was computed. Fig. 1 depicts the number of CpG sites on each gene. The differential methylation level of each gene was calculated through the mean.mean.β difference between normal skin (NS) and warts (W), the log2 of the mean quotient in β means across all CpG sites in a gene; and, using a limma statistical test, the adjusted combined p-value of all CpG sites in a gene (Al-Eitan et al., 2019a, Assenov et al., 2014, Ritchie et al., 2015). The Benjamini and Hochberg (B-H) procedure was employed to account for multiple testing. Based on the three aforementioned analyses, each gene was given a rank, and the combined rank score was computed as the maximum (lowest) rank among the three ranks. Genes which exhibit more differential methylation will have a smaller combined rank (Al-Eitan et al., 2020b, Assenov et al., 2014). Using the combined rank score, genes were sorted from smallest to largest, and the highest 1000 genes in terms of combined rank were selected for further analysis.
Fig. 1.
Distribution of the number of CpG sites per gene. The number of CpG sites (probes) that were considered for each gene in the Infinium MethylationEpic BeadChip platform is shown on the x-axis while the density of CpG sites is shown on the y-axis.
2.6. Gene ontology (GO) enrichment analysis
Using the gene ontology (GO) consortium, enrichment analysis for the (GO) terms associated with the highest-ranking 500 differentially methylated genes was carried out using GO consortium (The Gene Ontology Consortium, 2017).
2.7. Signaling network analysis
The signaling network of the highest-ranking 1000 differentially methylated genes was explored using the Signaling Network Open Resource 2.0 (Signor) (Perfetto et al., 2016). Only ‘direct’ interactions with a relaxed layout and a score of ‘0.0′ were included for analysis as a result of the great number of connections.
2.8. Validation of the top five differentially methylated genes
The 100 most DM genes were sorted according to the combined rank score and merged with the 100 most differentially expressed (DE) genes from a previous study carried out in our lab on the same sample set (Al-Eitan et al., 2020d). The top five overlapping genes were selected for further validation after examining both their expression and methylation levels.
3. Results
3.1. Samples clustering
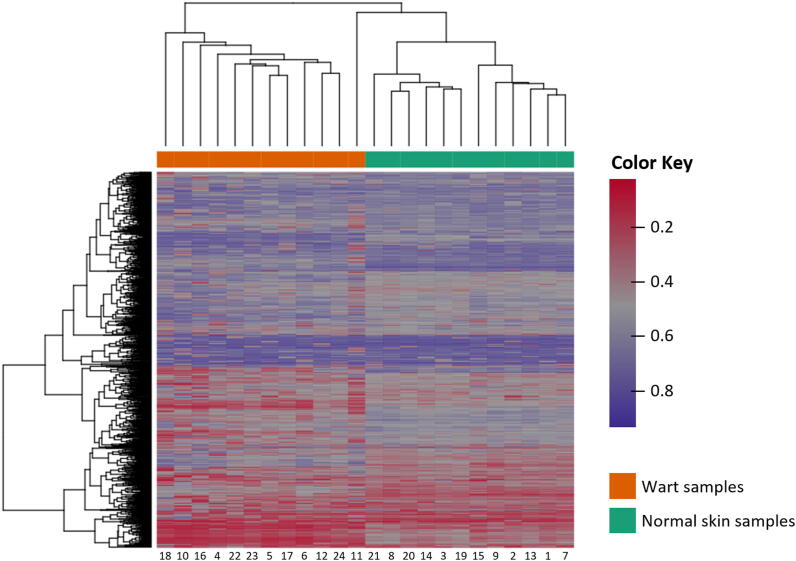
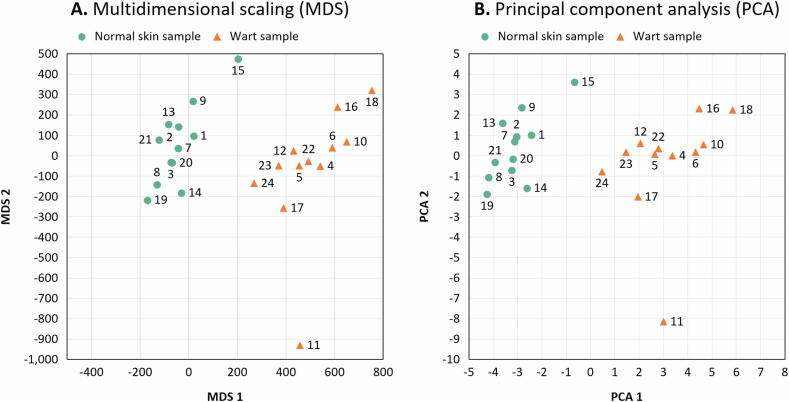
Samples were found to exhibit an expected pattern of hierarchical clustering based on the complete set of methylation values calculated for the 1000 highest-ranking genes in terms of differential methylation (Fig. 2). To further confirm this phenotypic difference in clustering, multidimensional scaling (MDS) (Fig. 3A) and principal component analysis (PCA) (Fig. 3B) were carried out on the dataset, showing that wart and normal skin samples were significantly different.
Fig. 2.
Hierarchical clustering of the 1,000 genes with the highest degree of variance across all samples. Complete linkage and Manhattan distance were used for clustering of samples. The patient identification number is shown on the bottom x-axis, while the normal skin (NS) and wart (W) samples are shown on the top x-axis. The scale’s color intensity correlates with the degree and direction of differential methylation (DM), whereby values of 0 (red color) and 1 (purple color) indicate decreased and increased methylation, respectively.
Fig. 3.
Scatter plots showing the sample coordinates after performing (A) Kruskal’s non-metric multidimensional scaling based on the matrix of average methylation levels and Manhattan distance and (B) principal component analysis.
3.2. Identification of differentially methylated genes
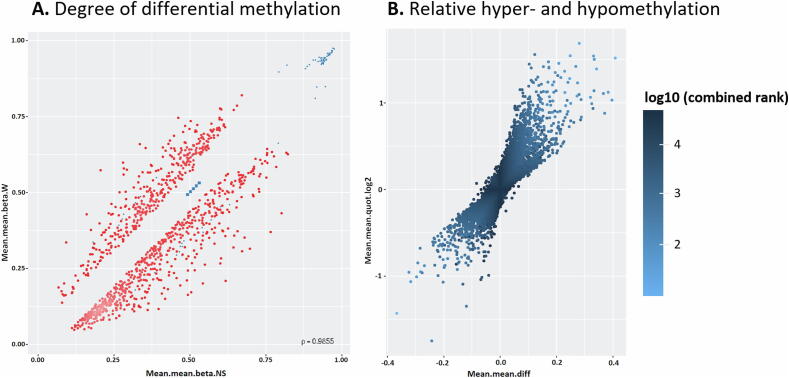
35,026 genomic identifiers (Ensembl release 75 data) passed the quality control and preprocessing steps, including some identifiers that did not map to gene symbols or were not assigned (NA). Genomic identifiers without gene symbols were then removed to obtain a total of 34,044 genes with known symbols. The list of differentially methylated genes in warts was limited to the highest-ranking 1,000 genes based on the combined ranking score. Using this scoring method, a list of 610 genes were found to be hypomethylated and 390 genes to be hypermethylated in warts (W) compared to normal skin (NS) with a mean β difference > 0.057 and < -0.056 (p-value =< 0.001 (adjusted p-value =<0.01)) (Fig. 4A). Of the 610 hypomethylated genes, the β difference ranged from −0.056 to −0.409. Of the 390 hypermethylated genes, the β difference ranged from 0.057 to 0.367. The log2 of the quotient in methylation between W and NS had a maximum value of 1.686 and minimum value of −1.751 (Fig. 4B). The 100 genes with the lowest combined rank score are presented in Table S1, and their categorical breakdown is illustrated in Fig. 5.
Fig. 4.
Identification of the 1,000 most differentially methylated (DM) genes between wart (W) and normal skin (NS) samples. (A) The scatterplot illustrates the mean of mean methylation (β) levels for NS (x-axis) and W (y-axis) samples. β levels lay between 0 (unmethylated) and 1 (methylated), with blue points indicating DM sites. (B) The volcano plot depicts relative hypomethylation (<0) and hypermethylation (>0) as measured by the mean of the mean fold difference (mean.mean.diff) (x-axis) and the log2 of the mean of mean quotient in methylation (mean.mean.quot.log2) (y-axis). The scale’s color intensity correlates with the combined rank score, whereby a lower score indicates more DM.
Fig. 5.
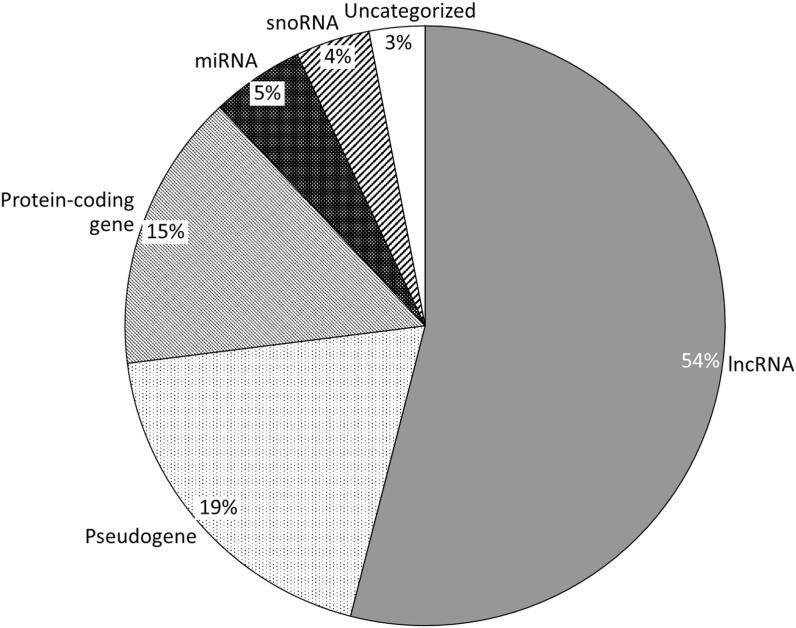
Category breakdown of the 100 most differentially methylated genes in warts (W) compared to normal skin (NS). The genes belonged to the following categories: RNA genes (65%), pseudogenes (19%), protein-coding genes (15%), and uncategorized genes (3%). The 65 RNA genes could be further classified into lncRNAs (54%), miRNAs (5%), and snoRNAs (4%).
3.3. Gene ontology (GO) enrichment analysis
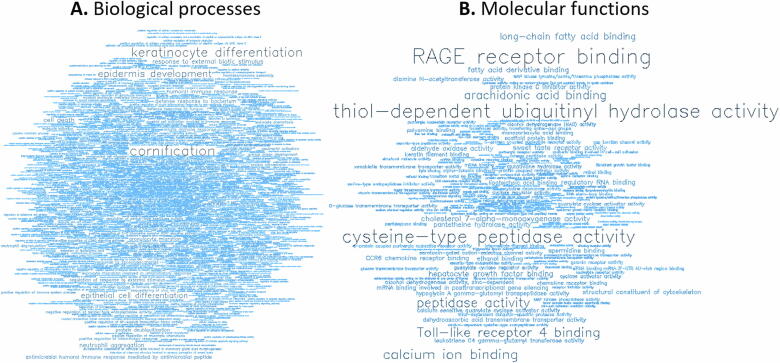
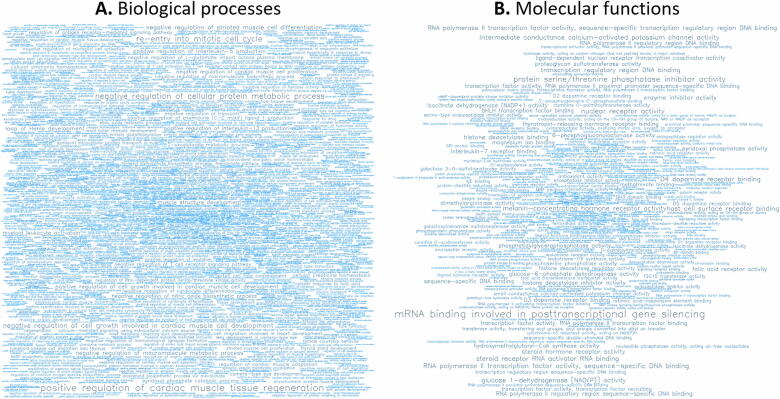
GO analysis of the 500 most hypermethylated (Fig. 6 and Tables S2 and S3) and the 500 most hypomethylated genes (Fig. 7 and Tables S4 and S5) revealed the level of representation of terms related to biological processes (BP) and molecular functions (MF).
Fig. 6.
For the 500 most hypermethylated genes, word clouds showing the significant (A) biological processes (BP) and (B) molecular functions (MF).
Fig. 7.
For the 500 most hypomethylated genes, word clouds showing the significant (A) biological processes (BP) and (B) molecular functions (MF).
3.4. Pathway analysis
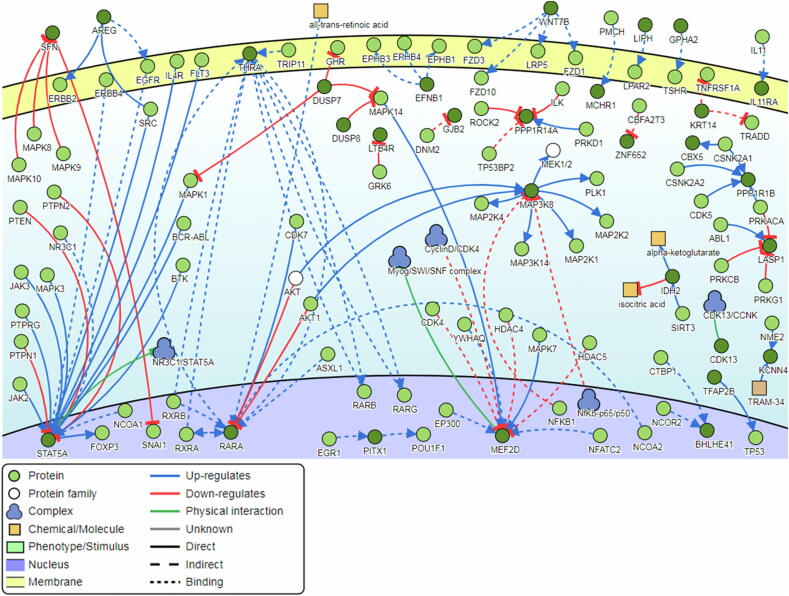
Pathway analysis of the top-ranking 1,000 differentially methylated genes showed that several genes were found to be common regulators of the gene network, with a minimum of 9 direct connectivities each (Fig. 8). Namely, these genes included the STAT5A, RARA, MEF2D, MAP3K8, and THRA genes.
Fig. 8.
Pathway signalling network generated from the 1000 most differentially methylated genes. Five genes (STAT5A, RARA, MEF2D, MAP3K8, and THRA) were found to have a minimum of 9 direct connectivities each.
3.5. Validation of the top five differentially methylated genes
The top five overlapping genes in terms of differential methylation and expression were the AREG, GJB2, S100A8, FABP7, and ZBED2 genes. The expression and methylation levels of these genes in HPV-induced warts and non-infected normal skin are listed in Table 1. Of the 5 genes selected for validation, 4 genes (AREG, GJB2, S100A8, and ZBED2) were hypomethylated and had decreased levels of expression in warts compared to normal skin. In contrast, the FABP7 gene was found to be hypermethylated and upregulated in warts compered to normal skin.
Table 1.
Methylation and expression levels of the 5 most differentially methylated genes in warts (W) compared to normal skin (NS).
| Gene | Methylation Level | Expression Level | ||||
|---|---|---|---|---|---|---|
| mean.mean β value (NS) 1 | mean.mean β value (W) 1 |
mean.mean β value (W-NS) 1 |
FDR | LogFC2 | FDR3 | |
| AREG | 0.618 | 0.209 | −0.408 | 7.87 × 10–8 | −3.393 | 1.38 × 10–9 |
| GJB2 | 0.516 | 0.244 | −0.272 | 1.29 × 10–7 | −3.404 | 3.11 × 10–10 |
| FABP7 | 0.255 | 0.453 | 0.198 | 4.62 × 10–7 | 4.427 | 1.60 × 10–5 |
| S100A8 | 0.317 | 0.15 | −0.167 | 3.47 × 10–7 | −8.412 | 1.07 × 10–8 |
| ZBED2 | 0.353 | 0.174 | −0.179 | 1.87 × 10–5 | −1.842 | 1.10 × 10–5 |
Mean.mean β value = mean of mean methylation levels across all sites in a region;
logFC = log fold change;
FDR = false discovery rate.
4. Discussion
The human papillomaviruses (HPVs) have evolved several epigenetic mechanisms to alter the gene expression and biology of host cells (Durzynska et al., 2017). Over the course of HPV infection, several epigenetic changes occur to both the host and viral genomes, including histone modifications as well as hypomethylation and hypermethylation of lncRNA and miRNA genes (Soto et al., 2017). DNA methylation is one of the most studied mechanisms of epigenetic change, and it is involved in the regulation of gene expression through its influence on transcriptional regulation (Barros and Offenbacher, 2009). Increased DNA methylation generally results in a loss of gene expression, but this relationship can fluctuate depending on biological context (Lim and Maher, 2010). In fact, a growing number of studies report that promoter hypermethylation results in gene activation (Smith et al., 2020). Similarly, methylation of the gene body is associated with a loss of expression in non-dividing and slowly dividing cells but not in actively dividing cells (Moore et al., 2013).
Aberrant methylation to the host genome has been extensively reported for high-risk HPV infection associated with cervical and oropharyngeal cancers, with certain methylation statuses acting as biomarkers for the disease (Clarke et al., 2018, Clarke et al., 2012, Marongiu et al., 2014). Furthermore, HPV-induced methylation of the host genome can act as a reliable diagnostic and prognostic tool in oropharyngeal squamous cell carcinoma (Boscolo-Rizzo et al., 2017). In contrast, a dearth of information exists regarding the impact of low-risk HPV infection on host genome methylation. Low-risk HPV infection of the basal epithelium, which is the only site of active cell division in the epithelium, results in the formation of warts (Münger et al., 2004). Since HPV infects actively dividing cells, hypermethylation of host genes in warts may not necessarily lead to gene silencing but could, in fact, result in increased gene expression (Moore et al., 2013).
In the present study, we investigated DNA methylation in HPV-induced common warts across entire gene regions, allowing us to identify a set of aberrantly methylated genes within different functional categories. Previously, we investigated the impact of wart formation on host DNA methylation in separate regulatory regions of the gene, including CpG islands (Al-Eitan et al., 2019a), promoters (Al-Eitan et al., 2020a), tiling regions (Al-Eitan et al., 2020b), and CpG sites (Al-Eitan et al., 2020c). Moreover, we have also reported a unique set of differentially expressed genes (Al-Eitan et al., 2020d) and microRNAs in common warts (Al-Eitan et al., 2019b). Each of the previously published works utilized a different methodological approach and focused on a specific gene region, yielding distinct and novel results. The function of a substantial number of the identified DM genes is yet to be elucidated upon in the scientific literature. To mitigate this issue, the identified genes were matched to their gene categories and, if applicable, their RNA classes to provide a holistic view of their potential roles in wart development.
4.1. Methylation status of long non-coding RNA (lncRNA) genes
The vast majority of the human genome is transcribed into long non-coding RNAs (lncRNAs), the latter of which comprise a diverse class of RNA molecules with transcript lengths exceeding 200 nucleotides (Yao et al., 2019). lncRNAs have been implicated as important players in epigenetic regulation, as they can act to directly or indirectly modulate gene methylation at CpG dinucleotides (Kung et al., 2013, Zhao et al., 2016). Moreover, lncRNAs play an integral role in regulating the biological processes of the skin, including those pertaining to keratinocyte differentiation (Kretz et al., 2013, Kretz et al., 2012, Li et al., 2017, Tang et al., 2020). In the context of HPV, dysregulation of certain lncRNAs has been found to be involved in HPV-associated cervical and squamous cell carcinomas (Kretz et al., 2013, Wang et al., 2018a, Wang et al., 2018b, Wang et al., 2018c). The findings of the present study showed that the AC005884, AL049646.2, AC126121.2, AP001790.1, and AC107959.3 lncRNA genes were among the topmost DM genes in warts compared to normal skin. However, only the AC107959.3 gene has been previously reported to be differentially expressed in hepatitis virus positive hepatocellular carcinoma (Huang et al., 2019).
4.2. Methylation status of microRNA (miRNA) genes
MicroRNAs (miRNAs) are a highly conserved group of short ncRNA that are significantly involved in the regulation of several biological processes, including the post-transcriptional regulation of gene expression (O’Brien et al., 2018). In the skin, miRNA biogenesis plays an important part in the development and differentiation of skin stem cells, and certain miRNAs have a significant impact on inflammatory disorders of the skin, including psoriasis and cancer (Singhvi et al., 2018). In the present study, MIR374B, MIR596, MIR1255B1, MIR26B, and MIR196A2 were found to be among the topmost DM genes in warts compared to normal skin. The MIR374B gene, the most differentially methylated miRNA and also known as hsa-MiR-374b-3p or hsa-MiR-374b-5p, promotes cell proliferation and aberrant glycosylation by targeting the phosphatase and tensin (PTEN) and Cosmc genes, respectively (Hu et al., 2015, Long et al., 2018). Additionally, MIR374B was reported to target the reversion inducing cysteine rich protein with Kazal motifs (RECK) and zinc finger E-box-binding homeobox 2 (ZEB2) genes in order to inhibit cellular migration and invasion in bladder cancer (Wang et al., 2018a, Wang et al., 2018b, Wang et al., 2018c). Dysregulation has been associated with several cancers as well as their degree of cellular chemoresistance (Sun et al., 2018). In healthy skin, MIR374B expression was found to be significantly downregulated in Han Chinese individuals compared to Uyghurs, suggesting that ethnicity plays a role in the miRNA profile of the skin (Wu et al., 2018).
4.3. Methylation status of small nucleolar RNA (snoRNA) genes
Found in all eukaryotes, the small nucleolar RNAs (snoRNAs) are a conserved group of ncRNA that are integral to the chemical modification and processing of other types of RNA, including small nuclear RNAs (snRNAs), ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs). snoRNAs can be divided into two main classes: the methylation-associated box C/D snoRNAs and the pseudouridylation-associated H/ACA box snoRNAs (Scott and Ono, 2011). Box C/D snoRNAs guide the 2′-O-methylation, a post-transcriptional modification, of the RNA ribose sugar, and altered expression of these snoRNAs have been observed in a number of different cancers and genetic diseases (Deogharia and Majumder, 2019). In the current study, the small nucleolar RNA, C/D Box 114–29 (SNORD114-29) gene was the third most hypomethylated gene in warts compared to normal skin. SNORD114-29, along with other members of the SNORD114 family, was found to be similarly hypomethylated in smoking-associated lung cancer tissue (Molina-Pinelo et al., 2018). Additionally, the SNORD114-22, SNORD70, and SNORD114-31 genes were similarly hypomethylated in warts compared to normal skin.
4.4. Methylation status of pseudogenes
Found abundantly throughout the genome, pseudogenes are non-functional segments of DNA that possess a high degree of homology to a protein-coding parent gene (Mighell et al., 2000). Although their function is yet to be definitively determined, pseudogenes have been found to be aberrantly methylated or deleted in a number of diseases and especially in human cancers (Poliseno, 2012, Poliseno et al., 2011, Poliseno et al., 2010, Yu et al., 2014). Pseudogenes have long been thought to be ‘junk’ DNA, but an increasing amount of research is showing that pseudogenes have the potential to regulate their parent genes (An et al., 2017, Pink et al., 2011). Our findings show that the AC069366.1, RNU4ATAC11P, and AC120057.1 pseudogenes were hypermethylated in warts, while the NANOGP3, AC106038.2, TPT1P2, SDC4P, PKMP3, and VN2R3P were hypomethylated. Table 2 shows the functions of the parent genes of the pseudogenes identified in this study. Many of the pseudogenes identified in this study were not previously reported to have relevant associations with disease. The AC069366.1 and AC120057.1 promoter regions were found to be hypermethylated and hypomethylated in HPV-induced warts, respectively (Al-Eitan et al., 2020a). In addition, aberrant promoter hypomethylation of SDC4P was found to be associated with survival in hepatocellular carcinoma patients, and it was similarly hypomethylated in skin affected by atopic dermatitis (Olisova et al., 2020, Zhong and Cen, 2017).
Table 2.
Functions of the parent genes for the most differentially methylated pseudogenes in warts.
| Pseudogene | Parent gene | Function of parent gene | Study |
|---|---|---|---|
| AC069366.1 | MSANTD3 | Novel putative human oncogene | (Barasch et al., 2017) |
| RNU4ATAC11P | RNU4ATAC | One of 5 components of the minor spliceosome | (Krøigård et al., 2016) |
| AC120057.1 | CRK | Integrates signals from a diverse array of sources to effect cellular tyrosine phosphorylation activity | (Birge et al., 2009) |
| NANOGP3 | NANOG | Critical factor for maintaining lack of differentiation in pluripotent cells | (Gawlik-Rzemieniewska and Bednarek, 2016) |
| AC106038.2 | PRELID3B | Regulates lipid accumulation on mitochondria | (Miliara et al., 2019) |
| TPT1P2 | TPT1 | Controls cell growth, proliferation and metabolism and is overexpressed in several types of human cancer | (Bae et al., 2017) |
| SDC4P | SDC4 | Plays a critical role in dendritic cell motility | (Polte et al., 2015) |
| PKMP3 | PKM | Key enzyme in glycolysis | (Zhang et al., 2019) |
| VN2R3P | VN2R | Putative pheromone receptor | (Rodriguez and Mombaerts, 2002) |
4.5. Methylation status of protein-coding genes
In warts, the amphiregulin (AREG), gap junction beta-2 protein (GJB2), chromosome 12 open reading frame 71 (C12orf71), AC020909.2, S100 calcium-binding protein A8 (S100A8), and zinc finger BED-type containing 2 (ZBED2) were hypomethylated in warts compared to normal skin, while the fatty acid binding protein 7 (FABP7) and cysteinyl leukotriene receptor 1 (CYSLTR1) genes were hypermethylated.
The AREG gene encodes the amphiregulin protein, which is an epidermal growth factor and a ligand of the epidermal growth factor receptor (Berasain and Avila, 2014). By far, amphiregulin was reported to be the most abundant epidermal growth factor receptor in cultured human keratinocytes (Stoll et al., 2010). Dysregulated AREG expression has been associated with hyperproliferative skin diseases, including actinic keratoses, psoriasis, and graft-versus-host disease (Bhagavathula et al., 2005, Holtan et al., 2018, Piepkorn, 1996). Furthermore, AREG hypomethylation was found to increase its expression in colorectal carcinoma (Lee et al., 2016). In an HPV context, AREG mRNA expression was reported to be lower in HPV-positive head-and-neck squamous cell carcinomas compared to those that were HPV-negative (Gao et al., 2016).
In the current study, the S100 calcium binding protein A8 (S100A8) and A9 (S100A9) genes were found to be hypomethylated in warts compared to normal skin. Additionally, GO enrichment analysis illustrated that the S100A8 and S100A9 proteins were involved in the Toll-like receptor 4 (TLR-4) pathway. The S100A8 and S100A9 proteins are expressed by neutrophils, monocytes, and macrophages during chronic inflammation as part of the calprotectin complex, the latter of which is secreted by cells during inflammatory stress (Pedersen et al., 2014). S100A8 expression plays an integral role in keratinocyte growth, proliferation, and response to wounds (Kerkhoff et al., 2012). Dysregulated S100A8 expression was reported to contribute to skin barrier dysfunction and squamous cell carcinoma development, and it was significantly hypomethylated in hepatocellular carcinomas as well (Funk et al., 2015, Iotzova-Weiss et al., 2015, Khammanivong et al., 2016, Liu et al., 2016, Wang et al., 2018a, Wang et al., 2018b, Wang et al., 2018c). In fact, expression of the calprotectin proteins was found to be highly upregulated in human psoriatic epidermis (Schonthaler et al., 2013). Lastly, global expression profiling of a stable HPV6B E7-transfected cell line found that S100A8 was among the most significantly enhanced genes (Zhang et al., 2018).
Mutations in the GJB2 gene, which encodes for the gap junction protein connexin 26, have been reported to be the cause of several syndromic skin diseases involving hyperkeratotic skin lesions (Iossa et al., 2012). In an HPV context, GJB2 was found to be downregulated in certain HPV-positive cancer sub-groups (Pyeon et al., 2007). On a similar note, the ZBED2 gene was recently revealed to be a modifier of epithelial lineage and an interferon inhibitor, and it was found to be significantly downregulated in HPV-positive cervical cancer in a Hong Kong population (Somerville et al., 2019, Wong et al., 2006). However, the FABP7 gene, which plays a role in fatty acid metabolism, transport, and uptake, was not previously reported to be associated with HPV infection, but it has been found to be involved in skin malignancies (Slipicevic et al., 2008).
Lastly, the CYSLTR1 gene, a G-protein coupled receptor, has been implicated in a number of autoimmune disorders (Jiang et al., 2007, Sokolowska et al., 2009). This gene is significantly expressed within the normal skin epidermis with an even higher expression in atopic eczema (Arriba-Méndez et al., 2008, Hussain et al., 2004). In fact, the CYSLTR1 gene plays an important role in allergic skin inflammation, namely via its involvement in hyperkeratosis and fibrosis (Oyoshi et al., 2012). Additionally, one of the most differentially methylated promoters in HPV-induced warts was found within the CYSLTR1 gene (Al-Eitan et al., 2020a).
4.6. Pathway analysis
The most common regulators of the 1000 most DM genes were revealed to be the STAT5A, RARA, MEF2D, MAP3K8, and THRA genes. The signal transducer and activator of transcription 5A (STAT5A) gene facilitates the cellular response to cytokines and hormones, thus regulating functions related to cell growth and proliferation as well as the immune and nervous systems (Kanai et al., 2014). STAT5A phosphorylation has been previously implicated in the promotion of HPV replication as well as high-risk HPV-mediated cervical cancer (Hong and Laimins, 2013, Sobti et al., 2010). Similarly, the retinoic acid receptor alpha (RARA) gene, which encodes for a transcription factor, was previously found to be downregulated in HPV-16-infected cell lines (Agarwal et al., 1996). In contrast, the mitogen-activated protein kinase kinase kinase 8 (MAP3K8) gene is a proto-oncogene that was reported to be over-expressed in HPV-positive oropharyngeal squamous cell carcinoma (Saba et al., 2015). However, another study found that MAP3K8 was upregulated in primary human keratinocytes but downregulated in HPV- human immortalized keratinocytes (De Schutter et al., 2013).
In a study previously published by our research group, signaling network analysis identified five different common regulators, namely AXIN1, GNB1, GRB2, NTRK1, and SKI, associated with the top DM CpG islands (Al-Eitan et al., 2019a). Although the same case cohort was used, the pathway analysis in the present study identified different common regulators due to the difference in approach to study the epigenetic modification in warts. Such differences point towards the magnitude of epigenetic changes that are initiated as a result of infection with HPV within the different regions of the gene. Our research potentially indicates that DNA methylation of CpG islands as well as non-CpG regions is involved in the etiology and development of HPV-induced common warts (Fuso, 2018, Han et al., 2011, Jang et al., 2017).
5. Conclusions
In the present study, the methylation status of HPV-induced warts was investigated on a genome-wide scale. To the best of the authors’ knowledge, this is the first study to determine the effects of low-risk HPV infection in the context of cutaneous warts on host cell methylation. Our findings might suggest that the genome of HPV-infected host cells is regulated by epigenetic mechanisms that contribute to wart pathogenesis and development. Although we may conclude that they arise during the wart formation process, it remains to be determined whether such epigenetic mechanisms are induced by HPV itself or by the host cell's response to infection.
Funding
This work was supported by the Deanship of Research at Jordan University of Science and Technology under grant number (Ref # 177/2017).
Declaration of Competing Interest
The authors declared that there is no conflict of interest.
Acknowledgments
The authors are grateful to all the participants of this study for their invaluable contribution. The authors would like also to express their gratitude to King Khalid University, Saudi Arabia, for providing administrative and technical support.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2020.10.050.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agarwal C., Chandraratna R.A.S., Teng M., Nagpal S., Rorke E.A., Eckert R.L. Differential regulation of human ectocervical epithelial cell line proliferation and differentiation by retinoid X receptor-and retinoic acid receptor-specific retinoids. Cell Growth Differ. 1996;7:521–530. [PubMed] [Google Scholar]
- Al-Eitan L.N., Alghamdi M.A., Tarkhan A.H., Al-Qarqaz F.A. Epigenome-wide analysis of common warts reveals aberrant promoter methylation. Int. J. Med. Sci. 2020;17:191–206. doi: 10.7150/ijms.39261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Alghamdi M.A., Tarkhan A.H., Al-Qarqaz F.A. Genome-wide tiling array analysis of HPV-induced warts reveals aberrant methylation of protein-coding and non-coding regions. Genes (Basel) 2020;11 doi: 10.3390/genes11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Alghamdi M.A., Tarkhan A.H., Al-Qarqaz F.A. Genome-wide identification of methylated CpG sites in nongenital cutaneous warts. BMC Med. Genomics. 2020;13:100. doi: 10.1186/s12920-020-00745-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Alghamdi M.A., Tarkhan A.H., Al-Qarqaz F.A. Genome-wide CpG Island methylation profiles of cutaneous skin with and without HPV infection. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Alghamdi M.A., Tarkhan A.H., Al-Qarqaz F.A. Gene Expression Profiling of MicroRNAs in HPV-Induced Warts and Normal Skin. Biomolecules. 2019;9:757. doi: 10.3390/biom9120757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eitan L.N., Tarkhan A.H., Alghamdi M.A., Al-Qarqaz F.A., Al-Kofahi H.S. Transcriptome analysis of HPV-induced warts and healthy skin in humans. BMC Med. Genomics. 2020;13:35. doi: 10.1186/s12920-020-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Aboud A.M., Nigam P.K. StatPearls Publishing; StatPearls: 2018. Wart (Plantar, Verruca Vulgaris, Verrucae) [Google Scholar]
- An Y., Furber K.L., Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J. Cell Mol. Med. 2017 doi: 10.1111/jcmm.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriba-Méndez S., Sanz C., Isidoro-García M., Pascual M., Ávila C., Dávila I., Lorente F. Analysis of 927T > C CYSLTR1 and –444A > C LTC4S polymorphisms in children with asthma. Allergol. Immunopathol. (Madr) 2008;36:259–263. doi: 10.1016/S0301-0546(08)75220-0. [DOI] [PubMed] [Google Scholar]
- Assenov Y., Müller F., Lutsik P., Walter J., Lengauer T., Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj P., Burch D. Human Papillomavirus Infection of the Skin. Arch. Pathol. Lab. Med. 2018;142:700–705. doi: 10.5858/arpa.2017-0572-RA. [DOI] [PubMed] [Google Scholar]
- Bae S.Y., Byun S., Bae S.H., Min D.S., Woo H.A., Lee K. TPT1 (tumor protein, translationally-controlled 1) negatively regulates autophagy through the BECN1 interactome and an MTORC1-mediated pathway. Autophagy. 2017;13:820–833. doi: 10.1080/15548627.2017.1287650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barasch N., Gong X., Kwei K.A., Varma S., Biscocho J., Qu K., Xiao N., Lipsick J.S., Pelham R.J., West R.B., Pollack J.R. Recurrent rearrangements of the Myb/SANTlike DNA-binding domain containing 3 gene (MSANTD3) in salivary gland acinic cell carcinoma. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros S.P., Offenbacher S. Epigenetics: Connecting environment and genotype to phenotype and disease. J. Dent. Res. 2009 doi: 10.1177/0022034509335868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berasain, C., Avila, M.A., 2014. Amphiregulin. Semin. Cell Dev. Biol. https://doi.org/10.1016/j.semcdb.2014.01.005. [DOI] [PubMed]
- Bhagavathula N., Nerusu K.C., Fisher G.J., Liu G., Thakur A.B., Gemmell L., Kumar S., Xu Z.H., Hinton P., Tsurushita N., Landolfi N.F., Voorhees J.J., Varani J. Amphiregulin and epidermal hyperplasia: Amphiregulin is required to maintain the psoriatic phenotype of human skin grafts on severe combined immunodeficient mice. Am. J. Pathol. 2005;166:1009–1016. doi: 10.1016/S0002-9440(10)62322-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge, R.B., Kalodimos, C., Inagaki, F., Tanaka, S., 2009. Crk and CrkL adaptor proteins: Networks for physiological and pathological signaling. Cell Commun. Signal. https://doi.org/10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed]
- Bosch, F.X., Lorincz, A., Muñoz, N., Meijer, C.J.L.M., Shah, K. V., 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. https://doi.org/10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed]
- Boscolo-Rizzo, P., Furlan, C., Lupato, V., Polesel, J., Fratta, E., 2017. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: Role of HPV and lifestyle factors. Clin. Epigenetics. https://doi.org/10.1186/s13148-017-0424-5. [DOI] [PMC free article] [PubMed]
- Bruggink S.C., de Koning M.N.C., Gussekloo J., Egberts P.F., ter Schegget J., Feltkamp M.C.W., Bavinck J.N.B., Quint W.G.V., Assendelft W.J.J., Eekhof J.A.H. Cutaneous wart-associated HPV types: Prevalence and relation with patient characteristics. J. Clin. Virol. 2012;55:250–255. doi: 10.1016/J.JCV.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Clarke M.A., Gradissimo A., Schiffman M., Lam J., Sollecito C.C., Fetterman B., Lorey T., Poitras N., Raine-Bennett T.R., Castle P.E., Wentzensen N., Burk R.D. Human Papillomavirus DNA Methylation as a Biomarker for Cervical Precancer: Consistency across 12 Genotypes and Potential Impact on Management of HPV-Positive Women. Clin. Cancer Res. 2018;24:2194–2202. doi: 10.1158/1078-0432.CCR-17-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, M.A., Wentzensen, N., Mirabello, L., Ghosh, A., Wacholder, S., Harari, A., Lorincz, A., Schiffman, M., Burk, R.D., 2012. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomarkers Prev. https://doi.org/10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed]
- Dankai W., Khunamornpong S., Siriaunkgul S., Soongkhaw A., Janpanao A., Utaipat U., Kitkumthorn N., Mutirangura A., Srisomboon J., Lekawanvijit S. Role of genomic DNA methylation in detection of cytologic and histologic abnormalities in high risk HPV-infected women. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0210289. e0210289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter T., Andrei G., Topalis D., Naesens L., Snoeck R. Cidofovir selectivity is based on the different response of normal and cancer cells to DNA damage. BMC Med. Genomics. 2013;6:1–17. doi: 10.1186/1755-8794-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deogharia M., Majumder M. Guide snoRNAs: Drivers or passengers in human disease? Biology (Basel) 2019 doi: 10.3390/biology8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durzynska, J., Lesniewicz, K., Poreba, E., 2017. Human papillomaviruses in epigenetic regulations. Mutat. Res. - Rev. Mutat. Res. https://doi.org/10.1016/j.mrrev.2016.09.006. [DOI] [PubMed]
- Egawa N., Egawa K., Griffin H., Doorbar J. Human papillomaviruses; Epithelial tropisms, and the development of neoplasia. Viruses. 2015 doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Dong J., Chang W., Cui M., Xu T. The Progress of Methylation Regulation in Gene Expression of Cervical Cancer. Int. J. Genomics. 2018;2018 doi: 10.1155/2018/8260652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk S., Mark R., Bayo P., Flechtenmacher C., Grabe N., Angel P., Plinkert P.K., Hess J. High S100A8 and S100A12 protein expression is a favorable prognostic factor for survival of oropharyngeal squamous cell carcinoma. Int. J. Cancer. 2015;136:2037–2046. doi: 10.1002/ijc.29262. [DOI] [PubMed] [Google Scholar]
- Fuso A. Non-CpG Methylation Revised. Epigenomes. 2018;2:22. doi: 10.3390/epigenomes2040022. [DOI] [Google Scholar]
- Gao J., Ulekleiv C.H., Halstensen T.S. Epidermal growth factor (EGF) receptor-ligand based molecular staging predicts prognosis in head and neck squamous cell carcinoma partly due to deregulated EGF- induced amphiregulin expression. J. Exp. Clin. Cancer Res. 2016;35 doi: 10.1186/s13046-016-0422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik-Rzemieniewska, N., Bednarek, I., 2016. The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol. Ther. https://doi.org/10.1080/15384047.2015.1121348. [DOI] [PMC free article] [PubMed]
- Graham S.V. Keratinocyte Differentiation-Dependent Human Papillomavirus Gene Regulation. Viruses. 2017;9 doi: 10.3390/v9090245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett, J.A., Azim Surani, M., 2013. DNA methylation dynamics during the mammalian life cycle. Philos. Trans. R. Soc. B Biol. Sci. https://doi.org/10.1098/rstb.2011.0328 [DOI] [PMC free article] [PubMed]
- Han H. NA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter. Hum. Mol. Genet. 2011;20:4299–4310. doi: 10.1093/hmg/ddr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan S.G., DeFor T.E., Panoskaltsis-Mortari A., Khera N., Levine J.E., Flowers M.E.D., Lee S.J., Inamoto Y., Chen G.L., Mayer S., Arora M., Palmer J., Cutler C.S., Arai S., Lazaryan A., Newell L.F., Jagasia M.H., Pusic I., Wood W.A., Renteria A.S., Yanik G., Hogan W.J., Hexner E., Ayuk F., Holler E., Bunworasate U., Efebera Y.A., Ferrara J.L.M., Pidala J., Howard A., Wu J., Bolaños-Meade J., Ho V., Alousi A., Blazar B.R., Weisdorf D.J., MacMillan M.L. Amphiregulin modifies the Minnesota acute graft-versus-host disease risk score: Results from BMT CTN 0302/0802. Blood Adv. 2018;2:1882–1888. doi: 10.1182/bloodadvances.2018017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Laimins L.A. The JAK-STAT Transcriptional Regulator, STAT-5, Activates the ATM DNA Damage Pathway to Induce HPV 31 Genome Amplification upon Epithelial Differentiation. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.A., Boulet G.A., Renoux V.M., Delvenne P.O., Bogers J.P.J. Mechanisms of cell entry by human papillomaviruses: An overview. Virol. J. 2010 doi: 10.1186/1743-422X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlihan C.F., Baisley K., Bravo I.G., Pavón M.A., Changalucha J., Kapiga S., De Sanjosé S., Ross D.A., Hayes R.J., Watson-Jones D. Human papillomavirus DNA detected in fingertip, oral and bathroom samples from unvaccinated adolescent girls in Tanzania. Sex. Transm. Infect. 2019;95:374–379. doi: 10.1136/sextrans-2018-053756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Bao H., Xu X., Zhou X., Qin W., Zeng C., Liu Z. Increased MIR-374b promotes cell proliferation and the production of aberrant glycosylated IgA1 in B cells of IgA nephropathy. FEBS Lett. 2015;589:4019–4025. doi: 10.1016/j.febslet.2015.10.033. [DOI] [PubMed] [Google Scholar]
- Huang Z.L., Li W., Chen Q.F., Wu P.H., Shen L.J. Eight key long non-coding RNAs predict hepatitis virus positive hepatocellular carcinoma as prognostic targets. World J. Gastrointest. Oncol. 2019;11:983–997. doi: 10.4251/wjgo.v11.i11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I., Kitagaki K., Businga T.R., Kline J.N. Expression of cysteinyl leukotriene receptor-1 in skin. J. Am. Acad. Dermatol. 2004;51:1032–1033. doi: 10.1016/j.jaad.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Iossa S., Marciano E., Franze A. GJB2 Gene Mutations in Syndromic Skin Diseases with Sensorineural Hearing Loss. Curr. Genomics. 2012;12:475–485. doi: 10.2174/138920211797904098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iotzova-Weiss G., Dziunycz P.J., Freiberger S.N., Läuchli S., Hafner J., Vogl T., French L.E., Hofbauer G.F.L. S100A8/A9 Stimulates Keratinocyte Proliferation in the Development of Squamous Cell Carcinoma of the Skin via the Receptor for Advanced Glycation-End Products. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0120971. e0120971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabłońska S., Majewski S., Obalek S., Orth G. Cutaneous warts. Clin. Dermatol. 1997;15:309–319. doi: 10.1016/S0738-081X(96)00170-8. [DOI] [PubMed] [Google Scholar]
- Jang H.S., Shin W.J., Lee J.E., Do J.T. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes (Basel). 2017;8 doi: 10.3390/genes8060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Borrelli L.A., Kanaoka Y., Bacskai B.J., Boyce J.A. CysLT2 receptors interact with CysLT1 receptors and down-modulate cysteinyl leukotriene dependent mitogenic responses of mast cells. Blood. 2007;110:3263–3270. doi: 10.1182/blood-2007-07-100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T., Seki S., Jenks J.A., Kohli A., Kawli T., Martin D.P., Snyder M., Bacchetta R., Nadeau K.C. Identification of STAT5A and STAT5B Target Genes in Human T Cells. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0086790. e86790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff C., Voss A., Scholzen T.E., Averill M.M., Zänker K.S., Bornfeldt K.E. Novel insights into the role of S100A8/A9 in skin biology. Exp. Dermatol. 2012;21:822–826. doi: 10.1111/j.1600-0625.2012.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khammanivong A., Sorenson B.S., Ross K.F., Dickerson E.B., Hasina R., Lingen M.W., Herzberg M.C. Involvement of calprotectin (S100A8/A9) in molecular pathways associated with HNSCC. Oncotarget. 2016;7:14029–14047. doi: 10.18632/oncotarget.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J., Johnston D., Kim G.E., Spitale R.C., Flynn R.A., Zheng G.X.Y., Aiyer S., Raj A., Rinn J.L., Chang H.Y., Khavari P.A. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M., Webster D.E., Flockhart R.J., Lee C.S., Zehnder A., Lopez-Pajares V., Qu K., Zheng G.X.Y., Chow J., Kim G.E., Rinn J.L., Chang H.Y., Siprashvili Z., Khavari P.A. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krøigård A.B., Frost M., Larsen M.J., Ousager L.B., Frederiksen A.L. Bone structure in two adult subjects with impaired minor spliceosome function resulting from RNU4ATAC mutations causing microcephalic osteodysplastic primordial dwarfism type 1 (MOPD1) Bone. 2016;92:145–149. doi: 10.1016/j.bone.2016.08.023. [DOI] [PubMed] [Google Scholar]
- Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: Past, present, and future. Genetics. 2013 doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., McGuffey E.J., Morris J.S., Manyam G., Baladandayuthapani V., Wei W., Morris V.K., Overman M.J., Maru D.M., Jiang Z.Q., Hamilton S.R., Kopetz S. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br. J. Cancer. 2016;114:1352–1361. doi: 10.1038/bjc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Xu W.W., Han L., Chan K.T., Tsao S.W., Lee N.P.Y., Law S., Xu L.Y., Li E.M., Chan K.W., Qin Y.R., Guan X.Y., He Q.Y., Cheung A.L.M. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.H.K., Maher E.R. DNA methylation: a form of epigenetic control of gene expression. Obstet. Gynaecol. 2010;12:37–42. doi: 10.1576/toag.12.1.037.27556. [DOI] [Google Scholar]
- Liu K., Zhang Y., Zhang C., Zhang Q., Li J., Xiao F., Li Y., Zhang R., Dou D., Liang J., Qin J., Lin Z., Zhao D., Jiang M., Liang Z., Su J., Gupta V.P., He M., Yang X. Methylation of S100A8 is a promising diagnosis and prognostic marker in hepatocellular carcinoma. Oncotarget. 2016;7:56798–56810. doi: 10.18632/oncotarget.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Z.W. MiR-374b promotes proliferation and inhibits apoptosis of human GIST cells by inhibiting PTEN through activation of the PI3K/Akt pathway. Mol. Cells. 2018;41:532–544. doi: 10.14348/molcells.2018.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marongiu L., Godi A., Parry J.V., Beddows S. Human Papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer. 2014;14:384. doi: 10.1186/1471-2407-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmurray H.R., Nguyen D., Westbrook T.F., Mcance D.J. Biology of human papillomaviruses. Int. J. Exp. Pathol. 2001;82:15–33. doi: 10.1046/j.1365-2613.2001.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mervis, J.S., McGee, J.S., 2020. DNA methylation and inflammatory skin diseases. Arch. Dermatol. Res. https://doi.org/10.1007/s00403-019-02005-9. [DOI] [PubMed]
- Mighell A.J., Smith N.R., Robinson P.A., Markham A.F. Vertebrate pseudogenes. FEBS Lett. 2000 doi: 10.1016/S0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- Milavetz, B.I., Balakrishnan, L., 2015. Viral Epigenetics, in: Methods in Molecular Biology (Clifton, N.J.). pp. 569–596. https://doi.org/10.1007/978-1-4939-1804-1_30. [DOI] [PMC free article] [PubMed]
- Miliara X., Tatsuta T., Berry J.L., Rouse S.L., Solak K., Chorev D.S., Wu D., Robinson C.V., Matthews S., Langer T. Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins. Nat. Commun. 2019;10:1–15. doi: 10.1038/s41467-019-09089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Pinelo S., Salinas A., Moreno-Mata N., Ferrer I., Suarez R., Andrés-León E., Rodríguez-Paredes M., Gutekunst J., Jantus-Lewintre E., Camps C., Carnero A., Paz-Ares L. Impact of DLK1-DIO3 imprinted cluster hypomethylation in smoker patients with lung cancer. Oncotarget. 2018;9:4395–4410. doi: 10.18632/oncotarget.10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013 doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhem E., Pinelis S. Treatment of nongenital cutaneous warts. Am. Fam. Physician. 2011;84:288–293. [PubMed] [Google Scholar]
- Münger K., Baldwin A., Edwards K.M., Hayakawa H., Nguyen C.L., Owens M., Grace M., Huh K. Mechanisms of Human Papillomavirus-Induced Oncogenesis. J. Virol. 2004;78:11451–11460. doi: 10.1128/jvi.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 2018 doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olisova O.Y., Kochergin N.G., Kayumova L.N., Zavarykina T.M., Dmitriev A.A., Asanov A.Y. Skin DNA methylation profile in atopic dermatitis patients: A case–control study. Exp. Dermatol. 2020;29:184–189. doi: 10.1111/exd.14064. [DOI] [PubMed] [Google Scholar]
- Oyoshi M.K., He R., Kanaoka Y., ElKhal A., Kawamoto S., Lewis C.N., Austen K.F., Geha R.S. Eosinophil-derived leukotriene C4 signals via type 2 cysteinyl leukotriene receptor to promote skin fibrosis in a mouse model of atopic dermatitis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4992–4997. doi: 10.1073/pnas.1203127109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L., Nybo M., Poulsen M.K., Henriksen J.E., Dahl J., Rasmussen L.M. Plasma calprotectin and its association with cardiovascular disease manifestations, obesity and the metabolic syndrome in type 2 diabetes mellitus patients. BMC Cardiovasc. Disord. 2014;14 doi: 10.1186/1471-2261-14-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetto L., Briganti L., Calderone A., Cerquone Perpetuini A., Iannuccelli M., Langone F., Licata L., Marinkovic M., Mattioni A., Pavlidou T., Peluso D., Petrilli L.L., Pirrò S., Posca D., Santonico E., Silvestri A., Spada F., Castagnoli L., Cesareni G. SIGNOR: a database of causal relationships between biological entities. Nucleic Acids Res. 2016;44:D548–D554. doi: 10.1093/nar/gkv1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepkorn M. Overexpression of amphiregulin, a major autocrine growth factor for cultured human keratinocytes, in hyperproliferative skin diseases. Am. J. Dermatopathol. 1996;18:165–171. doi: 10.1097/00000372-199604000-00010. [DOI] [PubMed] [Google Scholar]
- Pink R.C., Wicks K., Caley D.P., Punch E.K., Jacobs L., Carter D.R.F. Pseudogenes: Pseudo-functional or key regulators in health and diseasě. RNA. 2011 doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno, L., 2012. Pseudogenes: Newly discovered players in human cancer. Sci. Signal. https://doi.org/10.1126/scisignal.2002858 [DOI] [PubMed]
- Poliseno L., Haimovic A., Christos P.J., Saenz Vega Y, De Miera E.C., Shapiro R., Pavlick A., Berman R.S., Darvishian F., Osman I. Deletion of PTENP1 pseudogene in human melanoma. J, Invest. Dermatol. 2011 doi: 10.1038/jid.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polte T., Petzold S., Bertrand J., Schütze N., Hinz D., Simon J.C., Lehmann I., Echtermeyer F., Pap T., Averbeck M. Critical role for syndecan-4 in dendritic cell migration during development of allergic airway inflammation. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms8554. [DOI] [PubMed] [Google Scholar]
- Pyeon D., Newton M.A., Lambert P.F., Den Boon J.A., Sengupta S., Marsit C.J., Woodworth C.D., Connor J.P., Haugen T.H., Smith E.M., Kelsey K.T., Turek L.P., Ahlquist P. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, I., Mombaerts, P., 2002. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr. Biol. https://doi.org/10.1016/S0960-9822(02)00909-0. [DOI] [PubMed]
- Saba N.F., Wilson M., Doho G., DaSilva J., Benjamin Isett R., Newman S., Chen Z.G., Magliocca K., Rossi M.R. Mutation and Transcriptional Profiling of Formalin-Fixed Paraffin Embedded Specimens as Companion Methods to Immunohistochemistry for Determining Therapeutic Targets in Oropharyngeal Squamous Cell Carcinoma (OPSCC): A Pilot of Proof of Principle. Head Neck Pathol. 2015;9:223–235. doi: 10.1007/s12105-014-0566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler H.B., Guinea-Viniegra J., Wculek S.K., Ruppen I., Ximénez-Embún P., Guío-Carrión A., Navarro R., Hogg N., Ashman K., Wagner E.F. S100A8-S100A9 Protein Complex Mediates Psoriasis by Regulating the Expression of Complement Factor C3. Immunity. 2013;39:1171–1181. doi: 10.1016/j.immuni.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Scott M.S., Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011 doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapati, R., Senapati, N.N., Dwibedi, B., 2016. Molecular mechanisms of HPV mediated neoplastic progression. Infect. Agent. Cancer. https://doi.org/10.1186/s13027-016-0107-4. [DOI] [PMC free article] [PubMed]
- Singhvi, G., Manchanda, P., Krishna Rapalli, V., Kumar Dubey, S., Gupta, G., Dua, K., 2018. MicroRNAs as biological regulators in skin disorders. Biomed. Pharmacother. https://doi.org/10.1016/j.biopha.2018.09.090. [DOI] [PubMed]
- Slipicevic A., Jørgensen K., Skrede M., Rosnes A.K.R., Trøen G., Davidson B., Flørenes V.A. The fatty acid binding protein 7 (FABP7) is involved in proliferation and invasion of melanoma cells. BMC Cancer. 2008;8:276. doi: 10.1186/1471-2407-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., Sen S., Weeks R.J., Eccles M.R., Chatterjee A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends in Cancer. 2020 doi: 10.1016/j.trecan.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Smith, Z.D., Meissner, A., 2013. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. https://doi.org/10.1038/nrg3354. [DOI] [PubMed]
- Sobti R.C., Singh N., Hussain S., Suri V., Bharadwaj M., Das B.C. Deregulation of STAT-5 isoforms in the development of HPV-mediated cervical carcinogenesis. J. Recept. Signal Transduct. 2010;30:178–188. doi: 10.3109/10799891003786218. [DOI] [PubMed] [Google Scholar]
- Sokolowska M., Wodz-Naskiewicz K., Cieslak M., Seta K., Bednarek A.K., Pawliczak R. Variable expression of cysteinyl leukotriene type I receptor splice variants in asthmatic females with different promoter haplotypes. BMC Immunol. 2009;10:63. doi: 10.1186/1471-2172-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, T., Xu, Y., Wu, X., Vakoc, C., 2019. ZBED2 is an antagonist of Interferon Regulatory Factor 1 and modulates cell identity in pancreatic cancer. https://doi.org/10.1101/868141. [DOI] [PMC free article] [PubMed]
- Soto D., Song C., McLaughlin-Drubin M.E. Epigenetic alterations in human papillomavirus- associated cancers. Viruses. 2017 doi: 10.3390/v9090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S.W., Johnson J.L., Bhasin A., Johnston A., Gudjonsson J.E., Rittié L., Elder J.T. Metalloproteinase-mediated, context-dependent function of amphiregulin and hb-egf in human keratinocytes and skin. J, Invest. Dermatol. 2010;130:295–304. doi: 10.1038/jid.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Wang X., Sui G., Chen S., Yu M., Zhang P. Downregulation of miR-374b-5p promotes chemotherapeutic resistance in pancreatic cancer by upregulating multiple anti-apoptotic proteins. Int. J. Oncol. 2018;52:1491–1503. doi: 10.3892/ijo.2018.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Liang Y., Xie H., Yang X., Zheng G. Long non-coding RNAs in cutaneous biology and proliferative skin diseases: Advances and perspectives. Cell Prolif. 2020;53 doi: 10.1111/cpr.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaat W., Van Leeuwen R.W., Novianti P.W., Schuuring E., Meijer C.J.L.M., Van Der Zee A.G.J., Snijders P.J.F., Heideman D.A.M., Steenbergen R.D.M., Wisman G.B.A. Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics. 2018;13:769–778. doi: 10.1080/15592294.2018.1507197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Knebel Doeberitz M., Prigge E.-S. Role of DNA methylation in HPV associated lesions. Papillomavirus Res. 2019;7:180–183. doi: 10.1016/J.PVR.2019.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Suogang, Zhang G., Zheng W., Xue Q., Wei D., Zheng Y., Yuan J. MiR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci. Rep. 2018;38 doi: 10.1042/BSR20181436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Siwen, Song, R., Wang, Z., Jing, Z., Wang, Shaoxiong, Ma, J., 2018. S100A8/A9 in inflammation. Front. Immunol. https://doi.org/10.3389/fimmu.2018.01298.
- Wang X., Wang G., Zhang L., Cong J., Hou J., Liu C. LncRNA PVT1 promotes the growth of HPV positive and negative cervical squamous cell carcinoma by inhibiting TGF-β1. Cancer Cell Int. 2018;18 doi: 10.1186/s12935-018-0567-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.-F., Cheung T.-H., Tsao G.S.W., Lo K.W.K., Yim S.-F., Wang V.W., Heung M.M.S., Chan S.C.S., Chan L.K.Y., Ho T.W.F., Wong K.W.Y., Li C., Guo Y., Chung T.K.H., Smith D.I. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int. J. Cancer. 2006;118:2461–2469. doi: 10.1002/ijc.21660. [DOI] [PubMed] [Google Scholar]
- Wu X., Zhao Z., Ding Y., Xiang F., Kang X., Pu X. Differential expression of microRNAs in the normal skin of the Han and Uyghur populations in Xinjiang Province. Med. (United States) 2018;97 doi: 10.1097/MD.0000000000009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R.W., Wang, Y., Chen, L.L., 2019. Cellular functions of long noncoding RNAs. Nat. Cell Biol. https://doi.org/10.1038/s41556-019-0311-8. [DOI] [PubMed]
- Yong W.S., Hsu F.M., Chen P.Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016 doi: 10.1186/s13072-016-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Yao W., Gumireddy K., Li A., Wang J., Xiao W., Chen K., Xiao H., Li H., Tang K., Ye Z., Huang Q., Xu H. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol. Cancer Ther. 2014;13:3086–3097. doi: 10.1158/1535-7163.MCT-14-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Chen X., Zhou Q., Song Y., Sun S., Cheng H. Human gene expression microarray analysis of the HPV 6bE7-HaCaT stable cell line. Gene. 2018;657:60–68. doi: 10.1016/j.gene.2018.02.067. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Deng X., Liu Yahui, Liu Yuanda, Sun L., Chen F. PKM2, function and expression and regulation. Cell Biosci. 2019 doi: 10.1186/s13578-019-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sun H., Wang H. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016 doi: 10.1186/s13578-016-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D., Cen H. Aberrant promoter methylation profiles and association with survival in patients with hepatocellular carcinoma. Onco. Targets. Ther. 2017;10:2501–2509. doi: 10.2147/OTT.S128058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.