Abstract
Objective This paper presents an automatic, active learning-based system for the extraction of medical concepts from clinical free-text reports. Specifically, (1) the contribution of active learning in reducing the annotation effort and (2) the robustness of incremental active learning framework across different selection criteria and data sets are determined.
Materials and methods The comparative performance of an active learning framework and a fully supervised approach were investigated to study how active learning reduces the annotation effort while achieving the same effectiveness as a supervised approach. Conditional random fields as the supervised method, and least confidence and information density as 2 selection criteria for active learning framework were used. The effect of incremental learning vs standard learning on the robustness of the models within the active learning framework with different selection criteria was also investigated. The following 2 clinical data sets were used for evaluation: the Informatics for Integrating Biology and the Bedside/Veteran Affairs (i2b2/VA) 2010 natural language processing challenge and the Shared Annotated Resources/Conference and Labs of the Evaluation Forum (ShARe/CLEF) 2013 eHealth Evaluation Lab.
Results The annotation effort saved by active learning to achieve the same effectiveness as supervised learning is up to 77%, 57%, and 46% of the total number of sequences, tokens, and concepts, respectively. Compared with the random sampling baseline, the saving is at least doubled.
Conclusion Incremental active learning is a promising approach for building effective and robust medical concept extraction models while significantly reducing the burden of manual annotation.
Keywords: medical concept extraction, clinical free text, active learning, conditional random fields, robustness analysis
BACKGROUND AND SIGNIFICANCE
The goal of active learning (AL) is to maximize the effectiveness of the learning model while minimizing the number of annotated samples required. The main challenge is to identify the informative samples that guarantee the learning of such a model. 6
Settles and Craven 13 reported an extensive empirical evaluation of a number of AL selection criteria using different corpora for sequence labeling tasks. Information density (ID), sequence vote entropy, and least confidence (LC) were found to outperform the state-of-the-art in AL for sequence labeling in most corpora.
While the effectiveness of AL methods has been conclusively proven and demonstrated in many domains for tasks such as text classification, information extraction (IE), and speech recognition, 6 as Ohno-Machado et al1 highlighted, there are limited explorations of AL techniques in clinical and biomedical natural language processing (NLP) tasks. There are examples of using AL for classifying medical concepts according to their assertions 14,15 and coreference resolution, 16 both of which are 2 important elements of any clinical IE system. For assertion classification, Chen et al14 introduced a “model change” sampling-based algorithm which controls the changes of certain values from different models during the AL process. They found it performing better in terms of effectiveness and annotation rate than uncertainty sampling and ID selection criteria.
In the medical IE context, AL has also been used for de-identifying Swedish clinical records. 17 The most uncertain and the most certain sampling strategies were evaluated using the highest and lowest entropy, respectively. The evaluation showed that these methods outperformed other benchmarks, including a random sampling (RS) baseline. Figueroa et al18 analyzed the performance of distance-based and diversity-based algorithms as 2 AL methods, in addition to a combination of both, for the classification of smoking and depression status. The performance of the proposed methods in terms of accuracy and annotation rate was found to be strongly dependent on the data set diversity and uncertainty.
OBJECTIVE
The widespread use of e-Health technologies, in particular of electronic health records, offers many opportunities for data analysis. 1 The extraction of structured data from unstructured, free-text health documents (eg, clinical narratives) is essential to support such analysis, in particular for applications such as retrieving, reasoning, and reporting, eg, for cancer notification and monitoring from pathology reports. 2,3 This extraction process commonly consists of a concept extraction stage, ie, the identification of short-term sequences (eg, named entities, phrases, or others) from unstructured text written in natural language. These terms express meaningful concepts within a given domain, eg, medical problems, tests, and treatments ( figure 1 ) from the i2b2/VA 2010 data set. 4
Figure 1:
An example of input text and associated concepts from the i2b2/VA 2010 data set.
This IE process, however, is not straightforward. Challenges include the identification of concept instances that are referred to in ways not captured within current lexical resources and the presence of ambiguity, polysemy, synonymy (including acronyms), and word order variations. In addition to these challenges, the information presented in clinical narratives is often unstructured, ungrammatical, and fragmented, and language usage greatly differs from that of general free-text. Because of this, mainstream NLP technologies and systems often cannot be directly used in the health domain. 5 In fact, high quality manual annotations of large corpora are necessary for building robust statistical supervised machine learning (ML) classifiers. Obtaining these annotations is costly because it requires extensive involvement of domain experts (eg, pathologists or experienced clinical coders to annotate pathology reports) and linguists.
AL has been proposed as a feasible alternative to standard supervised ML approaches to reduce annotation costs. 6 We embrace this proposal, and in this article we investigate the use of active learning for medical concept extraction.
AL methods use supervised ML algorithms in an iterative process, where at each iteration, samples from the data set are automatically selected based on their “informativeness” to be annotated by an expert. This is effectively a human-in-the-loop process that allows to drastically reduce human involvement compared with the large amount of annotated data required upfront by standard supervised ML systems. Despite this interesting property, AL has not been fully explored for clinical IE, in particular for medical concept extraction. 7
The aim of this article is to extensively investigate the comparative performance of a fully supervised ML approach and an AL counterpart for the task of extracting medical concepts related to problems, tests, treatments (i2b2/VA 2010 task 4 ), and disorder mentions (ShARe/CLEF 2013, task 1 8 ). Specifically, the following hypotheses are validated: (1) AL achieves the same effectiveness as a supervised approach using much less annotated data, and (2) incremental AL results in more robust learnt models than standard AL regardless of selection criterion. Preliminary investigations suggest that a selected feature set and incremental learning could increase robustness. 9 However, it has not been examined across different settings and selection criteria.
To investigate our hypotheses, we use conditional random fields (CRFs) 10 as the supervised IE component. CRFs is a proven state-of-the-art technique for the task at hand. 4,11 Two AL selection criteria are investigated, namely uncertainty sampling-based 12 and information density (ID). 13 Annotated data from the i2b2/VA 2010 Clinical NLP Challenge and the ShARe/CLEF 2013 eHealth Evaluation Lab (task 1) are used for training the models and evaluating their performance.
MATERIALS AND METHODS
Tags for entity representation
Before applying ML algorithms to the free-text data, as shown in figure 1 , the annotated data need to be represented with an appropriate tagging format. We use the “BIO” format, where B refers to the “beginning” of an entity, I refers to the “inside” of an entity, and O refers to the “outside” of an entity. 19Figure 2 shows the BIO tag representation for the sentences from figure 1 .
Figure 2:
An example of BIO tag representation.
FEATURES FOR MACHINE LEARNING FRAMEWORKS
The following groups of features are used for both the fully supervised and the AL methods:
Linguistic and orthographical features (eg, regular expression patterns and part-of-speech tags)
Lexical and morphological features (eg, suffixes/prefixes and character n-gram)
Contextual features (eg, window of k words)
Semantic features (eg, SNOMED CT and UMLS semantic groups from Medtex, 20 a medical NLP toolkit)
Fully supervised approach
We use linear-chain CRFs as a supervised IE algorithm and as a base algorithm in the AL framework. Let and be an observation sequence and its corresponding label sequence, respectively. For example, sentence 2 in figure 2 would correspond to and . The posterior probability of given is described by the linear-chain CRFs model with a set of parameters :
| (1) |
where is the normalization factor. Each is the transition feature function between label states and on the sequence at position . The parameters represent the corresponding feature weights.
An important parameter when training CRFs is the Gaussian prior variance or regularization parameter. In our prior investigation, 9 we found that the optimal value for clinical data was 1.
Active learning
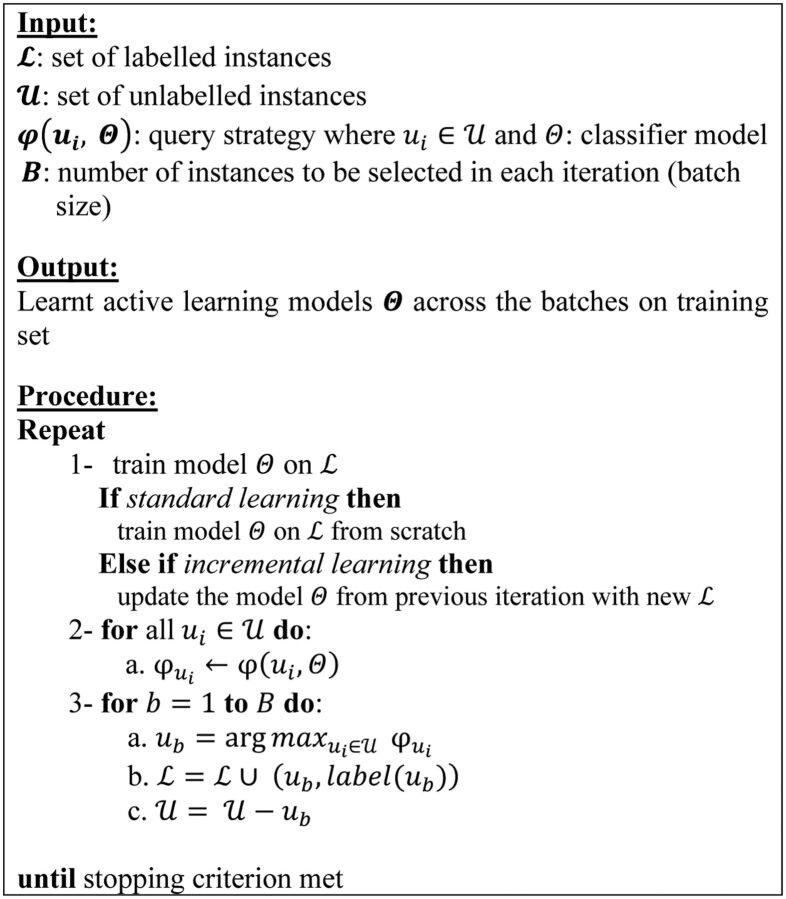
The AL framework described by the algorithm in figure 3 is characterized by 6 main elements:
Figure 3:
A generic pool-based AL algorithm.
The initial labeled set is usually a very small, randomly selected portion (eg, < ) of the whole data set;
A batch of instances is selected at each iteration of the AL algorithm;
The actual instances , in our case, correspond to a complete sequence or sentence, thus yielding a sequence-based AL method (rather than a token-based or document-based);
The stopping criterion identifies the condition to be met for terminating the AL process. As we aim to study how AL can contribute towards reducing the annotation effort compared to a supervised approach, we use supervised effectiveness as our target performance.
The selection criterion estimates the informativeness of an unlabeled instance based on the model .
Training the model can either be achieved by fully retraining the model using all labeled data at each step or incrementally by updating the model learnt in the preceding loop with new labeled instances.
The most critical part when designing an AL strategy is how to estimate the informativeness of each instance, ie, selecting an effective selection criterion.
Uncertainty sampling
One of the most common selection criteria is uncertainty sampling. 12 According to this paradigm, instances with the highest uncertainty are selected for labeling and inclusion in the labeled set used for training in the following iteration. We propose to use LC as one of our selection criteria. LC uses the confidence of the latest model in predicting the label of a sequence 21 :
| (2) |
The confidence of the CRFs model is estimated using the posterior probability described in Equation (1) , and is the predicted label sequence obtained with the Viterbi algorithm.
Information density
ID is an alternative selection criterion for AL. 13 The intuition behind ID is that the selection of instances that are both informative and representative lead to a better coverage of the data set characteristics. By considering also the representativeness of instances, along with their informativeness, outliers are less likely to be selected by the AL process. ID is computed according to the following:
| (3) |
where corresponds to the representativeness of instances. In this study, we use the LC ( Equation (2) ) to measure the informativeness of instances ( ). The average similarity between instance and all other sequences in the set of unlabeled instances indicates how representative the instance is—the higher the similarity, the more representative the instance is. Similarity between and another sequence is measured according to the cosine distance:
| (4) |
Where refers to the feature vector of instance , is calculated as the mean of the similarities between and all sequences across the unlabeled set:
| (5) |
Besides a fully supervised approach, common baselines for analyzing the benefits of the AL framework are RS and Longest Sequence (LS). Both baselines follow the steps of figure 3 , except for RS randomly selects instances and LS chooses instances with the longest length in each batch.
Incremental learning
In the standard AL approach, at each iteration, instances are selected from according to a selection criterion, manually labeled, and then added to the labeled set . A new model is built on independently of models built in previous iterations. In other words, a supervised model is built from (step 1 in figure 3 ) at each iteration.
The alternative is to use an incremental approach. Here all weights and values of the model learnt in previous iteration are maintained, and they are updated in the current iteration using the corresponding labeled set. It makes the training of models faster than the standard setting, leading to considerable reduction in processing time across the whole AL loop. Through the comparison of the standard and the incremental AL approaches, we aim to unveil the effect that maintaining models’ weights and values across iterations has on the robustness of the models themselves within the considered clinical IE tasks.
Data sets
For generating learning curves, we used the annotated train and test sets from the concept extraction task in the i2b2/VA 2010 NLP challenge 4 and ShARe/CLEF 2013 eHealth Evaluation Lab (task 1). 11 One of the i2b2/VA 2010 tasks was to extract medical problems, tests, and treatments from clinical reports. The training and testing sets included 349 and 477 reports, respectively.
Task 1 of the ShARe/CLEF 2013 eHealth Evaluation Lab was to extract and identify disorder mentions from clinical notes. The data set consisted of 200 training and 100 testing clinical notes.
Experimental settings
Our implementations of CRFs for supervised learning, RS, and incremental AL, including LC and ID, are based on the MALLET toolkit. 22 For AL approaches and RS, the initial labeled set is formed by randomly selecting 1% of training data. The batch size ( ) is set to 200 for i2b2/VA 2010 and 30 for ShARe/CLEF 2013 across all experiments, leading to a total of 153 and 91 batches, respectively. We simulate the human annotator in the AL process by using the annotations provided in the training portions of the 2 data sets; thus, these are treated as the input the annotator will provide to the AL algorithm.
In our evaluation, concept extraction effectiveness is measured by Precision, Recall, and F1-measure. Learning curves highlight the interaction between model effectiveness and the required annotation effort.
The point of intersection between the AL learning curve and the target supervised effectiveness is used to measure how much annotation effort is saved by an AL approach. We analyzed the results by considering the annotation rate (AR) for sequences, tokens, and concepts. This can be computed as the number of labeled annotation units (sequences, tokens, and concepts) used by AL for reaching this point over the total number of corresponding labeled annotation units used by the supervised method:
| (6) |
The lower the AR, the less annotation effort is required. Here we assume that every instance is considered as having the same annotation effort (uniform annotation effort). However, in reality, sentences could be short or long. Longer sentences with more entities would require more time for annotation. But shorter sentences without any entities could be skipped by annotators quickly, thus taking little time for annotating them. While this setting may not be fully representative of real-world use-cases, 23 actual annotation costs are not available for the considered data sets, and the literature lacks specific studies that consider annotation cost models for medical concept extraction. Modeling annotation cost is outside of the scope of this paper. However, our evaluation provides an indication of the reduction in annotation effort that AL could contribute.
In addition, we perform 10-fold cross validation experiments on the training data to analyze the robustness of AL and RS within the incremental settings in both data sets and across different selection criteria.
RESULTS
Incremental vs standard active learning
As discussed in prior work, 9 incremental AL framework with tuned Gaussian prior variance for CRFs (InALCE-Tun) leads to models that are more stable across batches and also achieves higher effectiveness compared with the nontuned standard AL framework (ALCE) and nontuned incremental AL framework (InALCE). We now investigate the annotation rate across these frameworks.
The results reported in table 1 suggest that InALCE reaches the same performance as the supervised approach in both data sets but with a faster learning rate than ALCE. Furthermore, by tuning the CRFs parameter in the incremental AL framework (InALCE-Tun), not only the supervised performance increases 9 but also the annotation rate decreases, which means lower annotation effort is required to reach the target performance. We then retain InALCE-Tun for the next experimental settings.
Table 1:
Annotation rate for ALCE and InALCE settings using least confidence as selection criterion
| SAR (%) | ||||
|---|---|---|---|---|
| ALCE | InALCE | InALCE-Tun | ||
| i2b2 2010 | Sup (F1 = 0.8018) | 30 | 23 | 15 |
| Sup-Tun (F1 = 0.8212) | NA | NA | 23 | |
| CLEF 2013 | Sup (F1 = 0.6579) | 44 | 35.5 | 19 |
| Sup-Tun (F1 = 0.6689) | NA | NA | 30 | |
Abbreviations: NA, not available; SAR, sequences annotation rate; ALCE, nontuned standard active learning framework; InALCE, nontuned incremental active learning framework; InALCE-Tun, tuned incremental active learning framework; Sup, supervised approach; Sup-Tun, tuned supervised approach.
Active learning selection criteria
So far, we have only considered the LC selection criterion. In this section, we compare its performance with that of the ID selection criterion, with RS and LS as baselines.
Figure 4 shows the performance of these with InALCE-Tun on i2b2/VA 2010 and ShARe/CLEF 2013, highlighting that the use of a different selection criteria does not have an impact on the robustness gained by using InALCE-Tun. In both data sets, we observe that LC and ID reach the target performance quicker than the RS and LS baselines. Furthermore, it can be noticed that these 2 criteria always outperform the baselines, and that there is no noticeable difference between LC and ID. The annotation rates from table 2 show that LC requires less annotation effort to reach the target performance.
Figure 4:
The performance of supervised (Sup), Random Sampling (RS), Longest Sequence (LS), and active learning approaches (LC and ID) in InALCE-Tun setting (a) i2b2/VA 2010 (b) ShARe/CLEF 2013.
Table 2:
Annotation rate for random sampling, longest sequence, information density, and least confidence
| SAR (%) | TAR (%) | CAR (%) | ||||
|---|---|---|---|---|---|---|
| i2b2 2010 | CLEF 2013 | i2b2 2010 | CLEF 2013 | i2b2 2010 | CLEF 2013 | |
| Random Sampling | 89 | 85 | 89 | 88 | 89 | 87.5 |
| Longest Sequence | 67 | 66 | 92 | 95 | 96 | 94 |
| Information Density | 24.5 | 30 | 45 | 59 | 54 | 77 |
| Least Confidence | 23 | 30 | 43 | 57 | 54 | 76 |
Abbreviations: SAR, sequences annotation rate; TAR, tokens annotation rate; CAR, concepts annotation rate.
ID is computationally costly compared to LC. Depending on the size of the data set, it may require a large amount of similarity calculations for all instances in the unlabeled set, which could be precomputed before running AL. While this would not be a problem in real-time AL systems, the results still suggest choosing LC a priori as a selection strategy because it always achieves a slightly lower AR.
Effectiveness of AL beyond the target performance
Results in table 3 consider the batch in which AL approaches achieve the highest F1-measure, which is generally beyond the target (supervised) performance (where the AL learning curve and the target intersect). These results demonstrate that LC can outperform the supervised method using 44% of the whole training data for i2b2/VA 2010 and using 48% of the whole training data for the ShARe/CLEF 2013 task. The highest performance rates reported in table 3 suggest that LC requires less training data than ID to achieve the highest performance in i2b2/VA 2010. The same finding is however not confirmed by the ShARe/CLEF 2013 results, for which no difference is observed.
Table 3:
The highest performance of active learning methods
| i2b2/VA 2010 | ShARe/CLEF 2013 | |||||||
|---|---|---|---|---|---|---|---|---|
| P | R | F1 | SAR (%) | P | R | F1 | SAR (%) | |
| Sup | 0.8378 | 0.8053 | 0.8212 | NA | 0.7865 | 0.5819 | 0.6689 | NA |
| ID | 0.8429 | 0.8114 | 0.8268 | 47 | 0.7934 | 0.5953 | 0.6803 | 48 |
| LC | 0.8444 | 0.8112 | 0.8275 | 44 | 0.7911 | 0.5965 | 0.6803 | 48 |
Abbreviations: NA, not available; SAR, sequences annotation rate; P, precision; R, recall; F1, F1-measure; Sup, supervised approach; ID, information density; LC, least confidence .
Robustness Analysis
To analyze the robustness of AL models in the incremental setting (InALCE-Tun), we perform a 10-fold cross validation on both i2b2/VA 2010 and ShARe/CLEF 2013 training sets. In these experiments, the training set is split in 10 random sets; 9 are used as labeled training data and 1 as testing data. This process is then iterated by varying which fold is used for testing.
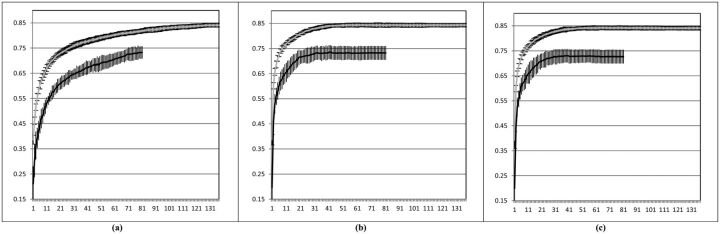
AL is applied throughout the training data and the performance of the learnt model at each batch is averaged across the testing folds. Figure 5 shows the learning curves of InALCE-Tun with different selection criteria across the 2 data sets; these are obtained considering at each batch the mean performance of the learnt model and its variance across the cross-validation folds. AL models built across batches in the cross validation setting are robust, as they show small variance across folds.
Figure 5:
Ten-fold cross validation results across the batches on i2b2/VA 2010 (gray curve) and ShARe/CLEF 2013 (black curve) data sets. The horizontal axis corresponds to the number of batches used for training and the vertical axis reports F1-measure values. Bars indicating the standard deviation across the folds are reported for each batch along the learning curves (a) RS, (b) ID, (c) LC.
DISCUSSION
The results of our empirical investigation demonstrate that AL strategies clearly have a role to play in reducing the burden of annotation for high-quality medical concept extraction. We found that the AL selection criteria always perform better than the baselines (RS and LS) in terms of effectiveness and annotation rate. While RS does not reach the target supervised learning performance much before having trained on all batches, it is interesting to note that it does come within 10% of the target performance after only 26 (5273 instances) out of 153 and 30 (912 instances) out of 91 iterations in i2b2/VA 2010 and ShARe/CLEF 2013, respectively. This suggests that the data sets used in our study present patterns that are often repeated between instances. It is also important, however, to remember that the training instances are sentences; thus, on average, after 26 to 30 iterations there could be nearly 10 sentences from each document in the training set. Repeated patterns, especially repeated concepts themselves, are to be expected within patient records, and therefore it should be expected that the entire training set presents redundancies.
Furthermore, it only takes AL 7 iterations (1400 instances) in i2b2/VA 2010 and 6 iterations (200 instances) in ShARe/CLEF 2013 to reach an F1-measure value within 10% of that reached by the supervised CRFs model. This is a very important result in contexts where one may be able to balance the effectiveness against annotation effort, eg, when performing concept extraction to build a knowledge base.
Our comparison of various AL methods shows that, in the considered task, the LC selection criterion outperforms the ID criterion. This suggests that the probability distribution of the data in clinical narratives is representative of the characteristics of the data. In addition, we verified that an incremental AL approach (InALCE-Tun) provides higher robustness than the standard AL approach (ALCE) and it requires less labeled data to reach the target performance.
Despite ID being regarded as the state-of-the-art in AL, 14,18 this criterion was not able to outperform the LC approach within the medical concept extraction task considered in this paper. We speculate that this is because of the high similarity found in clinical narratives. The ID method tends to select samples from dense regions of the data to avoid selecting outliers; thus, samples that are more similar to other samples in the data set are more likely to be selected.
Figure 6 shows the distribution of full duplicates in the i2b2/VA 2010 data set based on the sequence length. We found that 35% of the total number of sequences in the data set was exactly replicated. As shown in figure 6 , 71% of full duplicates are sequences with lengths between 1 and 3, which are less likely to contain the target concepts. To investigate if there is any effect of full duplicates on AL performance, the experiments were replicated with a precondition in the steps of the AL process, which prevented the algorithm from selecting full duplicates. The annotation rates are reported in table 4 against the same data set and target performance as in table 2 .
Figure 6:
The distribution of full duplicate sequences based on their length in the i2b2/VA 2010 data set .
Table 4:
Annotation rate for random sampling, longest sequence, information density, and least confidence on i2b2/VA 2010 data set when full duplicates are not allowed to be selected in the active learning process
| SAR (%) | TAR (%) | CAR (%) | |
|---|---|---|---|
| Random Sampling | 58 | 75 | 82 |
| Longest Sequence | 56 | 84 | 91 |
| Information Density | 24 | 46 | 56 |
| Least Confidence | 23 | 44 | 55 |
Abbreviations: SAR, sequences annotation rate; TAR, tokens annotation rate; CAR, concepts annotation rate.
Results show that ignoring full duplicates have no significant effect on LC and ID performance in terms of annotation rates. It is not surprising as LC and ID are likely to select almost the same informative instances as in previous settings. Indeed, LC selects sequences containing labels where the model has low confidence, and an already seen sequence would immediately have high probabilities associated to its labels.
Although full duplicates would potentially have been selected by ID due to their high similarity, the probability component in the ID function ( Equation (3) ) avoids choosing those instances as the model is confident enough about their labels.
The differences for RS results from table 2 are due to the fact that the probability of selecting full duplicates for RS is now zero, which means useful samples are more likely to be selected at each iteration.
We also found that a fully supervised model trained on a duplicate-free version of the data set yielded almost the same performance (F1 measure = 0.8224) as on the full training set (F1 measure = 0.8212), which suggests that CRFs models do not make use of repeated sequences.
We analyzed the errors performed in the considered concept extraction task by considering the confusion matrices obtained from the classification results (reported in supplementary appendix A ), and we observed that in both data sets the largest amount of errors is found to be the misclassification of a target entity into a nontarget entity (eg, “problem , ” “test , ” or “treatment” entities misclassified as “others” in i2b2/VA 2010). In this data set, the target problem presents the least classification errors; this is consistent across all approaches. The error analysis using confusion matrices also suggested that the AL model learnt up to the target performance is similar (in terms of classification errors) to the model learnt by the supervised approach on the whole data set, suggesting AL does not overfit the data.
In summary, we can conclude that (1) AL can reach at least the same effectiveness of supervised learning for medical concept extraction while using less training data, and (2) the IE models learnt by AL are not overfitted as they appear to lead to similar errors to those from the models learnt by the fully supervised approach.
CONCLUSION
This paper presented a simulated study of AL for medical concept extraction. We have empirically demonstrated that AL can be highly effective for reducing the effort of manual annotation while building reliable models. We demonstrated this by comparing the effectiveness of fully supervised CRFs against 2 AL approaches and 2 baselines. The evaluation based on the i2b2/VA 2010 NLP challenge and the ShARe/CLEF 2013 eHealth Evaluation Lab (task 1) data sets showed that AL (specifically when using the LC selection criterion) achieves the same effectiveness of supervised learning using 54% (i2b2/VA 2010) and 76% (ShARe/CLEF 2013) of the total number of concepts in training data. We also showed that incremental learning leads to more reliable models within the AL framework.
While this research contributes a very important first step in introducing AL for medical concept extraction, further work is required to examine other selection criteria and develop a cost model for evaluation of AL.
CONTRIBUTORS
This article is a part of MK’s PhD thesis. She developed the ideas and wrote all the coding. LS, GZ, and AN supervised, analyzed, and discussed the results of MK’s work. All authors have contributed in writing and revising the manuscript.
COMPETING INTERESTS
None.
Supplementary Material
Acknowledgments
We would like to thank the anonymous reviewers for their helpful feedback.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/ .
REFERENCES
- 1. Ohno-Machado L Nadkarni P Johnson K . Natural language processing: algorithms and tools to extract computable information from EHRs and from the biomedical literature . J Am Med Inform Assoc. 2013. ; 20 ( 5 ): 805 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nguyen A Moore J Lawley M et al. . Automatic extraction of cancer characteristics from free-text pathology reports for cancer notifications . Stud Health Technol Inform. 2011. : 117 – 124 . [PubMed] [Google Scholar]
- 3. Zuccon G Wagholikar AS Nguyen AN et al. . Automatic classification of free-text radiology reports to identify limb fractures using machine learning and the SNOMED CT ontology . AMIA Summit Clin Res Inform. 2013. ; 2013 : 300 – 304 . [PMC free article] [PubMed] [Google Scholar]
- 4. Uzuner Ö South BR Shen S et al. . 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text . J Am Med Inform Assoc. 2011. ; 18 ( 5 ): 552 – 556 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nadkarni PM Ohno-Machado L Chapman WW . Natural language processing: an introduction . J Am Med Inform Assoc. 2011. ; 18 ( 5 ): 544 – 551 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Settles B. Active Learning . Synthesis Lectures on Artificial Intelligence and Machine Learning . Morgan & Claypool Publishers; ; 2012. . [Google Scholar]
- 7. Skeppstedt M . Annotating named entities in clinical text by combining pre-annotation and active learning . Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics (ACL 2013) . Sofia, Bulgaria, 2013:74–80 . [Google Scholar]
- 8. Suominen H Salanterä S Velupillai S et al. . Overview of the ShARe/CLEF eHealth Evaluation Lab 2013 . In: Forner P, Müller H, Paredes R, Rosso P, Stein B , eds. Information Access Evaluation. Multilinguality, Multimodality, and Visualization . Berlin, Heidelberg: : Springer; ; 2013. : 212 – 231 . [Google Scholar]
- 9. Kholghi M Sitbon L Zuccon G et al. . Factors influencing robustness and effectiveness of conditional random fields in active learning frameworks . In: Proceedings of the 12th Australasian Data Mining Conference (AusDM 2014) . Brisbane, Australia: : Queensland University of Technology: Australian Computer Society Inc; 2014; . [Google Scholar]
- 10. Lafferty JD McCallum A Pereira FCN . Conditional random fields: probabilistic models for segmenting and labeling sequence data . In: Proceedings of the Eighteenth International Conference on Machine Learning (ICML) . San Francisco, CA: : Morgan Kaufmann Publishers Inc; ; 2001. : 282 – 289 . [Google Scholar]
- 11. Pradhan S Elhadad N South B et al. . Task 1: ShARe/CLEF ehealth evaluation lab 2013 . In: Proceedings of CLEF 2013 Evaluation Labs and Workshops: Working Notes . Valencia, Spain: CLEF; 2013 . [Google Scholar]
- 12. Lewis DD Catlett J . Heterogenous uncertainty sampling for supervised learning . In: Proceedings of the 18th International Conference on Machine Learning . Williamstown, MA: : Morgan Kaufmann Publishers Inc; ; 1994. : 148 – 156 . [Google Scholar]
- 13. Settles B Craven M . An analysis of active learning strategies for sequence labeling tasks . In: Proceedings of the Conference on Empirical Methods in Natural Language Processing ; Waikiki, Honolulu, Hawaii: Association for computational Linguistics; 2008:1070–1079 . [Google Scholar]
- 14. Chen Y Mani S Xu H . Applying active learning to assertion classification of concepts in clinical text . J Biomed Inform. 2012. ; 45 ( 2 ): 265 – 272 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosales R Krishnamurthy P Rao RB . Semi-supervised active learning for modeling medical concepts from free text . In: Proceedings of the Sixth International Conference on Machine Learning and Applications . Cincinnati, OH: : IEEE Computer Society; ; 2007. : 530 – 536 . [Google Scholar]
- 16. Zhang H-T Huang M-L Zhu X-Y . A unified active learning framework for biomedical relation extraction . J Comput Sci Technol. 2012. ; 27 ( 6 ): 1302 – 1313 . [Google Scholar]
- 17. Boström H Dalianis H . De-identifying health records by means of active learning . Recall (micro). 2012. ; 97 ( 97.55 ): 90 – 97 . [Google Scholar]
- 18. Figueroa RL Zeng-Treitler Q Ngo LH et al. . Active learning for clinical text classification: is it better than random sampling? J Am Med Inform Assoc. 2012. ; 19 ( 5 ): 809 – 816 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sang EFTK Veenstra J . Representing text chunks . In: Proceedings of the Ninth Conference on European Chapter of the Association for Computational Linguistics . Bergen, Norway: : Association for Computational Linguistics; ; 1999. : 173 – 179 . [Google Scholar]
- 20. Nguyen AN Lawley MJ Hansen DP et al. . A simple pipeline application for identifying and negating SNOMED clinical terminology in free text . In: Proceedings of the Health Informatics Conference (HIC) . Canberra, Australia: : Health Informatics Society of Australia (HISA);2009:188–193 . [Google Scholar]
- 21. Culotta A McCallum A. Reducing labeling effort for structured prediction tasks . In: Proceedings of the National Conference on Artificial Intelligence (AAAI) . Pittsburgh, Pennsylvania: AAAI Press; 2005:746–751 . [Google Scholar]
- 22. McCallum AK. MALLET: A Machine Learning for Language Toolkit . Secondary MALLET: A Machine Learning for Language Toolkit. 2002. http://mallet.cs.umass.edu . [Google Scholar]
- 23. Settles B Craven M Friedland L . Active learning with real annotation costs . In: Proceedings of the NIPS Workshop on Cost-Sensitive Learning . Whistler, BC, Canada: : Neural Information Processing Systems (NIPS) ; 2008. : 1 – 10 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.