Abstract
Background
Genome-wide association studies have convincingly shown that single nucleotide polymorphisms (SNPs) in HFE and TMPRSS6 are associated with iron parameters. It was commonly thought that these associations could be explained by the intermediate effect on hepcidin concentration. A recent study in an isolated Italian population, however, concluded that these associations were not exclusively dependent on hepcidin values. We report here the second study to investigate the role of hepcidin in the associations between common variants in HFE and TMPRSS6 with iron parameters.
Methods
We extracted 101 SNPs in HFE and TMPRSS6 from genome-wide imputed SNP data of 1832 individuals from the general population (Nijmegen Biomedical Study). Single locus and haplotype associations with serum iron parameters and hepcidin were studied using linear regression analyses.
Results
We found that HFE rs1800562 and TMPRSS6 rs855791 are the main determinants of HFE and TMPRSS6 related variation in serum iron, ferritin, transferrin saturation, and total iron binding capacity. These SNPs are associated with the ratios hepcidin/ ferritin (p<1×10−5) and hepcidin/transferrin saturation (p<1×10−3), but not with serum hepcidin (p>0.2). Adjustment for hepcidin or the ratio hepcidin/ferritin did not decrease the strength of the SNP–iron parameter associations.
Conclusions
Our results do not support an intermediate role for hepcidin in the SNP–iron parameter associations, which confirms previous findings, and indicate a pleiotropic SNP effect on the hepcidin ratios and the iron parameters. Taken together, this suggests that there might be other, yet unknown, serum hepcidin independent mechanisms which play a role in the association of HFE and TMPRSS6 variants with serum iron parameters.
INTRODUCTION
Genome-wide association studies (GWAS) have shown that at a population level single nucleotide polymorphisms (SNPs) in the haemochromatosis gene (HFE) and in the transmembrane serine protease 6 gene (TMPRSS6) are associated with ferritin, iron, transferrin, and transferrin saturation (TS) (ie, iron parameters). These associations have been found for the SNPs rs1800562 in HFE (p.Cys282Tyr), rs855791 in TMPRSS6 (p.Ala736Val), and rs4820268 in TMPRSS6 (p.Asp521Asp).1–6 The proteins encoded by HFE and TMPRSS6, Hfe and matriptase 2 (MT2) respectively, have been suggested to play a role in the transcriptional regulation of the hepatic peptide hormone hepcidin, key regulator of iron homeostasis.7–12 It was commonly thought that the reported GWAS associations between the HFE and TMPRSS6 SNPs and iron parameters could be explained by this intermediate effect on hepcidin concentration. However, serum hepcidin concentrations were not measured in these GWAS, preventing a definite evaluation of this assumption. Recently, Traglia and colleagues3 analysed serum hepcidin concentrations, measured by a mass spectrometry based method,13 in 1657 related individuals from the Val Borbera genetic isolate in northern Italy. They explored relationships between hepcidin and a set of anthropometric, haematologic and iron parameters and performed a GWAS for hepcidin. In the same paper, they also focused on the association of two common variants HFE rs1800562 and TMPRSS6 rs855791 with iron, erythrocyte parameters and hepcidin values in 1545 genotyped individuals. They reported that their study allowed them to conclude that associations between these SNPs and iron parameters were not exclusively dependent on hepcidin values. This unexpected finding has not been replicated in other populations yet.
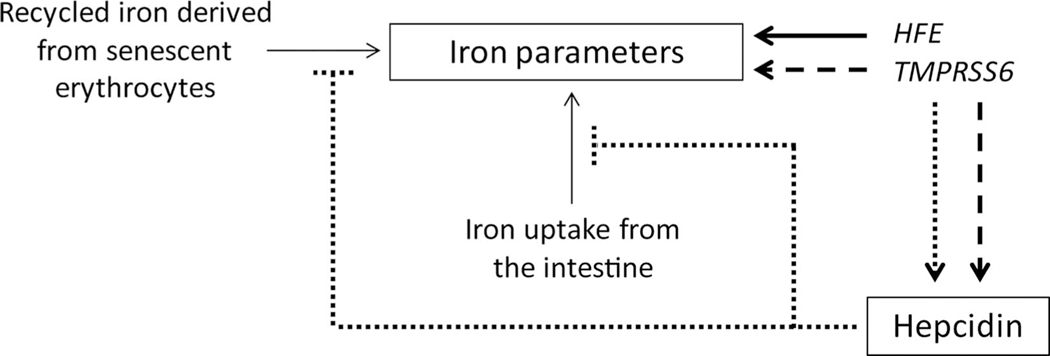
In this study, we aim to evaluate the role of hepcidin in the association between HFE and TMPRSS6 related SNP variation and iron parameters in a second, independent population. We used data from the Nijmegen Biomedical Study (NBS) to analyse the associations between common variants in and surrounding the HFE and TMPRSS6 genes and iron parameters and hepcidin on a population level. The first objective of our study was to replicate the associations previously found in the iron GWAS and to determine which SNPs in both genes are main determinants of iron parameters in our population. These analyses revealed that out of the studied SNPs, HFE rs1800562 and TMPRSS6 rs855791 were most strongly associated with the iron parameters. Secondly, we focused on these SNPs and evaluated the role of serum hepcidin in the associations of HFE and TMPRSS6 variants with iron parameters. We also included ratios of hepcidin to ferritin and TS given the known dependence of hepcidin expression on stored iron and circulating iron, respectively.14–19 We (1) considered hepcidin and its ratio to ferritin as intermediate variables in the association between the SNPs and iron parameters; (2) explored the presence of pleiotropy, that is, whether the SNPs both independently affect hepcidin and the iron parameters; and (3) evaluated the presence of an effect on iron parameters only (figure 1).
Figure 1.
Hypothetical roles of serum hepcidin in the associations of haemochromatosis gene (HFE) and the transmembrane serine protease 6 gene (TMPRSS6) variants with serum iron parameters. (1) Hepcidin and its ratio to ferritin might be intermediate in the association between HFE and TMPRSS6 variants and iron parameters (dotted arrow). (2) The variants might both independently affect hepcidin and the iron parameters (dashed arrows), that is, presence of pleiotropy. (3) The variants may affect the iron parameters only and not hepcidin (bold arrow).
METHODS
Study population
This study was performed in participants from the NBS, a well phenotyped Dutch population based cohort. Details of the NBS have been described before.20 For this study we used the subset of 1832 NBS participants that was selected to serve as controls in GWAS.21 Questionnaire data on age, use of iron supplements, body mass index (BMI), presence of anaemia, and pregnancy, and measurements of hepcidin, iron, ferritin, TS and total iron binding capacity (TIBC), liver enzyme alanine aminotransferase (ALT), creatinine, and C-reactive protein (CRP) were available. The iron parameters were measured as described before.22 Serum hepcidin was measured with an in-house developed and validated competitive enzyme linked immunosorbent assay (ELISA).22,23
Genotyping and selection of SNPs
The NBS participants were genotyped with the Illumina HumanHapCNV370-Duo BeadChip21; density of genetic variants was increased by imputation using the CEU HapMap Phase II data as reference.
Genotype data for the SNPs within the genes HFE and TMPRSS6 and the 10 kB surrounding regions were extracted for the purpose of this study, resulting in the inclusion of 35 SNPs for HFE and 66 SNPs for TMPRSS6 (see online supplementary table S1).
Haplotype analyses were applied to uncover allelic interactions. We included only non-synonymous SNPs and SNPs known to influence expression of HFE or TMPRSS6 based on expression databases (expression quantitative trait loci (eQTL)): the non-synonymous SNPs rs855791 (p.Ala736Val) and rs2235324 (p.Lys253Glu) and the eQTL rs2160906 for TMPRSS6, and the non-synonymous SNPs rs1800562 (p.Cys282Tyr) and rs1799945 (p.His63Asp) and the eQTL rs198853 for HFE.
Statistical analysis
The variables hepcidin, ferritin, and the ratios hepcidin/ferritin and hepcidin/TS were skewed towards higher values and therefore log-transformed to normalise their distributions.eOutliers, defined as values that differed more than three times the SD from the mean, were reduced to mean±3 SD (maximal number of outliers per trait: 26).
Two different subsets were created to investigate whether results for the whole cohort were influenced by extreme values on variables evidently influencing hepcidin concentration. Subset 1 was based on exclusion of persons with CRP >10 mg/L or ferritin <30 μg/L in agreement with the study of Traglia et al.3 Subset 2 was selected based on the same exclusion criteria as previously22: pregnancy at time of blood sampling, ALT >50 U/L, CRP >10 mg/L, estimated glomerular filtration rate (eGFR) <60 mL/min/1.73 m2, use of iron supplements, presence of anaemia, or BMI >30 kg/m2.
Association analyses were performed using Plink V.1.07 (http://pngu.mgh.harvard.edu/purcell/plink/).24 The associations between the SNPs and the traits were evaluated using linear regression analyses based on the genotypic model and adjusted for age, gender, and time of blood sampling, since these variables are independent determinants of serum hepcidin.22 The resulting regression coefficients express the mean change in the independent variable for the heterozygous and minor homozygous genotype or haplotype relative to the reference genotype or haplotype. In case of log-transformation of the independent variables, regression coefficients express the mean change in the log-transformed variable relative to the reference genotype or haplotype.
Due to multiple testing issues, the nominal significance level of 0.05 is not sufficient to maintain an overall study false positive rate of 5%. Application of a Bonferroni correction for the number of tested SNPs (ie, 35+66=101) would lead to a p value threshold of significance of 5×10−4.
A full description of the methods used for this study can be found online (see online supplementary methods).
RESULTS
Characteristics of the study population
The mean age of the 1832 individuals was 62 years. Additional characteristics of the individuals included in the study are shown in table 1. Application of the exclusion criteria resulted in the inclusion of 1505 individuals in subset 1 and 1177 individuals in subset 2 (see online supplementary table S2). Characteristics of the subsets and the whole cohort were similar, except for the proportion of females (51% in the whole cohort vs 47% in subset 1 and 44% in subset 2) and geometric mean hepcidin and ferritin concentration in subset 1 (6.4 nM and 107.2 μg/L in the whole cohort and 7.7 nM and 131.5 μg/L in subset 1, respectively).
Table 1.
Characteristics of the total study population (N=1832)
| N* | % | Mean (SD) | |
|---|---|---|---|
| Gender | |||
| Males | 906 | 49 | NA |
| Females | 926 | 51 | NA |
| Age, years | 1832 | 100 | 61.5 (10.3) |
| Time of blood sampling | |||
| Before 12:00 | 378 | 21 | NA |
| Between 12:00 and 17:00 | 1172 | 64 | NA |
| After 17:00 | 274 | 15 | NA |
| Unknown | 8 | 0 | NA |
| Hepcidin, nM† | 1832 | 100 | 6.4 (2.6) |
| Ferritin, μg/L† | 1830 | 100 | 107.2 (2.6) |
| Ratio hepcidin/ferritin, nmoles/μg† | 1830 | 100 | 59.4 (1.9) |
| Ratio hepcidin/TS, nM/%† | 1812 | 99 | 0.23 (2.6) |
| Iron, μM | 1812 | 99 | 17.2 (5.6) |
| TS, % | 1812 | 99 | 29.6 (10.3) |
| TIBC, μM | 1812 | 99 | 59.2 (9.0) |
N indicates number; NA, not applicable.
Numbers are different from the total number of included persons because of missing values.
The variables hepcidin, ferritin, ratio hepcidin/ferritin, and ratio hepcidin/TS were log-transformed, and therefore geometric mean and SD are given.
TIBC, total iron binding capacity; TS, transferrin saturation.
Associations of common variants in HFE and TMPRSS6 with ferritin, iron, TS, and TIBC
Single SNP association analyses revealed that the SNP HFE rs1800562 was the strongest associate of ferritin, iron, TS, and TIBC of all variants in and surrounding HFE included in our study (p between 1×10−18 and 1×10−3) (see online supplementary table S3). For TMPRSS6, rs855791 was most strongly associated with both iron and TS of all variants tested (p=3.4×10−12 and 8.5×10−14, respectively). In addition, this SNP was one of the strongest associates of TIBC and ferritin, although not significantly associated (p=0.13 and 0.19, respectively) (see online supplementary table S3). Besides these two SNPs, other common variants in HFE and TMPRSS6 showed associations (using the stringent Bonferroni corrected significance threshold of p<5×10−4) with iron and TS: 10 SNPs for HFE and iron, 11 SNPs for HFE and TS, 11 SNPs for TMPRSS6 and iron, and 22 SNPs for TMPRSS6 and TS. However, all of these associations for TMPRSS6 and most of the associations for HFE were dependent on TMPRSS6 rs855791 and HFE rs1800562, respectively, as shown by conditional analyses (see online supplementary table S4). Only rs1799945 (p. His63Asp), rs6918586, rs198855, and rs198851 (all three in flanking regions) in HFE were significantly associated with both iron and TS after conditioning on HFE rs1800562, but their strength of association did not approach that of HFE rs1800562.
Evaluation of allelic interaction between non-synonymous and eQTL variants via haplotype analysis (see online supplementary tables S5 and S6) suggested the presence of a haplotype effect for HFE that was independent of the marginal effect of HFE rs1800562. Still, this SNP was the greatest contributor to the haplotype effect. There was no evidence for allelic interaction within TMPRSS6. These results confirm HFE rs1800562 and TMPRSS6 rs855791 as the most important variants within HFE and TMPRSS6, respectively, in their ability to affect the iron phenotypes.
Table 2 shows the associations between TMPRSS6 rs855791 and HFE rs1800562 with the iron parameters. Both SNPs are c.G>A SNPs, with a minor allele frequency (MAF) for the A allele of 0.455 for TMPRSS6 rs855791 and 0.063 for HFE rs1800562 in the total study population. The SNPs showed the strongest association with iron and TS. The minor allele A of TMPRSS6 rs855791 showed decreased iron concentration (β (95% CI) AG vs GG −1.5 (−2.1 to −0.9); AA vs GG −2.5 (−3.2 to −1.8)) and TS (β (95% CI) AG vs GG −2.9 (−4.0 to −1.9); AA vs GG −5.0 (−6.3 to −3.7)), while the minor allele A of HFE rs1800562 was associated with both increased iron concentrations (β (95% CI) AG vs GG 2.3 (1.5 to 3.0); AA vs GG 10.7 (5.5 to 15.9)) and TS (β (95% CI) AG vs GG 5.4 (4.0 to 6.8); AA vs GG 22.6 (13.2 to 32.0)). Ferritin and TIBC were associated with HFE rs1800562, but not with TMPRSS6 rs855791. The A allele of HFE rs1800562 is associated with an increase of ferritin and a decrease of TIBC, respectively.
Table 2.
Associations between rs855791 in the transmembrane serine protease 6 gene (TMPRSS6) and rs1800562 in the haemochromatosis gene (HFE) with iron parameters for the total study population (N=1832)
| Additionally adjusted for: | Total N* | p Value | AG | AA | |||
|---|---|---|---|---|---|---|---|
| N | β AG vs GG (95% CI) | N | β AA vs GG (95% CI) | ||||
| TMPRSS6 rs855791† | |||||||
| Log(hepcidin)‡, nM | NA | 1824 | 2.02E-01 | 928 | 0.01 (−0.03 to 0.05) | 365 | 0.05 (−0.01 to 0.10) |
| Log(ferritin), μg/L‡ | NA | 1822 | 1.86E-01 | 926 | −0.03 (−0.07 to 0.01) | 365 | −0.04 (−0.09 to 0.01) |
| Log(hepcidin) | 1822 | 1.36E-05 | 926 | −0.04 (−0.06 to −0.01) | 365 | −0.07 (−0.10 to −0.04) | |
| Log(hepcidin/ferritin)‡, nmoles/μg | NA | 1822 | 8.60E-06 | 926 | 0.04 (0.01 to 0.07) | 365 | 0.08 (0.05 to 0.12) |
| Log(hepcidin/TS)‡, nM/% | NA | 1804 | 3.42E-05 | 919 | 0.05 (0.01 to 0.09) | 358 | 0.12 (0.07 to 0.17) |
| Iron, μM | NA | 1804 | 3.36E-12 | 919 | −1.50 (−2.06 to −0.93) | 358 | −2.53 (−3.24 to −1.82) |
| Log(hepcidin) | 1804 | 6.71E-13 | 919 | −1.51 (−2.08 to −0.95) | 358 | −2.60 (−3.31 to −1.90) | |
| Log(hepcidin/ferritin) | 1804 | 7.90E-11 | 919 | −1.42 (−1.98 to −0.86) | 358 | −2.37 (−3.08 to −1.66) | |
| TS, % | NA | 1804 | 8.47E-14 | 919 | −2.92 (−3.96 to −1.89) | 358 | −4.96 (−6.26 to −3.66) |
| Log(hepcidin) | 1804 | 1.48E-15 | 919 | −2.98 (−3.99 to −1.96) | 358 | −5.21 (−6.48 to −3.94) | |
| Log(hepcidin/ferritin) | 1804 | 1.54E-12 | 919 | −2.81 (−3.84 to −1.77) | 358 | −4.71 (−6.01 to −3.41) | |
| TIBC, μM | NA | 1804 | 1.27E-01 | 919 | 0.74 (−0.20 to 1.68) | 358 | 1.16 (−0.02 to 2.33) |
| Log(hepcidin) | 1804 | 2.57E-02 | 919 | 0.82 (−0.07 to 1.71) | 358 | 1.51 (0.40 to 2.63) | |
| Log(hepcidin/ferritin) | 1804 | 7.87E-02 | 919 | 0.81 (−0.13 to 1.75) | 358 | 1.30 (0.11 to 2.48) | |
| HFE rs1800562† | |||||||
| Log(hepcidin)‡, nM | NA | 1824 | 3.89E-01 | 217 | 0.00 (−0.06 to 0.05) | 4 | −0.27 (−0.66 to 0.12) |
| Log(ferritin), μg/L‡ | NA | 1822 | 2.01E-04 | 216 | 0.04 (−0.01 to 0.10) | 4 | 0.69 (0.33 to 1.04) |
| Log(hepcidin) | 1822 | 2.08E-15 | 216 | 0.05 (0.01 to 0.08) | 4 | 0.88 (0.66 to 1.11) | |
| Log(hepcidin/ferritin)‡, nmoles/μg | NA | 1822 | 1.35E-07 | 216 | −0.04 (−0.08 to −0.01) | 4 | −0.65 (−0.90 to −0.41) |
| Log(hepcidin/TS)‡, nM/% | NA | 1804 | 5.73E-04 | 212 | −0.07 (−0.13 to −0.02) | 4 | −0.58 (−0.96 to −0.20) |
| Iron, μM | NA | 1804 | 1.39E-11 | 212 | 2.26 (1.50 to 3.02) | 4 | 10.71 (5.53 to 15.88) |
| Log(hepcidin) | 1804 | 5.70E-12 | 212 | 2.26 (1.51 to 3.02) | 4 | 11.09 (5.94 to 16.24) | |
| Log(hepcidin/ferritin) | 1804 | 3.33E-10 | 212 | 2.18 (1.42 to 2.93) | 4 | 9.33 (4.15 to 14.52) | |
| TS, % | NA | 1804 | 7.00E-18 | 212 | 5.36 (3.98 to 6.75) | 4 | 22.60 (13.17 to 32.04) |
| Log(hepcidin) | 1804 | 3.20E-19 | 212 | 5.37 (4.02 to 6.73) | 4 | 23.93 (14.68 to 33.18) | |
| Log(hepcidin/ferritin) | 1804 | 1.85E-16 | 212 | 5.24 (3.87 to 6.62) | 4 | 20.58 (11.10 to 30.06) | |
| TIBC, μM | NA | 1804 | 1.17E-06 | 212 | −3.05 (−4.30 to −1.80) | 4 | −9.58 (−18.13 to −1.02) |
| Log(hepcidin) | 1804 | 6.81E-08 | 212 | −3.06 (−4.25 to −1.88) | 4 | −11.51 (−19.61 to −3.41) | |
| Log(hepcidin/ferritin) | 1804 | 3.44E-07 | 212 | −3.13 (−4.38 to −1.88) | 4 | −10.94 (−19.54 to −2.33) | |
Associations are adjusted for age, gender, time of blood sampling, and additionally for serum hepcidin or the ratio hepcidin/ferritin.
N indicates number; β AG vs GG, regression coefficient for AG genotype versus GG genotype; β AA versus GG, regression coefficient for AA genotype versus GG genotype.
For TMPRSS6 rs855791 (p.Ala736Val), minor allele is A with frequency 0.455 in the whole cohort. Therefore, genotype GG is used as the reference genotype.
For HFE rs1800562 (p.Cys282Tyr), minor allele is A with frequency 0.063 in the whole cohort. Therefore, genotype GG is used as the reference genotype.
Numbers are different from the total number of included persons because of missing values.
Both SNPs are genotyped.
The dependent variables hepcidin, ferritin, hepcidin/ferritin, and hepcidin/TS were log-transformed. Therefore, the regression coefficients express the changes in each log-transformed variable that are associated with each genotype relative to the reference genotype.
SNP, single nucleotide polymorphisms; TIBC, total iron binding capacity; TS, transferrin saturation.
Results for the two subsets were comparable to the results observed for the total study population, indicating that our findings are not driven by extremes on the exclusion variables (see online supplementary tables S7 and S8).
Role of hepcidin in the associations of HFE rs1800562 and TMPRSS6 rs855791 with the iron parameters
Results of the association analyses of HFE rs1800562 and TMPRSS6 rs855791 with serum hepcidin, the ratio of hepcidin to ferritin and the ratio of hepcidin to TS are presented in table 2 (see online supplementary tables S7 and S8 for results in the subsets). The SNPs were not associated with hepcidin (p>0.2) but were by far the strongest associates of the ratios out of all variants tested in our study (see online supplementary table S9). TMPRSS6 rs855791 and HFE rs1800562 were associated with an increase and decrease, respectively, in both log hepcidin/ferritin (TMPRSS6 rs855791: β (95% CI) AG vs GG 0.04 (0.01 to 0.07), AA vs GG 0.08 (0.05 to 0.12); HFE rs1800562: β (95% CI) AG vs GG −0.04 (−0.08 to −0.01), AA vs GG −0.65 (−0.90 to −0.41)) and log hepcidin/TS (TMPRSS6 rs855791: β (95% CI) AG vs GG 0.05 (0.01 to 0.09), AA vs GG 0.12 (0.07 to 0.17); HFE rs1800562: β (95% CI) AG vs GG −0.07 (−0.13 to −0.02), AA vs GG −0.58 (−0.96 to −0.20)). Stratification of the study population by both HFE rs1800562 and TMPRSS6 rs855791 and subsequent calculation of mean hepcidin concentrations per stratum indicated the presence of statistical interaction, but our sample size was not sufficient to reach statistical significance (see online supplementary table S10).
The associations of TMPRSS6 rs855791 and HFE rs1800562 with ferritin, iron, TS, and TIBC were not dependent on serum hepcidin: regression coefficients for the associations did not change after inclusion of serum hepcidin concentrations in the regression models (table 2). p Values for the associations of the SNPs with iron, TS, and TIBC before and after adjustment for serum hepcidin were similar, but smaller for the associations of the SNPs with log ferritin after adjustment for serum hepcidin. Inclusion of the hepcidin/ferritin ratio as covariate in the regression models did not change the associations either and only decreased the p values for the associations of the SNPs with log ferritin (table 2). Identical observations were done for subset 1 and 2 (see online supplementary tables S7 and S8). We did not correct the associations for the hepcidin/TS ratio because TS was calculated by dividing serum iron by TIBC.
DISCUSSION
Our results showed that HFE rs1800562 and TMPRSS6 rs855791 are the strongest associates of these genes for iron parameters in our study population. These SNPs and their correlated SNP variants also emerged from six previously published GWAS on serum iron, transferrin, TS, and ferritin.1–6 We found that serum hepcidin was not statistically significantly associated with HFE rs1800562 or TMPRSS6 rs855791 nor with any other SNP in HFE and TMPRSS6. However, the ratios of hepcidin to ferritin and hepcidin to TS did show association with the two SNPs. Adjustment for hepcidin did not result in a decrease of the strength of the associations between the SNPs and the iron parameters, neither did adjustment for the ratio of hepcidin to ferritin in the SNP association analyses for iron, TS and TIBC. Hence, our data do not support an intermediate role of hepcidin in the SNP–iron parameter associations nor do they support a pleiotropic effect of the HFE and TMPRSS6 SNPs on iron parameters and hepcidin (figure 1). However, we did find evidence for an independent, pleiotropic effect of the HFE and TMPRSS6 SNPs on iron parameters and ratios of hepcidin to ferritin and TS.
Our findings confirm the results found by Traglia et al in an isolated Italian population.3 They replicated associations of HFE rs1800562 with serum iron, transferrin and TS and of TMPRSS6 rs855791 with serum iron and TS in their cohort of 1545 related individuals, and reported a borderline genome-wide significant association for serum ferritin and HFE rs1800562. TMPRSS6 rs855791 association with serum ferritin was only nominally significant. In agreement with our results, Traglia et al did not find an association of these SNPs with hepcidin, and use of hepcidin as covariate in their association analysis of the SNPs with the iron parameters did not change the associations. In contrast, we observed both for the total study population and for the subsets a remarkable stronger association between ferritin and the SNPs after adjustment for serum hepcidin, which was not observed in the data of Traglia et al. Nevertheless, estimates of regression coefficients were comparable between our study and the study of Traglia et al.3 Finally, Traglia et al reported that HFE rs1800562 and TMPRSS6 rs855791 were associated with the ratio of hepcidin to ferritin in subset 1. We observed this association both in our total study population and in our two subsets. Traglia et al did not study the ratio of hepcidin to TS.3
Our results and the results of Traglia et al3 are in contrast to the generally accepted idea that HFE and TMPRSS6 affect hepcidin transcription, thereby adapting the hepcidin expression in response to the systemic iron concentration. Indeed, there is only limited evidence from animal and in vitro studies for a direct relation between the Hfe protein and (intracellular) iron homeostasis in different cell types.16,25–28 For example, it was shown that HFE mutations can directly affect iron accumulation in hereditary haemochromatosis macrophages, independently of the presence of hepcidin.25,26 On the other hand, evidence for HFE and TMPRSS6 affecting hepcidin transcription is abundant. Mice experiments and observational studies in humans have shown that defects in HFE result in insufficient expression of hepcidin in the liver and lower serum hepcidin concentrations relative to the body iron stores, leading to the iron storage disorder hereditary haemochromatosis.7–11 Mutations in TMPRSS6 have been associated with inappropriately high urine and serum hepcidin concentration for the setting of systemic iron deficiency, which has been suggested to cause iron refractory iron deficiency anaemia. Furthermore, a recent in vitro study by Nai et al showed that the G allele of TMPRSS6 rs855791 inhibits hepcidin more efficiently than the A allele.29 In this same publication, it was also reported that hepcidin was significantly lower in TMPRSS6 rs855791 GG homozygotes than in AA homozygotes in normal subjects after exclusion of iron deficient individuals (serum ferritin <30 ng/mL) and individuals with clinically relevant inflammatory conditions (CRP >1 mg/dL).29 We and Traglia et al3 did not observe an association between hepcidin and TMPRSS6 rs855791 in our data, even though we also excluded iron deficient individuals and individuals with clinically relevant inflammatory conditions in subset 1. In addition, the difference in results between Nai et al29 and Traglia et al3 is striking, because Nai et al used a selection of unrelated individuals (N=545) of the same population used by Traglia et al. For HFE rs1800562, van Dijk et al11 demonstrated in a small population of first degree family members of clinically diagnosed HFE rs1800562 homozygous probands that hepcidin was lower in HFE rs1800562 AA homozygotes compared to AG heterozygotes and GG homozygotes combined (p<0.01); however, we did not find a significant association between hepcidin and HFE rs1800562 in our sample of the general population. On the other hand, our findings for the ratios of hepcidin to ferritin and hepcidin to TS corroborate the results of both Nai et al29 and van Dijk et al.11
Reasons that may have masked an intermediate role of hepcidin in the association between the two SNP variants and iron parameters in our study and that of Traglia et al3 include the reliance on measurement of hepcidin in serum and the potential omission of environmental and genetic factors that may cause variation in serum hepcidin concentrations independent of iron regulation. In other words, the hepcidin as measured in our study (by ELISA) and that of Traglia et al (by mass-spectrometry) may not be a correct reflection of the hepcidin that is intermediate in the Hfe and MT2 regulation of iron parameters. This could also be true for TS as a proxy for circulating iron, as TS does not necessarily reflect the concentration of the (putative) hepcidin signalling form of the molecule—that is, differic transferrin.30 Besides, we used serum concentrations of hepcidin as well as serum concentrations of iron and ferritin, but the associations that we studied might be cell type or tissue specific. Hepcidin is predominantly produced in hepatocytes31–33 whereas the Hfe protein is expressed in a whole range of tissues and cell types34,35 and MT2 is expressed primarily in the liver.36 The identified associations between the HFE and TMPRSS6 variants and ratios of hepcidin to ferritin and hepcidin to TS—a better reflection of the long term balance between hepcidin and body iron status—may support this hypothesis. Nevertheless, adjustment for the ratio of hepcidin to ferritin in the association analyses for the SNPs with iron, TS, and TIBC did not change the effects of the SNPs on the iron parameters either, indicating an independent, pleiotropic effect of the SNPs on both the hepcidin ratios and the iron parameters.
In summary, we can confirm that HFE rs1800562 and TMPRSS6 rs855791 are the main determinants of HFE and TMPRSS6 related variation in iron, ferritin, TS, and TIBC in the general population. These SNPs are not associated with serum hepcidin itself, but do influence ratios reflecting hepcidin relative to circulating iron and iron stores, measured by the iron parameters TS and ferritin, respectively. Our study can confirm that serum hepcidin, whether corrected for iron stores or not, is not the intermediate variable in the associations of the SNPs with the iron parameters, thereby confirming the results of Traglia et al.3 In addition, our data indicate that the SNPs exert a pleiotropic effect on both the hepcidin ratios and the iron parameters. Taken together, our study points to a direct SNP effect on certain serum iron parameters, rather than an indirect effect entirely dependent upon a change in serum hepcidin concentrations. We call for additional functional studies in a controlled setting that allow further elucidation of the role of hepcidin in the associations between the SNPs and iron parameters, which will contribute to our understanding of the underlying mechanisms and eventually facilitate the development of interventions for iron disorders.
Supplementary Material
Acknowledgements
The Nijmegen Biomedical Study is a population based survey conducted at the Department of Epidemiology, Biostatistics and HTA, and the Department of Laboratory Medicine of the Radboud University Medical Centre. Principal investigators of the Nijmegen Biomedical Study are LALM Kiemeney, M den Heijer, ALM Verbeek, DW Swinkels and B Franke. We would like to thank our colleagues from the Department of Laboratory Medicine, hepcidinanalysis.com and the Department of Endocrinology for their excellent technical assistance. In addition, we thank Doorlène van Tienoven for performing the serum hepcidin measurements. This work was sponsored by the Stichting Nationale Computer faciliteiten (National Computing Facilities Foundation, NCF) for the use of supercomputer facilities, with financial support from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research, NWO). This work was performed within a PhD project supported by the Nijmegen Centre for Evidence Based Practice, Radboud University Medical Centre, Nijmegen, The Netherlands.
Footnotes
Competing interests None.
Ethics approval Radboud University Medical Centre Institutional Review Board.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Benyamin B, Ferreira MA, Willemsen G, Gordon S, Middelberg RP, McEvoy BP, Hottenga JJ, Henders AK, Campbell MJ, Wallace L, Frazer IH, Heath AC, de Geus EJ, Nyholt DR, Visscher PM, Penninx BW, Boomsma DI, Martin NG, Montgomery GW, Whitfield JB. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet 2009;41:1173–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, Palotie A, Peltonen L, Martin NG, Montgomery GW, Whitfield JB, Visscher PM. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet 2009;84:60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traglia M, Girelli D, Biino G, Campostrini N, Corbella M, Sala C, Masciullo C, Vigano F, Buetti I, Pistis G, Cocca M, Camaschella C, Toniolo D. Association of HFE and TMPRSS6 genetic variants with iron and erythrocyte parameters is only in part dependent on serum hepcidin concentrations. J Med Genet 2011;48:629–34. [DOI] [PubMed] [Google Scholar]
- 4.Middelberg RP, Ferreira MA, Henders AK, Heath AC, Madden PA, Montgomery GW, Martin NG, Whitfield JB. Genetic variants in LPL, OASL and TOMM40/ APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med Genet 2011;12:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, Porcu E, Pattaro C, Busonero F, Zanon A, Maschio A, Melville SA, Grazia Piras M, Longo DL, Guralnik, Hernandez D, Bandinelli S, Aigner E, Murphy AT, Wroblewski V, Marroni F, Theurl, Gnewuch C, Schadt E, Mitterer M, Schlessinger D, Ferrucci L, Witcher DR, Hicks AA, Weiss G, Uda M, Pramstaller PP. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum Mol Genet 2011;20:1232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Roy CN, Yao W, Matteini A, Semba RD, Arking D, Walston JD, Fried LP, Singleton A, Guralnik J, Abecasis GR, Bandinelli S, Longo DL, Ferrucci L. A genome-wide association analysis of serum iron concentrations. Blood 2010;115:94–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, Sly WS, Fleming RE. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis 2002;29:361–6. [DOI] [PubMed] [Google Scholar]
- 8.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet 2003;361:669–73. [DOI] [PubMed] [Google Scholar]
- 9.Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet 2003;34:102–7. [DOI] [PubMed] [Google Scholar]
- 10.Nicolas G, Viatte L, Lou DQ, Bennoun M, Beaumont C, Kahn A, Andrews NC, Vaulont S. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet 2003;34:97–101. [DOI] [PubMed] [Google Scholar]
- 11.van Dijk BA, Laarakkers CM, Klaver SM, Jacobs EM, van Tits LJ, Janssen MC, Swinkels DW. Serum hepcidin levels are innately low in HFE-related haemochromatosis but differ between C282Y-homozygotes with elevated and normal ferritin levels. Br J Haematol 2008;142:979–85. [DOI] [PubMed] [Google Scholar]
- 12.Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The serine protease TMPRSS6 is required to sense iron deficiency. Science 2008;320:1088–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swinkels DW, Girelli D, Laarakkers C, Kroot J, Campostrini N, Kemna EH, Tjalsma H. Advances in quantitative hepcidin measurements by time-of-flight mass spectrometry. PLoS ONE 2008;3:e2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corradini E, Meynard D, Wu Q, Chen S, Ventura P, Pietrangelo A, Babitt JL. Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)-SMAD signaling pathway in mice. Hepatology 2011;54:273–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell 2010;142:24–38. [DOI] [PubMed] [Google Scholar]
- 16.Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, Roth MP, Nemeth E, Ganz T. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology 2011;53:1333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, Migas MC, Waheed A, Britton RS, Fleming RE. Ferritin upregulates hepatic expression of bone morphogenetic protein 6 and hepcidin in mice. Am J Physiol Gastrointest Liver Physiol 2012;302:G1397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med 2012;366:348–59. [DOI] [PubMed] [Google Scholar]
- 19.Finberg KE. Iron-refractory iron deficiency anemia. Semin Hematol 2009;46:378–86. [DOI] [PubMed] [Google Scholar]
- 20.Hoogendoorn EH, Hermus AR, de Vegt F, Ross HA, Verbeek AL, Kiemeney LA, Swinkels DW, Sweep FC, den Heijer M. Thyroid function and prevalence of anti-thyroperoxidase antibodies in a population with borderline sufficient iodine intake: influences of age and sex. Clin Chem 2006;52:104–11. [DOI] [PubMed] [Google Scholar]
- 21.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, Blondal T, Witjes JA, Vermeulen SH, Hulsbergen-van de Kaa CA, Swinkels DW, Ploeg M, Cornel EB, Vergunst H, Thorgeirsson TE, Gudbjartsson D, Gudjonsson SA, Thorleifsson G, Kristinsson KT, Mouy M, Snorradottir S, Placidi D, Campagna M, Arici C, Koppova K, Gurzau E, Rudnai P, Kellen E, Polidoro S, Guarrera S, Sacerdote C, Sanchez M, Saez B, Valdivia G, Ryk C, de Verdier P, Lindblom A, Golka K, Bishop DT, Knowles MA, Nikulasson S, Petursdottir V, Jonsson E, Geirsson G, Kristjansson B, Mayordomo JI, Steineck G, Porru S, Buntinx F, Zeegers MP, Fletcher T, Kumar R, Matullo G, Vineis P, Kiltie AE, Gulcher JR, Thorsteinsdottir U, Kong A, Rafnar T, Stefansson K. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet 2008;40:1307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galesloot TE, Vermeulen SH, Geurts-Moespot AJ, Klaver SM, Kroot JJ, van Tienoven D, Wetzels JF, Kiemeney LA, Sweep FC, den Heijer M, Swinkels DW. Serum hepcidin: reference ranges and biochemical correlates in the general population. Blood 2011;117:e218–25. [DOI] [PubMed] [Google Scholar]
- 23.Kroot JJ, Laarakkers CM, Geurts-Moespot AJ, Grebenchtchikov N, Pickkers P, van Ede AE, Peters HP, van Dongen-Lases E, Wetzels JF, Sweep FC, Tjalsma H, Swinkels DW. Immunochemical and mass-spectrometry-based serum hepcidin assays for iron metabolism disorders. Clin Chem 2010;56:1570–9. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacolot S, Yang Y, Paitry P, Ferec C, Mura C. Iron metabolism in macrophages from HFE hemochromatosis patients. Mol Genet Metab 2010;101:258–67. [DOI] [PubMed] [Google Scholar]
- 26.Makui H, Soares RJ, Jiang W, Constante M, Santos MM. Contribution of Hfe expression in macrophages to the regulation of hepatic hepcidin levels and iron loading. Blood 2005;106:2189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem 2008;283:21462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos P, Guy E, Chen N, Proenca CC, Gardenghi S, Casu C, Follenzi A, Van Rooijen N, Grady RW, de Sousa M, Rivella S. Enhanced erythropoiesis in Hfe-KO mice indicates a role for Hfe in the modulation of erythroid iron homeostasis. Blood 2011;117:1379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nai A, Pagani A, Silvestri L, Campostrini N, Corbella M, Girelli D, Traglia M, Toniolo D, Camaschella C. TMPRSS6 rs855791 modulates hepcidin transcription in vitro and serum hepcidin levels in normal individuals. Blood 2011;118:4459–62. [DOI] [PubMed] [Google Scholar]
- 30.DiRusso SC, Check IJ, Hunter RL. Quantitation of apo-, mono-, and diferric transferrin by polyacrylamide gradient gel electrophoresis in patients with disorders of iron metabolism. Blood 1985;66:1445–51. [PubMed] [Google Scholar]
- 31.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 2000;480:147–50. [DOI] [PubMed] [Google Scholar]
- 32.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 2001;276:7811–19. [DOI] [PubMed] [Google Scholar]
- 33.Kroot JJ, Tjalsma H, Fleming RE, Swinkels DW. Hepcidin in human iron disorders: diagnostic implications. Clin Chem 2011;57:1650–69. [DOI] [PubMed] [Google Scholar]
- 34.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R Jr, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 1996;13:399–408. [DOI] [PubMed] [Google Scholar]
- 35.Parkkila S, Niemela O, Britton RS, Fleming RE, Waheed A, Bacon BR, Sly WS. Molecular aspects of iron absorption and HFE expression. Gastroenterology 2001;121:1489–96. [DOI] [PubMed] [Google Scholar]
- 36.Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J Biol Chem 2002;277:37637–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.