Abstract
Objective Traditional Chinese medicine (TCM) is a unique and complex medical system that has developed over thousands of years. This article studies the problem of automatically extracting meaningful relations of entities from TCM literature, for the purposes of assisting clinical treatment or poly-pharmacology research and promoting the understanding of TCM in Western countries.
Methods Instead of separately extracting each relation from a single sentence or document, we propose to collectively and globally extract multiple types of relations (eg, herb-syndrome, herb-disease, formula-syndrome, formula-disease, and syndrome-disease relations) from the entire corpus of TCM literature, from the perspective of network mining. In our analysis, we first constructed heterogeneous entity networks from the TCM literature, in which each edge is a candidate relation, then used a heterogeneous factor graph model (HFGM) to simultaneously infer the existence of all the edges. We also employed a semi-supervised learning algorithm estimate the model’s parameters.
Results We performed our method to extract relations from a large dataset consisting of more than 100 000 TCM article abstracts. Our results show that the performance of the HFGM at extracting all types of relations from TCM literature was significantly better than a traditional support vector machine (SVM) classifier (increasing the average precision by 11.09%, the recall by 13.83%, and the F1-measure by 12.47% for different types of relations, compared with a traditional SVM classifier).
Conclusion This study exploits the power of collective inference and proposes an HFGM based on heterogeneous entity networks, which significantly improved our ability to extract relations from TCM literature.
Keywords: traditional Chinese medicine, relation extraction, heterogeneous entity networks, collective inference, factor graph model
INTRODUCTION
The essential philosophy of traditional Chinese medicine (TCM) is holism, emphasizing the regulation of the integrity of the human body as well as the interaction between individuals and their environment, which provides a distinctive methodology and approach for diagnosing and treating disease. 1 TCM has attracted more and more attention worldwide as an alternative to modern medicine. Hundreds of thousands of TCM researchers have made great efforts to modernize TCM and integrate it with modern medicine. A large number of TCM research articles are published every year. Meanwhile, large-scale analyses of the large body of TCM literature has become an interesting research area in recent years, because such analyses can exploit the collective knowledge of TCM researchers and, in turn, add to the body of medical knowledge.
On the other hand, TCM is a very complex medical system in which multiple types of entities are involved, such as “herb,” “formula” (a composition that consists of certain herbs), “symptom,” and “syndrome” (“zheng” in Mandarin Chinese, a complex pattern of signs and symptoms, which is used as a holistic summary of a patient’s status). 2 Multiple types of intricate relations can exist between these heterogeneous entities, such as composition relations between herbs and formulae, treatment relations between formulae and syndromes, effectiveness relations between herbs and syndromes, and association relations between syndromes and diseases. Establishing these relations is the goal of TCM research. Every researcher contributes his or her discoveries to the TCM knowledge base to form a large-scale, multi-source, and unstructured pool of natural language text data.
In this article, we study the problem of extracting relations from this pool of TCM data. More specifically, given a set of published scientific TCM documents, our goal is to identify all the relations between the instances of different types of entities in those documents. One of the main objectives of relation extraction from TCM literature is to help generate scientific hypotheses and clinical guidelines for practical diagnoses and treatments. 3 Specifically, the knowledge of all TCM researchers (found in the general body of TCM literature) can be integrated into one pool of data, the most significant associations between entities can be extracted from that pool, and these associations can be used to assist clinical treatment or poly-pharmacology research. In addition, the extracted relations may promote the understanding of TCM in Western countries.
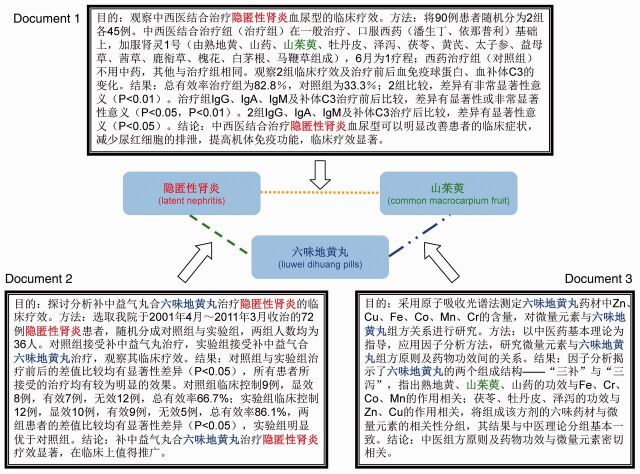
Relation extraction from TCM data is somewhat more complicated than relation extraction from biomedical data. The main challenge of relation extraction from TCM data is the complexity of the TCM system itself. Multiple types of interwoven relations can exist between tens of thousands of heterogeneous TCM entities, so it is not advisable to extract a single type of relation independently of the others. Figure 1 gives an example of extracting multiple types of correlated relations between heterogeneous TCM entities. In addition, the vast majority of TCM literature is written in Chinese, a language in which the sentences have no spaces between words and, therefore, word segmentation is needed to automatically divide sentences into words. Errors in word segmentation obstructs feature generation in the relation extraction process.
Figure 1:
An example of extracting multiple types of correlated relations between heterogeneous traditional Chinese medicine (TCM) entities. Words in red font indicate a disease, words in blue font indicate a formula, and words in green font indicate an herb. The relations extracted from different documents are correlated with one another through their common entities.
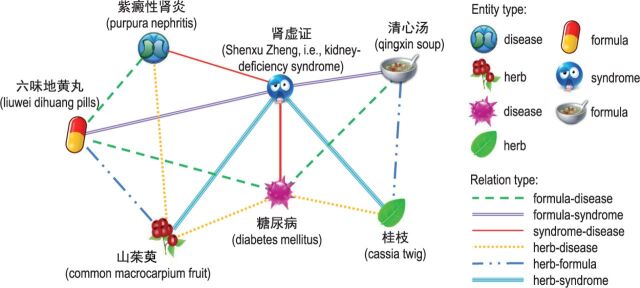
In this article, we propose a novel approach to relation extraction, to collectively and globally extract relations from the entire corpus of TCM literature from the perspective of network mining. In this approach, we first construct heterogeneous entity networks from the TCM literature. Specifically, we take all types of TCM entities that occur in the literature as nodes and create an edge for each pair of heterogeneous entities co-occurring in the same document. All the edges are treated as candidate relations to be identified. Figure 2 gives a simple example of a heterogeneous TCM network. We then propose a unified graphical model, called the heterogeneous factor graph model (HFGM), to simultaneously infer the labels of all the candidate relations by employing the concept of collective inference. 4,5
Figure 2:
An example of a simple heterogeneous traditional Chinese medicine (TCM) network. Different icons and line styles indicate different types of TCM entities and candidate relations, respectively, which are closely correlated with one another. Many triadic closures are formed in the network.
To evaluate the performance of our proposed method, we collected a dataset consisting of more than 100 000 article abstracts from a Chinese publication database and randomly annotated a sample of the relations found therein. We trained and evaluated our HFGM on this partially labelled dataset in a semi-supervised way. Our results demonstrate that our proposed method performs very well at extracting multiple types of relations, including herb-syndrome, herb-disease, formula-syndrome, formula-disease, and syndrome-disease relations.
RELATED WORK
Relation extraction has recently attracted increasing interest in the information extraction community, and many related methods have been successfully applied in the medical field. 6 Compared with open information extraction, 7–9 which tries to find all potentially useful facts and extract as many relations as possible from a large and diverse corpus in which no relation type is specified in advance, medical relation extraction only focuses on deriving the most salient prespecified types of relations from a single domain.
Many techniques have been proposed for biomedical relation extraction, in which a variety of biomedical relations (such as the interactions between proteins, genes, phenotypes, biological targets, and diseases) have been the subject of relation extraction tasks. The simplest method of biomedical relation extraction is to calculate the co-occurring frequency of the entity pairs. 10 This method commonly results in high recall but low precision. 11,12 Other researchers use part-of-speech rules 13 defined by domain experts 14 or derived from an annotated corpus 15 to describe the linguistic patterns of particular relations, a method that exhibits high precision but low recall. Classification-based approaches are also commonly used to identify biomedical relations. Roberts et al. 16 and Rink et al. 17 both describe a supervised classification system for detecting various clinical relations. Bundschus et al. 18 used conditional rand fields to identify disease-treatment and gene-disease relations. Abach and Zweigenbaum 19 propose a hybrid approach that combines patterns defined by domain experts as well as a support vector machine (SVM) classifier for extracting relations between diseases and treatments. Syntactic structures are also thought to assist relation extraction. Miyao et al. 20 performed deep parsing to annotate predicate-argument structures in order to identify and retrieve relational concepts from MEDLINE abstracts. Fundel et al. 21 produced dependency trees and generated syntactic rules to identify gene and protein associations. Similarly, Rinaldi et al. 22 used dependency trees to support the process of querying interactions between genes and proteins. Kernel methods have also been employed in the relation extraction process. 23–25
Related works on relation extraction from the TCM literature are scarce. One of the pioneering works on this subject is a study by Wu et al., 26 in which the authors used a bootstrapping method to extract syndrome-disease associations from a corpus of data. Based on this study, Zhou et al. 27 developed an integrative data mining system, called MeDisco/3S, to identify relations between syndromes and genes. Fang et al. 28 integrated the association information between entities in both TCM and modern medicine literature into a database system named TCMGeneDIT, in which a co-occurrence-based method and a rule-based method are used to extract different types of relations. In a recent work by Xue et al., 29 the authors used a TCM integrated database (TCMID), which contains the most comprehensive information on TCM entities and relations to date, and employed co-occurrence to collect relations between herbs, ingredients, and targets from TCM articles published in Chinese.
All the above-mentioned methods assume that the labels of all relations are independently and identically distributed. However, these heterogeneous relations may be dependent on one another, through their common entities. For example, if we have identified one relation between herb A and formula B and another relation between formula B and syndrome C, then it is very likely that there is a relation between herb A and the syndrome C. This is a kind of transitive property among relations. If we can adequately model the widespread dependencies in the literature, it is likely that these dependencies will greatly help us to identify relations. However, the previous methods cannot capture such dependencies.
In this article, we propose a novel HFGM to incorporate all the data gathered from the body of TCM literature into a unified framework, for better identifying TCM relations. A factor graph 30 is a type of probabilistic graphical model that provides an elegant way of representing graphical structure with more emphasis on the factorization of the distribution of data. Many modified factor graph models have been proposed and successfully used in social network analyses, such as those measuring social influence, 31 mining social relationships, 32 inferring social ties, 33,34 and predicting reciprocal interactions. 35
METHODS
Data Collection and Annotation
We collected abstracts of published articles on TCM from a Chinese publication database, then used four authoritative terminology dictionaries to detect TCM entities in the text of these abstracts. Next, we generated candidate relations between all co-occurring heterogeneous entities. Finally, a sample of relations was labelled by domain experts.
Terminology Dictionaries
Extracting relations from text first requires recognizing instances of the entities. Studying entity recognition is not within the scope of this article. Several relatively complete TCM terminology dictionaries have been published in online TCM databases such as TCMonline and TCMID. TCMonline ( http://www.cintcm.com ) is the earliest online TCM database system, built by the Institute of Information on Traditional Chinese Medicine at the China Academy of Chinese Medical Science in 1984. TCMID ( http://www.megabionet.org/tcmid ), which was built by the Institute of Biomedical Sciences at East China Normal University within the last few years, is a comprehensive database that provides information on and bridges the gap between TCM and modern life sciences. We collected dictionaries directly from these databases to detect the instances of entities in TCM literature.
We collected four TCM entity dictionaries by copying and merging the names of the four types of entities from both the TCMonline and TCMID databases: an herb dictionary that contains 8082 herbs, a formula dictionary that contains 39 932 formulae, a syndrome dictionary that contains 2209 syndromes, and a disease dictionary that contains 3316 Medical Subject Headings (MeSH) ( http://www.nlm.nih.gov/mesh ) disease terms.
TCM Literature
We collected a corpus of data from the China National Knowledge Infrastructure (CNKI) ( http://www.cnki.net ), which is one of the largest online Chinese publication databases. The collected corpus of data contains the abstracts of all 106 150 papers published in the 114 most popular Chinese TCM journals over the past 5 years, which almost covers all the aspects of TCM research. We used the four terminology dictionaries mentioned above to detect entities that occurred in the corpus of data, and found 3024 herbs, 4957 formulae, 1126 syndromes, and 1650 diseases. We then generated the candidate relations between the co-occurring heterogeneous entity pairs and identified: 11 197 herb-syndrome candidate relations, 11 755 herb-disease candidate relations, 9659 formula-syndrome candidate relations, 7882 formula-disease candidate relations, and 9645 syndrome-disease candidate relations. Note that we did not extract herb-formula relations, because all the formulae in the dictionary are classical TCM formulae, and their relations with herbs have already been well-defined.
Data Annotation
For training and quantitatively evaluating our proposed model, we randomly labelled a small fraction (ie, 10%) of each type of candidate relation. We asked three TCM experts (respectively denoted as Kang, Tang, and Zhan, according to their family names), who specialize in Chinese materia medica and clinical TCM, to annotate the data. For each candidate relation to be labelled, the three domain experts read the corresponding papers’ abstracts to identify whether the two end-entities were really related or had just co-occurred by chance. The statistics of our dataset are summarized in Table 1 , and the entire dataset is available online at http://arnetminer.org/TCMRelExtr .
Table 1:
Statistics of the Dataset
| Relation Type | Number of Unique Candidate Relations | ||
|---|---|---|---|
| Labelled | Unlabelled | ||
| Positive | Negative | ||
| Herb-Syndrome | 538 | 582 | 10 077 |
| Herb-Disease | 534 | 642 | 10 579 |
| Formula-Syndrome | 392 | 574 | 8693 |
| Formula-Disease | 377 | 411 | 7094 |
| Syndrome-Disease | 431 | 532 | 8681 |
We used Kappa statistics to measure inter-annotator agreements. As shown in Table 2 , the Cohen’s Kappa 36 scores between any two annotators and the Fleiss’s Kappa 37 scores among the three annotators are mostly above 0.7, indicating substantial agreement. After that, we employed the majority rule to decide the final label of each candidate relation.
Table 2:
Inter-annotator Kappa Agreements
| Relation Type | Cohen’s κ Score | Fleiss’s κ Score | ||
|---|---|---|---|---|
| Kang-Tang | Tang-Zhan | Kang-Zhan | Kang-Tang-Zhan | |
| Herb-Syndrome | 0.7199 | 0.7537 | 0.7236 | 0.7032 |
| Herb-Disease | 0.8271 | 0.8657 | 0.7994 | 0.7789 |
| Formula-Syndrome | 0.7983 | 0.8431 | 0.8323 | 0.8264 |
| Formula-Disease | 0.6849 | 0.8176 | 0.8124 | 0.7735 |
| Syndrome-Disease | 0.7460 | 0.8867 | 0.8411 | 0.8110 |
Problem Definition
In this paper, we propose extracting relations in the context of heterogeneous TCM networks. So, we first give the definition of heterogeneous TCM networks, then present the problem formulation.
We use and to denote the set of types of TCM entities and relations, respectively. In this paper, we have and , where , , , and represent the entities “herbs,” “formulae,” “syndromes,” and “diseases,” respectively, and , , , , , and represent the relations “herb-formula,” “herb-syndrome,” “herb-disease,” “formula-syndrome,” “formula-disease,” and “syndrome-disease,” respectively.
Heterogeneous TCM Networks : Let be a set of TCM entities of type and be a set of TCM relations of type . We define a heterogeneous TCM network as a graph , where , and is a set of attribute matrices. Each matrix is associated with the edges of type , where is the number of attributes of type , each row of matrix corresponds to an edge, each column corresponds to an attribute, and an element denotes the value of the k -th attribute of edge .
TCM Relation Extraction Problem : Given a heterogeneous TCM network, , then our objective is to learn a function to predict the label of candidate relations between TCM entities, ie,
Where is the label space of the problem.
In this work, our goal is to identify the correctness (ie, reliability) of each candidate relation; therefore, we have , where the label of 1 means an edge is reliable and 0 means it is unreliable.
Heterogeneous Factor Graph Model
Model Framework
Given a heterogeneous TCM network , we use to indicate the label of edge of type . Let and . Our objective is to then estimate the values of , and we can use a joint posterior probability to model its distribution. Here, denotes all forms of network information. This joint probability indicates that the labels of the edges depend not only on the local attributes associated with each edge, but also on the structure of the network.
A factor graph provides a way to factorize the “global” joint probability as a product of “local” factors, each of which depends on a subset of variables in the graph. To represent the dependencies between the labels and the attributes and the correlations among the labels , we can define the following two categories of local factors:
Evidence factors , which are used to capture the dependencies between the labels of edges and their attributes. For instance, an evidence factor represents the dependency of the label of an edge on its attributes .
Compatibility factors , which are used to capture the compatibility among the labels of edges. We use triadic closures in heterogeneous TCM networks to construct compatibility factors. Triadic closure 38 is one of the fundamental processes of linking information in a network and has been applied in many aspects of social network mining, such as in inferring social ties 34 as well as social roles and statuses. 39 We use to denote the set of types of triadic closures, where , , , and represent “herb-formula-syndrome” relations, “herb-formula-disease” relations, “herb-syndrome-disease” relations, and “formula-disease-syndrome” relations, respectively. Let be a triadic closure of type and its corresponding subset of labels; then, the compatibility factor indicates the correlations among the labels in .
Then, the joint probability can be factorized as follows:
| (1) |
Where is the subset of edges of type , and is the subset of triadic closures of type .
The factors and can be instantiated by exponential-linear functions:
| (2) |
| (3) |
Where and are local normalization factors; is the feature function of the m -th attribute associated with edge of type , is its weight, and is the total number of attributes of edges of type . Equation 3 indicates that we define feature functions for each triadic closure of type , and is the weight of the n -th feature function .
The joint probability defined in Equation 1 can be rewritten as:
| (4) |
Where is a global normalization factor and are the parameters to be estimated.
Figure 3 gives the graphical representation of an HFGM. The dotted ellipse at the bottom of the figure encloses the constructed heterogeneous TCM network, in which a node represents a TCM entity of a certain type. The dotted ellipse in the middle of the figure encloses the set of candidate relations, each of which corresponds to an edge in the input network. The dotted ellipse at the top of the figure encloses the factor graph generated from the input network, in which the colored ovals represent variables (labels) corresponding to the candidate relations, and the squares represent factors. The green ovals represent the known labels that are taken as supervised information, while the red ovals represent the unknown labels to be predicted. The black squares represent the evidence factors between variables and their attributes, while the blue squares represent the compatibility factors of triadic closures.
Figure 3:
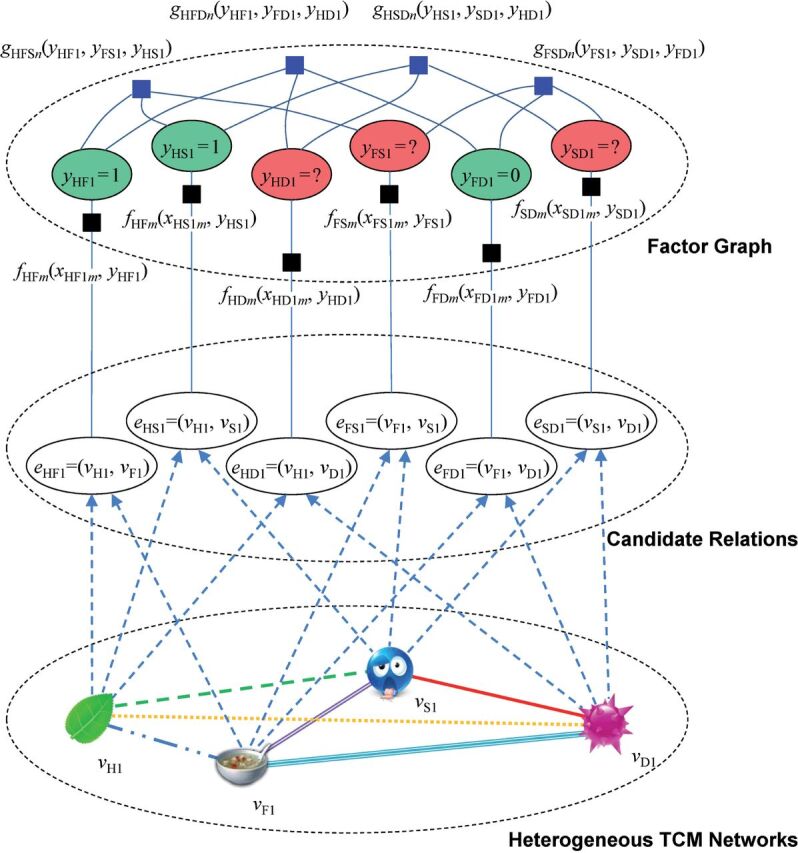
Graphical representation of a heterogeneous factor graph model (HFGM). Here, represents a traditional Chinese medicine (TCM) entity of a certain type (eg, represents an herb), represents an edge (a candidate relation) of a certain type (eg, represents an herb-formula relation), represents the corresponding variable (label) of an edge (eg, represents the label of the edge ), represents a feature function for evidence factors defined on the observed attributes, and represents a feature function for compatibility factors defined among latent labels. The labels , , and are known in advance and are taken as supervised information, while the labels , , and are unknown and yet-to-be-predicted.
Features in Consideration
In general, the features of evidence factors should be able to reflect prior knowledge of the labels of edges. In this study, we employed three categories of features for evidence factors:
Co-occurring frequency : This is the simplest feature, which represents the instances in which the two end-entities of a relation co-occur in the same document.
Lexical context : Intuitively, the context surrounding the two end-entities is very important for identifying a relation. We used ICTCLAS (Institute of Computing Technology, Chinese Lexical Analysis System; http://ictclas.org ) to segment the Chinese documents into words, and took the surrounding words of each entity as features. Six surrounding words were collected for each instance of an entity, three before and three after the instance. After removing the infrequent words, 9784 distinct words remained. We then defined a feature function for each of the words, to indicate the frequency that each word appeared around the two end-entities of a relation.
Semantic distance : Determining the latent semantic relatedness of the two end-entities may also be helpful for identifying a relation. For calculating semantic distance, we first need to represent the semantic meanings of each entity. Distributed vector representations facilitate learning word meanings from large collections of text. Each word is learned as a distinct pattern of continuous values over a single, large vector, with each dimension corresponding to a latent topic. We can then measure the semantic relatedness among words in terms of distances in the resulting vector space. We used word2vec 40 ( https://code.google.com/p/word2vec ), an efficient tool for computing the vector representations of words by employing deep-learning approaches, to calculate the semantic vectors of TCM entities from our corpus of data, and, with this tool, we generated a 200-dimension vector for each entity. We then defined a feature function on each dimension by using the absolute value of the difference between the values of the dimensions of the two end-entities.
Except for the above three categories of features, syntactic structure (ie, dependency relation) is another type of information that is useful for relation extraction. However, being able to determine syntactic structure requires that the two end-entities co-occur within one single sentence. So, we did not take syntactic structure into account in this study.
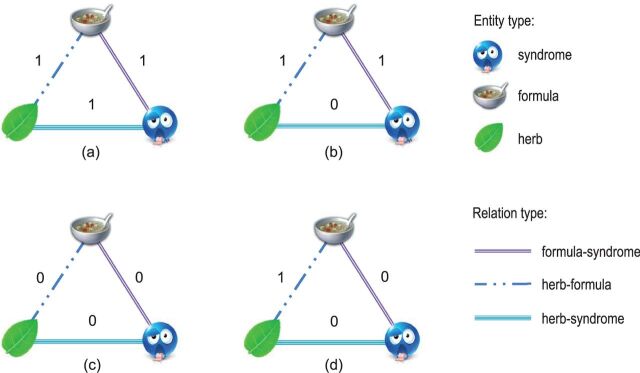
The feature functions of compatibility factors should be able to reflect dependencies among the labels of edges. We defined one category of feature functions for compatibility factors based on the transitive property of triadic closures. Figure 4 gives an illustration of the transitive property, in which the labels of three edges form a triad closure. According to the transitive property, we can define the feature function as follows:
Figure 4:
Illustration of the transitive property of triadic closures. ( a ) Complies with the transitive property, because all the three labels are equal to 1; ( b ) violates the transitive property, because only two labels are equal to 1; for ( c ) and ( d ) the transitive property does not apply, because only one label or no labels are equal to 1.
Learning and Inferring the HFGM
We propose using a semi-supervised learning algorithm to estimate the parameters of the model (see Supplementary Appendix A ), which enables us to predict the labels of unknown edges based on the estimated parameters (see Supplementary Appendix B ).
RESULTS
We trained and evaluated our proposed HFGM on the dataset we collected and annotated. Five types of relations – herb-syndrome, herb-disease, formula-syndrome, formula-disease, and syndrome-disease – were extracted from the TCM literature we analyzed.
The HFGM model was implemented in C++, and all experiments were conducted on a server running Windows Server 2008, with an Intel Xeon CPU E7-4820 2.00 GHz processor and 256 GB of memory. The entire semi-supervised learning and inference process took about 4.5 h.
A traditional classification approach was employed as the baseline, in which the co-occurring frequency, lexical context, as well as the aforementioned semantic distance were taken as the classification features. Because the number of features involved in this approach was very large, we used an SVM 41 ( www.csie.ntu.edu.tw/∼cjlin/libsvm ) as the basic classifier.
In order to let the basic classifier utilize the relations’ compatibility information, we also ran an iterative classification algorithm on our relation extraction task. Compared with probabilistic graphical models (eg, factor graph models), iterative classification provides a simple and approximate way of implementing the concept of collective inference, by graphing interdependent variables and taking the inferred neighboring values of a variable as known information to assist with iteratively inferring the variable’s value. We designed an iterative SVM classifier in our experiments that takes the results of the basic SVM classifier as the initiate values of all the labels, then iteratively updates the label of each relation by simultaneously using its observed features and inferred neighboring label values, until the labels no longer change. The key challenge of iterative classification is transforming the neighboring label values into regular features in order to capture the labels’ compatibility information. In this study, we also defined iterative features based on the transitive property of triadic closures. Specifically, we calculated the numbers of triadic closures with different sums (ie, 0, 1, 2, and 3, as shown in Figure 4 ) that were formed by each relation with its neighboring relations.
We performed a five-fold cross-validation to evaluate the performance of our model. Table 3 shows the performance of the HFGM as compared with that of two other approaches. Our results show that the HFGM is more efficient at extracting relations from the TCM literature compared with the basic SVM classifier (increasing precision by ∼10–12%, recall by ∼12–15%, and the F1-measure by ∼11–14%, for different types of relations) and the iterative SVM classifier (increasing precision by ∼6–8%, recall by ∼7–10%, and the F1-measure by ∼7–11%, for different types of relations). We performed a t -test between the performances of these three approaches with regard to the extraction of different types of relations and found that the difference between them is extremely statistically significant ( P < 0.001 for all precision, recall, and F1-measures).
Table 3:
Performance of TCM Relation Extraction by Different Approaches (%)
| Relation Type | HS | HD | FS | FD | SD | Average | |
|---|---|---|---|---|---|---|---|
| Basic SVM | Precision | 78.89 | 79.13 | 80.12 | 81.04 | 77.72 | 79.30 |
| Recall | 72.34 | 74.59 | 72.32 | 73.08 | 73.22 | 73.15 | |
| F1 | 75.47 | 76.79 | 76.02 | 76.85 | 75.40 | 76.09 | |
| Iterative SVM | Precision | 83.35 (+4.46) | 83.1 (+3.97) | 84.33 (+4.21) | 85.55 (+4.51) | 81.88 (+4.16) | 83.54 (+4.24) |
| Recall | 77.66 (+5.32) | 79.51 (+4.92) | 77.75 (+5.43) | 78.41 (+5.33) | 78.34 (+5.12) | 78.36 (+5.21) | |
| F1 | 80.4 (+4.93) | 81.27 (+4.48) | 80.91 (+4.89) | 81.82 (+4.97) | 80.07 (+4.67) | 80.87 (+4.78) | |
| HFGM | Precision | 90.94 (+12.05) | 89.48 (+10.35) | 90.81 (+10.69) | 91.07 (+10.03) | 89.87 (+12.15) | 90.39 (+11.09) |
| Recall | 86.93 (+14.59) | 87.34 (+12.75) | 85.69 (+13.37) | 88.25 (+15.17) | 86.87 (+13.65) | 86.98 (+13.83) | |
| F1 | 88.89 (+13.42) | 88.40 (+11.60) | 88.18 (+12.16) | 89.64 (+12.78) | 87.86 (+12.94) | 88.56 (+12.47) | |
FD, formula-disease relation; FS, formula-syndrome relation; HD, herb-disease relation; HFGM, heterogeneous factor graph model; HS, herb-syndrome relation; SD, syndrome-disease relation; SVM, support vector machine; TCM, traditional Chinese medicine.
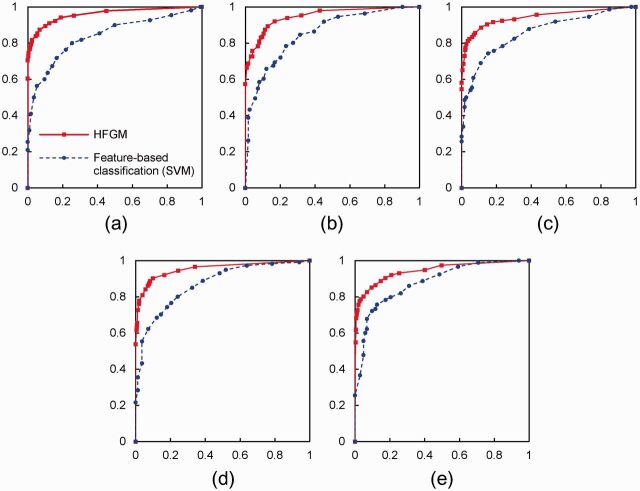
To further determine the effectiveness of our approach, we plotted receiver operating characteristic curves of the basic SVM and HFGM approaches (as shown in Figure 5 ), where the y -axis represents the rate of predicated positive labels in all the positive samples, and the x -axis represents the rate of predicted positive labels in all the negative samples. It is clear that the HFGM significantly outperforms the basic SVM classifier on extracting all types of relations.
Figure 5:
Receiver operating characteristic curves of different approaches for extracting ( a ) herb-syndrome, ( b ) herb-disease, ( c ) formula-syndrome, ( d ) formula-disease, and ( e ) syndrome-disease relations. The y -axis represents the true positive rate and the x -axis represents the false positive rate.
DISCUSSION
After performing an in-depth analysis of some specific instances, we found that our HFGM significantly improves the accuracy of relation extraction in the following cases (in which traditional classifiers have difficult identify relations):
The context is very short. For a candidate relation, we took the co-occurring frequency of the two end-entities appearing in the same documents and the frequencies of their surrounding words as the classification features, so if a relation only appears once in a single, short document, then the context information will not be enough to identify the relation.
The context contains confusing information . In some TCM treatment experiments, one or more control groups are used to compare the effectiveness of different treatments, which often misleads traditional classifiers to extract some relations from the control groups that are not actually present.
Several different studies are reported on in the same document. Occasionally, several studies will be covered within a single article or even a single sentence. An example of this would be if an author wrote, “the herb A is curative to the disease B; the herb C is curative to the disease D.” In this example, it would be difficult for traditional classifiers to distinguish the correct relations (A–B and C–D) from the incorrect relations (A–C, A–D, B–C, and B–D), which may lead traditional classifiers to extract some incorrect relations from the document.
The name of an entity is a polysemous word. Some Chinese names for TCM entities have other meanings. For instance, the term “
 ” refers to a syndrome called “Tai Yang” in the TCM domain, but it means “the sun” in most contexts, and the term “
” refers to a syndrome called “Tai Yang” in the TCM domain, but it means “the sun” in most contexts, and the term “
 ” refers to dehydration in a medical context, but can also mean “the physical process of dewatering.” Such polysemous terms complicate the process of identifying candidate relations.
” refers to dehydration in a medical context, but can also mean “the physical process of dewatering.” Such polysemous terms complicate the process of identifying candidate relations.
Our HFGM utilizes the correlations between all types of relations to overcome the above difficulties by employing collective inference in the context of heterogeneous entity networks, thus greatly improving the model’s ability to extract TCM relations. The effectiveness of the HFGM demonstrates the existence of correlations among different types of relations. In addition, using the transitive property of triadic closures to model the dependencies among the labels of edges in the network is a reasonable and practicable approach.
However, some of our results can be improved upon, and some of the approaches we employ can be expanded upon in the future. Firstly, some other types of important TCM entities, such as symptoms, are not incorporated into our model. This is because there is not a standard or unified terminology glossary for TCM symptoms, so entity recognition techniques are needed to detect the instances of symptom entities in text. If we can bring such entities into our unified model in the future, then more types of relations can be extracted. Secondly, many biomedical discoveries, such as known disease-target and ingredient-target relations or research on the integration of Chinese and Western medicine (eg, established herb-ingredient relations), can be used as prior knowledge in our model and are expected to further improve the model’s performance.
Another challenge is that the computational complexity of learning the HFGM is very high, because multiple rounds of approximate inferences are required over the entire dataset (see Algorithm 1 of the online supplementary data ). Consequently, we need to develop efficient learning approaches. Tang et al. 33 has proposed a parallel algorithm to learn factor graph models, which can be used for reference in our future research. In addition, other approximation techniques, such as the pseudolikelihood measure, 42,43 may also be used in our collective inference methods.
Our approach also be directly applied to relation extraction in the field of biomedical text mining. We can construct heterogeneous networks between biomedical entities (eg, proteins, genes, phenotypes, biological targets, diseases, drugs, treatments) gathered from biomedical literature or clinical records, then employ the HFGM to extract biomedical relations in the context of these heterogeneous biomedical networks.
The HFGM model proposed in this article is only suitable for heterogeneous entity networks that contain at least three kinds of entities, because we use triadic closures formed by three kinds of entities to construct compatibility factors in the model.
Another limitation of the current version of our model is that it can only extract one class of relations between the same two types of entities at the same time, because we treat the relation extraction problem as a binary classification problem in this study. For instance, the annotated dataset in this study contains only “herb-treatment-disease” relations, so the learned model also can only extract “herb-treatment-disease” relations. However, if the user wants to extract “herb-hasSideEffect-disease” relations, they may use the data that contains annotated “herb-hasSideEffect-disease” relations to re-train the model.
CONCLUSION
In this article, we examine the problem of automatically extracting meaningful entity relations from TCM literature and propose an HFGM that exploits the power of collective inference in the context of heterogeneous entity networks to simultaneously and globally extract all types of relations (eg, herb-syndrome, herb-disease, formula-syndrome, formula-disease, and syndrome-disease relations) from the entire corpus of TCM data. We propose using a semi-supervised learning algorithm to estimate the parameters of the model. The results of our analysis of a professionally annotated dataset show that our approach is superior to traditional classification methods in extracting multiple types of relations from TCM literature.
Supplementary Material
Acknowledgments
This work was mainly done while the first author was visiting KU Leuven, Belgium. We also would like to thank three traditional Chinese medicine experts from the China Academy of Chinese Medical Sciences, Shihuan Tang, Zhilai Zhan, and Liping Kang, for their hard work on data annotation. In addition, we very much appreciate the Chinese word segmentation tool (ICTCLAS), the word semantic vector tool (word2vec), and the support vector machine tool (LIBSVM) that we used in this study.
CONTRIBUTORS
The work was a collaboration between all the authors. H.W., X.Z., Q.M., and L.L. constructed the guidelines for data annotation. H.W., M.F.M., W.L., and J.T. designed the methods and experiments. H.W. and J.T. wrote the code and carried out the experiments. H.W., M.F.M., Q.M., and J.T. analyzed the data, interpreted the results, and drafted the paper. All the authors have made valuable contributions to revising and approving the manuscript.
FUNDING
The work is supported by National High-tech R&D Program (No. 2014AA015103), National Basic Research Program of China (No. 2014CB340500, 2012CB316006), National Natural Science Foundation of China (No. 61222212, 61103065, 61035004), NSFC-ANR (No. 61261130588), and co-funding by Tsinghua University and KU Leuven.
COMPETING INTERESTS
None.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at Supplementary Data .
REFERENCES
- 1. Qiu Z . Traditional medicine: a culture in the balance . Nature. 2007. ; 448 : 126 – 128 . [DOI] [PubMed] [Google Scholar]
- 2. Jiang B Liang X Chen Y et al. . Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome . Sci Rep. 2012. ; 2 : 936 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou X Peng Y Liu B . Text mining for traditional Chinese medical knowledge discovery: a survey . J Biomed Inform. 2010. ; 43 : 650 – 660 . [DOI] [PubMed] [Google Scholar]
- 4. Jensen D Neville J Gallagher B . Why collective inference improves relational classification . Proceedings of the 10th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD'04) . 2004. : 593 – 598 . [Google Scholar]
- 5. Xiang R Neville J . Collective inference for network data with copula latent Markov networks . Proceedings of the 6th ACM International Conference on Web Search and Data Mining (WSDM'13) . 2013. : 647 – 656 . [Google Scholar]
- 6. Aggarwal CC Zhai CX , ed. Mining Text Data . New York, NY: : Springer; ; 2012. . [Google Scholar]
- 7. Banko M Cafarella MJ Soderland S et al. . Open information extraction from the Web . Proceedings of the 20th International Joint Conference on Artificial Intelligence (IJCAI'07) . 2007. : 2670 – 2676 . [Google Scholar]
- 8. Banko M Etzioni O . The tradeoffs between open and traditional relation extraction . Proceedings of the 46th Annual Meeting of the Association for Computational Linguistics (ACL'08) . 2008. : 28 – 36 . [Google Scholar]
- 9. Fader A Soderland S Etzioni O . Identifying relations for open information extraction . Proceedings of the 2011 Conference on Empirical Methods in Natural Language Processing (EMNLP'11) . 2011. : 1535 – 1545 . [Google Scholar]
- 10. Chen ES Hripcsak G Xu H et al. . Automated acquisition of disease-drug knowledge from biomedical and clinical documents: an initial study . JAMIA. 2008. ; 15 : 87–98 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnickel T Weston J Collobert R et al. . Large scale application of neural network based semantic role labeling for automated relation extraction from biomedical texts . PLoS ONE. 2009. ; 4 : e6393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramani AK Bunescu RC Mooney RJ et al. . Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome . Genome Biol. 2005. ; 6 : R40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ono T Hishigaki H Tanigami A et al. . Automated extraction of information on protein-protein interactions from the biological literature . Bioinformatics. 2001. ; 17 : 155 – 161 . [DOI] [PubMed] [Google Scholar]
- 14. Šarić J Jensen LJ Ouzounova R et al. . Extraction of regulatory gene/protein networks from MEDLINE . Bioinformatics. 2006. ; 22 : 645 – 650 . [DOI] [PubMed] [Google Scholar]
- 15. Hakenberg J Plake C Leser U . LLL'05 challenge: genic interaction extraction-identification of language patterns based on alignment and finite state automata . Proceedings of the ICML-2005 Workshop on Learning Language in Logic (LLL'05) . 2005. : 38 – 45 . [Google Scholar]
- 16. Roberts A Gaizauskas R Hepple M . Extracting clinical relationships from patient narratives . Proceedings of the ACL-2008 Workshop on Current Trends in Biomedical Natural Language Processing (BioNLP'08) . 2008. : 10 – 18 . [Google Scholar]
- 17. Rink B Harabagiu S Roberts K . Automatic extraction of relations between medical concepts in clinical texts . JAMIA. 2011. ; 18 : 594 – 600 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bundschus M Dejori M Stetter M et al. . Extraction of semantic biomedical relations from text using conditional random fields . BMC Bioinformatics. 2008. ; 9 : 207 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abacha AB Zweigenbaum P . A hybrid approach for the extraction of semantic relations from MEDLINE abstracts . Proceedings of the 12th International Conference on Computational Linguistics and Intelligent Text Processing (CICLing'11), Part II . 2011. : 139 – 150 . [Google Scholar]
- 20. Miyao Y Ohta T Masuda K et al. . Semantic retrieval for the accurate identification of relational concepts in massive textbases . Proceedings of the 21st International Conference on Computational Linguistics and the 44th Annual Meeting of the Association for Computational Linguistics (COLING ACL'06) . 2006. : 1017 – 1024 . [Google Scholar]
- 21. Fundel K Küffner R Zimmer R . RelEx – Relation extraction using dependency parse trees . Bioinformatics. 2007. ; 23 : 365 – 371 . [DOI] [PubMed] [Google Scholar]
- 22. Rinaldi F Schneider G Kaljurand K et al. . Mining of relations between proteins over biomedical scientific literature using a deep-linguistic approach . Artif Intell Med. 2007. ; 39 : 127 – 136 . [DOI] [PubMed] [Google Scholar]
- 23. Airola A Pyysalo S Bjorne J et al. . All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning . BMC Bioinformatics. 2008. ; 9 : S2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S Yoon J Yang J . Kernel approaches for genic interaction extraction . Bioinformatics. 2008. ; 24 : 118 – 126 . [DOI] [PubMed] [Google Scholar]
- 25. Miwa M Sætre R Miyao Y et al. . Protein-protein interaction extraction by leveraging multiple kernels and parsers . Int J Med Inform. 2009. ; 78 : e39 – e46 . [DOI] [PubMed] [Google Scholar]
- 26. Wu Z Zhou X Liu B et al. . Text mining for finding functional community of related genes using TCM knowledge . Proceedings of the 8th European Conference on Principles and Practice of Knowledge Discovery in Databases (PKDD'04) . 2004. : 459 – 470 . [Google Scholar]
- 27. Zhou X Liu B Wu Z et al. . Integrative mining of traditional Chinese medicine literature and MEDLINE for functional gene networks . Artif Intell Med. 2007. ; 41 : 87 – 104 . [DOI] [PubMed] [Google Scholar]
- 28. Fang YC Huang HC Chen HH et al. . TCMGeneDIT: a database for associated traditional Chinese medicine, gene and disease information using text mining . BMC Complement Altern Med. 2008. ; 8 : 58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xue R Fang Z Zhang M et al. . TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis . Nucleic Acids Res. 2013. ; 41 : D1089 – D1095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kschischang FR Frey BJ Loeliger HA . Factor graphs and the sum-product algorithm . IEEE Trans Inf Theory. 2001. ; 47 : 498 – 519 . [Google Scholar]
- 31. Tang J Sun J Wang C et al. . Social influence analysis in large-scale networks . Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD'09) . 2009. : 807 – 816 . [Google Scholar]
- 32. Wang C Han J Jia Y et al. . Mining advisor-advisee relationships from research publication networks . Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD'10) . 2010. : 203 – 212 . [Google Scholar]
- 33. Tang W Zhuang H Tang J . Learning to infer social ties in large networks . Proceedings of the 2011 European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML/PKDD'11) . 2011. : 381 – 397 . [Google Scholar]
- 34. Tang J Lou T Kleinberg J . Inferring social ties across heterogeneous networks . Proceedings of the 5th ACM International Conference on Web Search and Data Mining (WSDM'12) . 2012. : 743 – 752 . [Google Scholar]
- 35. Lou T Tang J Hopcroft J et al. . Learning to predict reciprocity and triadic closure in social networks . ACM Trans Knowl Discov Data. 2013. ; 7 : 5 . [Google Scholar]
- 36. Cohen J . A coefficient of agreement for nominal scales . Educ Psychol Meas. 1960. ; 20 : 37 – 46 . [Google Scholar]
- 37. Fleiss JL . Measuring nominal scale agreement among many raters . Psychol Bull. 1971. ; 76 : 378 – 382 . [Google Scholar]
- 38. Romero DM Kleinberg JM . The directed closure process in hybrid social-information networks, with an analysis of link formation on Twitter . Proceedings of the AAAI International Conference on Weblogs and Social Media (ICWSM’10) . 2010. : 138 – 145 . [Google Scholar]
- 39. Zhao Y Wang G Yu PS et al. . Inferring social roles and statuses in social networks . Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD'13) . 2013. : 695 – 703 . [Google Scholar]
- 40. Mikolov T Sutskever I Chen K et al. . Distributed representations of words and phrases and their compositionality . Proceedings of the 27th Annual Conference on Neural Information Processing Systems (NIPS'13) . 2013. : 3111 – 119 . [Google Scholar]
- 41. Chung C Lin CJ . LIBSVM: a library for support vector machines . ACM Trans Intell Syst Technol. 2011. ; 2 : 27 . [Google Scholar]
- 42. Besag J . Statistical analysis of non-lattice data . The Statistician. 1975. ; 24 : 179 – 195 . [Google Scholar]
- 43. Wan H Lin Y Wu Z et al. . A community-based pseudolikelihood approach for relationship labeling in social networks . Proceedings of the 2011 European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML/PKDD'11) . 2011:491 – 505 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.