Abstract
Introduction.
Salivary metabolite profiles are altered in adults with HIV compared to their uninfected counterparts. Less is known about youth with HIV and how oral disorders that commonly accompany HIV infection impact salivary metabolite levels.
Objective:
As part of the Adolescent Master Protocol multi-site cohort study of the Pediatric HIV/AIDS Cohort Study (PHACS) network we compared the salivary metabolome of youth with perinatally-acquired HIV (PHIV) and youth HIV-exposed, but uninfected (PHEU) and determined whether metabolites differ in PHIV vs. PHEU.
Methods:
We used three complementary targeted and discovery-based liquid chromatography-tandem mass spectrometry (LC-MS/MS) workflows to characterize salivary metabolite levels in 20 PHIV and 20 PHEU youth with and without moderate periodontitis. We examined main effects associated with PHIV and periodontal disease, and the interaction between them.
Results:
We did not identify differences in salivary metabolite profiles that remained significant under stringent control for both multiple between-group comparisons and multiple metabolites. Levels of cadaverine, a known periodontitis-associated metabolite, were more abundant in individuals with periodontal disease with the difference being more pronounced in PHEU than PHIV. In the discovery-based dataset, we identified a total of 564 endogenous peptides in the metabolite extracts, showing that proteolytic processing and amino acid metabolism are important to consider in the context of HIV infection.
Conclusion.
The salivary metabolite profiles of PHIV and PHEU youth were overall very similar. Individuals with periodontitis particularly among the PHEU youth had higher levels of cadaverine, suggesting that HIV infection, or its treatment, may influence the metabolism of oral bacteria.
Keywords: Targeted metabolomics, mass spectrometry, HIV infection, HAART, periodontal disease, biomarkers
Introduction
Human immunodeficiency virus (HIV) infection increases the risk of many opportunistic infections including those associated with oral and periodontal disorders (Lloyd 1996; Ryder et al. 2012). Oral manifestations are frequently the first clinical presentation of HIV infection and periodontal disorders are among the most prevalent oral diseases in HIV-infected adults (Reddy 2007). HIV-infected youth who were perinatally infected have a high risk of developing HIV- and antiretroviral therapy-related periodontal disease and other comorbidities later in life (Ryder et al. 2020). Only a small number of studies have examined the relationship between periodontal health and HIV in perinatally HIV-infected (PHIV) youth (Howell et al. 1996; Vaseliu et al. 2005). Our team recently compared the prevalence and severity of periodontal disease between youth with perinatally-acquired HIV (PHIV) and youth HIV-exposed, but uninfected (PHEU) enrolled in the Adolescent Master Protocol (AMP) multi-site cohort study of the Pediatric HIV/AIDS (PHACS) network (Ryder et al. 2017). We hypothesized that HIV will distort the composition of the oral microbiome which in turn will result in altered host inflammatory response. In our study, PHIV youth had fewer health-associated taxa, suggesting that HIV infection or its treatment contribute to oral dysbiosis (Starr et al. 2018). Monitoring alterations in gingival health that typically precede more advanced periodontal disease manifestations could help in managing the risk for progression to significant periodontal disease. Oral metabolites reflect the interplay between host and microbes in the oral environment and therefore characterization of metabolite levels can provide valuable insights into underlying pathophysiological processes. Advances in mass spectrometry-based profiling techniques have enabled the large-scale detection and quantitation of salivary components (Álvarez-Sánchez et al. 2012; Cuevas-Córdoba, Santiago-García 2014; Zhang et al. 2012). Prior studies have examined the salivary metabolome in the context of a diverse set of disorders and utilized analytical approaches including capillary electrophoresis-MS (Sugimoto et al. 2010; Tsuruoka et al. 2013), gas chromatography (GC)-MS (Kageyama et al. 2015; Kuboniwa et al. 2016; Mueller et al. 2014), liquid chromatography (LC)-MS (Barnes et al. 2014; Liebsch et al. 2019; Wang et al. 2014; Wei et al. 2011; Zheng et al. 2012), nuclear magnetic resonance (NMR) spectroscopy (Neyraud et al. 2020; Romano et al. 2018) or a multi-platform-approach (Dame et al. 2015).
Human saliva serves many biological functions, such as mucosa lubrication, enzymatic digestion of nutrients, protection of dental substance, as well as prevention of bacterial and viral infections (Mandel, 1987; Spielmann and Wong, 2011). It consists of a large number of organic and inorganic components, such as electrolytes, DNA, proteins, peptides, mucus, and small metabolites, whose composition is influenced by the varying environmental, pathological and physiological state of an individual. To date, there has been only one study examining the effect of HIV infection and highly-active antiretroviral therapy (HAART) on oral metabolites in adults. Analysis of oral washes by LC/MS and GC/MS detected elevated levels of mainly amino acids while carbohydrate metabolites were mainly decreased (Ghannoum et al. 2013). In plasma, initiation of antiretroviral therapy has been shown to primarily lead to persistent changes in phospholipid metabolism (Peltenburg et al. 2018).
The purpose of this study was to examine, for the first time, the associations between salivary metabolites and perinatal HIV infection in youth adjusted for oral health status (e.g. healthy vs. moderate periodontitis). To this end, we utilized three complementary LC-MS/MS approaches for the simultaneous detection and quantitation of metabolites in clinical saliva samples (Figure 1): 1) targeted multiple-reaction monitoring (MRM) LC-MS/MS for the absolute quantitation of metabolites selected a priori as representative compounds from central metabolic pathways; 2) data-independent acquisition (DIA, also known as SWATH) for non-targeted quantitation and 3) data-dependent acquisition (DDA) for the identification and relative quantitation of unknown metabolites associated with HIV infection and periodontitis.
Figure 1.
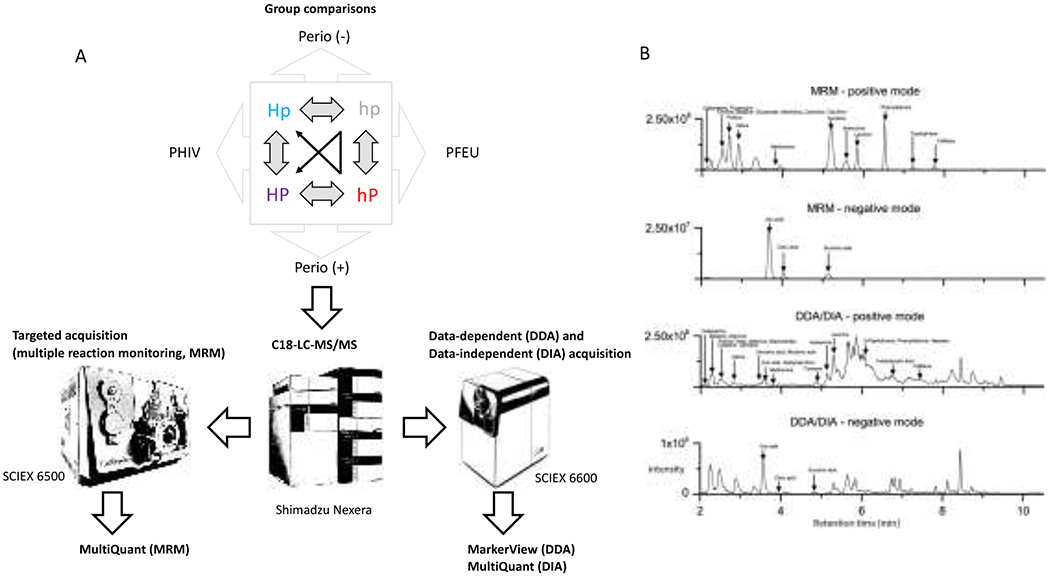
A.) Schematic representation of the targeted (MRM) and discovery metabolomics workflows (DDA and DIA). B.) Representative total ion chromatograms of MRM- and DDA/DIA-LC-MS/MS analyses in positive and negative ionization modes. Elution profiles of major salivary metabolites are indicated.
Material and Methods
Study design and population.
The Oral Health Protocol is a cross-sectional, observational sub-study within the Adolescent Master Protocol (AMP) of PHACS (https://phacsstudy.org) funded by the National Institutes of Health. The AMP is an ongoing prospective cohort study at 14 clinical US sites designed to determine the impact of HIV infection and antiretroviral therapy (ART) on PHIV youth. The AMP includes a comparison group of PHEU youth, for whom the mothers had HIV and were exposed to ART in utero (Siberry et al. 2011; Van Dyke et al. 2011). In this cross-sectional study, 209 PHIV and 126 PHEU participants were enrolled as reported in detail elsewhere (Moscicki et al. 2016). Institutional review boards (IRBs) at clinical sites and the Harvard T.H. Chan School of Public Health approved the study. Parents/legal guardians provided written informed consent for their child’s participation. Youth consented/assented per local IRB guidelines. A subset of 40 study subjects matched by age and sex was selected for this study: 10 PHIV and 10 PHEU youth with moderate periodontitis according to CDC/AAP classifications, and 10 PHIV and 10 PHEU youth without periodontal disease (Table 1 and Supplemental Table 1). The PHEU youth represent an important comparison group since they are exposed to HIV-infected mothers and ART in utero similarly to PHIV youth, avoiding potential confounding by these factors.
Table 1.
Summary of participant’s clinical parameters.
| HAART | Periodontitis 1 | Age (Yrs) | Sex | CD4 count (cells/mL) Median, [IQR] | Viral load (percent unsuppressed (>400 copies/ml)) | ||
|---|---|---|---|---|---|---|---|
| hp | PHEU Perio (−) | - | absent | 14.3 ± 1.9 | 5 male, 5 female | - | - |
| Hp | PHIV Perio (−) | + | absent | 15.1 ± 3.3 | 3 male, 7 female | 744 [561.3, 1145] | 20 |
| HP | PHIV Perio (+) | + | moderate | 17.6 ± 1.8 | 3 male, 7 female | 653.5 [580, 883.8] | 20 |
| hP | PHEU Perio (+) | - | moderate | 15.2 ± 2.9 | 5 male, 5 female | - | - |
according to CDC/AAP classifications
Sample Collection.
Regularly scheduled annual AMP visits included physical examination, and chart reviews for medication, diagnoses, CD4 counts and viral load (VL). The one time Oral Health study visit was within three months of an AMP visit, oral mucosal, dental, and periodontal examinations and collection of oral specimens were performed by study-trained dentists (Moscicki et al. 2016). Prior to the dental examination, unstimulated whole saliva (1-3 mL) was collected over a 5 min period and kept on ice (Navazesh 1993). Saliva samples were centrifuged at 12,000×g for 10 min at 4 °C to remove cells and the supernatants were aliquoted and stored at −80°C within 4 h of collection.
Chemicals.
LC-MS-grade acetonitrile, methanol, isopropanol and water were purchased from Fisher Scientific (Hampton, NH, USA). Formic acid (> 99% purity) was supplied by Thermo Scientific (Waltham, MA, USA). All substance standards were obtained from Fisher Chemical and Sigma-Aldrich (St. Louis, MO).
Preparation of calibration standards for quantification.
Calibration standards were prepared by dilution of a stock solution containing 52 metabolite standards with 0.1% formic acid in water. The range of concentrations used for each calibration standard is provided in Supplemental Table 2. All stock solutions were aliquoted and stored at −20 °C until used. MultiQuant 3.0.1 software (SCIEX) was used to extract peak areas from the chromatographic data. Quantification was achieved for each analyte by using linear regression analysis of peak areas versus the concentration of the analytes.
Sample Processing.
Saliva samples (100 μL) were deproteinized using cold methanol; after incubation at −20 °C for 2 h, samples were centrifuged (16,000×g for 10 min at 4 °C) and supernatants evaporated to dryness in a SpeedVac and reconstituted in 100 μL 0.1% formic acid. To remove insoluble material, the centrifugation step was repeated prior to LC-MS/MS analysis.
Targeted LC-MS/MS-analysis by multiple-reaction monitoring (MRM).
Targeted quantitation experiments were performed at the Forsyth Center for Salivary Diagnostics on a SCIEX QTRAP 6500 triple quadrupole mass spectrometer (SCIEX, Framingham, MA, USA) equipped with an Ion Drive Turbo V ESI source hyphenated to a Shimadzu Nexera XR HPLC system (Shimadzu, Kyoto, Japan). A Poroshell 120 EC-C18 (4.6x100 mm, 2.7-micron particle size) column (Agilent Technologies, Santa Clara, CA, USA) guarded by a Security Guard Ultra C18 pre-column (Phenomenex, Torrance, CA, USA) was used for HPLC separation. The mass spectrometer was optimized for the detection of each individual analyte to obtain maximum intensity (polarity, collision energy, precursor and product ion selection) and the parameters are summarized in Supplemental Table 2. Injection volumes were 5 μL. After one min at 2% solvent B (0.1% formic acid in acetonitrile), analytes were separated by a linear gradient up to 50% solvent B over 7.5 min. The column was subsequentially washed for 1 min at 90% solvent B and re-equilibrated with 98% solvent A (0.1% formic acid) over 2.5 min. The flow rate was 0.4 mL/min and the column was heated to 40 °C. The ESI source nebulizer and heater nitrogen gas was adjusted to a flow of 65 (arbitrary units), while the curtain gas was maintained at 30 (arbitrary units). The ion source temperature was maintained at 650 °C with a voltage of 5200 V in positive and −4500 V in negative mode. Samples were analyzed in randomized order. A pooled saliva quality control sample was prepared by combining aliquots of equal volume of all study samples. An external quality control sample was prepared containing a subset of the targeted metabolites (Supplemental Table 2). Both QC samples were analyzed at predefined intervals during and between analysis batches to monitor the stability of the analytical system and detect potential batch effects. Inter- and intra-batch coefficients of variance of <20% were used as QC criteria.
Discovery LC-MS/MS-analysis using data-dependent acquisition (DDA).
DDA-LC-MS/MS analyses were performed on a SCIEX QTOF 6600 mass spectrometer (SCIEX, Framingham, MA, USA) equipped with a DuoSpray ESI source (SCIEX, Framingham, MA, USA) hyphenated to a Shimadzu Nexera XR HPLC system (Shimadzu, Kyoto, Japan) and controlled by the SCIEX Analyst TF 1.7 software. For DDA experiments, each cycle included a 100 msec TOF-MS scan (m/z range 50-1000) followed by the acquisition of up to 20 MS/MS-spectra at an intensity threshold of 300 counts (collision energy set at 20V with a 10V spread in high sensitivity acquisition mode). The ESI source was operated with source gas 1 and 2 adjusted to a flow of 60 (arbitrary units), while the curtain gas was maintained at 25 (arbitrary units). The ion source temperature was maintained at 650 °C with a voltage of 5200 V in positive and −4500 V in negative mode. Chromatographic separation was performed using the LC-method described for the MRM-LC-MS/MS analysis; acquisitions were performed in positive and negative ionization mode.
Discovery LC-MS/MS-analysis using data-independent acquisition (DIA).
DIA-LC-MS/MS analyses were performed on the SCIEX QTOF 6600 mass spectrometer hyphenated to a Shimadzu Nexera XR HPLC system (Shimadzu, Kyoto, Japan) as described for the DDA-mode, with the exception that in DIA mode, the duty cycle included a single MS-scan (96 msec; m/z range 50-1000) followed by a series of product ion scans (41.2 msec each) across 28 isolation windows of m/z width 26.
Data processing and statistical analysis.
MRM- and DIA-data were processed using MultiQuant 3.0.1 software (SCIEX) to detect and quantify the targeted compounds using the respective transition settings listed for each analyte in Supplemental Table 2. External calibration curves were used for the absolute quantification of salivary metabolites. Skyline 3.5 software (University of Washington) was used to extract the peak areas of selected precursor ions of the DDA-data set. DDA-data for the untargeted, discovery metabolomics analysis were processed (noise filtering, peak detection and chromatographic alignment) using MarkerView 1.2.1 software (SCIEX) and peaks above the noise threshold (>50 signal/noise) and within the mass tolerance (20 ppm) and retention time tolerance (0.3 min) were exported and processed using a custom R-script. Global normalization of samples was performed by scaling each sample to have the same 90th-percentile of peak areas (which is more robust to outlier peaks than normalizing to the sum of intensities). Normalized peak areas were then log2 transformed, after adding a small offset – here taken to be the 2nd percentile of all positive peak areas – to smooth the transition between zero and nonzero areas in log space. Because technical sources of variability may not be adequately accounted for by the global normalization – due e.g. to some sample runs having diminished signal for certain m/z ranges or retention times – we used surrogate variable analysis (Leek et al. 2012) to account for latent sources of variability. The surrogate variables (3-5 depending on the dataset) were determined automatically with the R package sva (method = “irw”), relative to a base model with factors for the main effects of PHIV vs. PHEU status and periodontal disease status and their interaction, and an additive factor for sex. The base model was then expanded to include the surrogate variables as additive covariates, and the R package limma (Ritchie et al. 2015) was used to compute log2-fold changes, empirical Bayes-moderated t-statistics and p-values for the two main effects and their interaction (function eBayes with trend=T), and a moderated F-statistic as an omnibus test for these three contrasts (function topTable)(Phipson et al. 2016). To aid in the interpretation of the main effects and their interaction, these statistics were also computed for the four simple effects – the effect of PHIV vs. PHEU status separately for individuals with and without periodontal disease, and the effect of periodontal disease separately for PHIV and PHEU individuals; as these four contrasts are linear combinations of the other three, their inclusion does not affect the omnibus F-test. False discovery rates (FDRs) were calculated using the Benjamini and Hochberg method (Benjamini, Hochberg 1995). Metabolite features that were caused by isotope peaks, as judged by accurate molecular masses and retention times, were removed for simplification of the final list of metabolites. Raw metabolomics data were deposited in the Metabolights repository (Haug et al. 2013).
Comparison of dynamic range and limits of detection.
To determine the dynamic range and limits of detection for the three acquisition methods, we prepared a dilution series of a metabolite standard mix containing 52 metabolites and data were collected in MRM, DDA, and DIA-mode. Limit of detection for metabolites was in the femtomolar range, showing a linear response across five orders of magnitude with correlation coefficients varying between 0.98 and 1 (Supplemental Figure 1). DDA (MS1-based) quantitation was in some instances negatively influenced by signals of co-eluting isobaric compounds that prevented the accurate extraction of MS1 peak areas, for example cadaverine (m/z 103.12, positive mode). The reliance of MRM and DIA-methods on MS2-peak extraction circumvented this shortcoming and allowed for the highly sensitive and specific extraction of the cadaverine product ion (m/z 86.1, positive mode) instead. Methionine (m/z precursor ion: 150.06, m/z product ion: 133.0, positive mode) was quantified across a broad dynamic range in DDA and MRM-mode but showed saturation at higher concentrations in DIA mode. The detectability of citric acid (m/z precursor ion: 191.02, m/z product ion: 85.0, negative mode) was almost identical across all three acquisition methods (Supplemental Figure 1a). To test the reproducibility of the three acquisition methods, we analyzed our panel of targeted metabolites three times per day on three consecutive days. The coefficients of variation for intraday reproducibility ranged from 4.5% for cadaverine to 11.4% for methionine in MRM-mode, 4.9% for citric acid to 18.3% for methionine in DIA-mode, and 7.5% for citric acid to 67.2% for methionine in DDA-mode (Supplemental Figure 1b). Similarly, the best interday reproducibility was observed for the MRM-measurements ranging from 5.5% for citric acid to 19.6% for methionine, followed by the DIA-measurements ranging from 10.1% for citric acid to 20% for methionine and DDA-measurements ranging from 9.8% for citric acid to 62.9% for methionine. Methionine measurements probably displayed the highest variability due to its propensity to oxidize. Supplemental Figure 1c shows the abundance distribution of the targeted metabolites quantified in the MRM, DDA and DIA-LC-MS/MS methods, respectively. Citric acid, methionine and cadaverine are highlighted. Interestingly, their ranked abundance measurements differed across the three acquisition methods, likely because MRM and DIA peak area measurements occur on the MS2 fragment ions, whereas the DDA method extract MS1 precursor ion signals. The difference between the observed intensity of the MRM and DIA measurements can be explained by the narrow mass filtering employed by the MRM-method for both precursor and product ion, whereas the wider precursor windows utilized in the DIA method could allow for precursor ion suppression. In total, the MRM-acquisition method provided the largest dynamic range of peak abundances.
Metabolite identification.
For metabolite features that displayed the most significant abundance differences between patient cohorts, their accurate molecular masses, isotopic distributions, MS/MS fragmentation and retention times were used to predict their elemental composition using the PeakView 2.2 software (SCIEX) and to identify them using the SCIEX Accurate Mass Metabolite Spectral Library and the ChemSpider database through the LibraryView 1.01 software framework (SCIEX). Remaining unknown compounds were queried against the METLIN (Tautenhahn et al. 2012) and Human Metabolome Database (Wishart et al. 2009) or individually submitted for de novo peptide identification using the PEAKS software package (version 8.0) (Ma et al. 2003).
Peptide identification.
Raw DDA data files were processed using PEAKS for database search-based peptide identification and quantitation using the following parameters: scans merged over 0.5 min retention time windows with a 20-ppm precursor m/z error tolerance; precursor masses were corrected; centroiding, deisotoping and deconvolution was performed automatically. The following parameters were used for searching against the UniProt/SwissProt human database (Release 2017_03): parent mass error tolerance 20 ppm; fragment mass error tolerance 0.1 Da; non-specific cleavage; pyro-glu (Q), oxidation (M), deamidation (NQ) as variable modifications. Identified peptides were matched to original metabolite feature tables using R.
Results and Discussion
Three complementary LC-MS/MS workflows (MRM, DIA and DDA) were utilized to characterize the saliva metabolome in PHIV vs. PHEU youth in the presence or absence of periodontal disease (Figure 1a). Metabolite ion chromatograms were more complex in positive than in negative ionization mode across the three LC-MS/MS-acquisition methods (Figure 1b). Absolute quantities measured for 52 representative compounds of central metabolic pathways by MRM were overall similar between PHIV and PHEU groups with and without periodontal disease (Supplemental Table 3). The top five metabolites in the MRM dataset, ranked according to their p-values from the F-test, are shown in Table 2 along with their corresponding values in the DDA and DIA datasets. Note that the rankings of the metabolites by p-value differed between acquisition methods, but in Table 2 they are all ordered based on ranks for the MRM dataset for consistency. Statistical tests were performed on log2 transformed intensities, and the coefficients in Tables 2 and 3 represent estimated log2(fold-changes) between groups – or in the case of the interaction term a difference in log2(fold-changes) – as detailed in Table 2. In the discussion that follows, log2(fold-changes) are converted to ordinary fold-changes by base-2 exponentiation, and a fold-change r <1 is referred to as an s-fold decrease, with s = 1/r. The top ranked metabolite in the MRM dataset, and also the DIA dataset, was cadaverine, which was on average 2.1 times as abundant in individuals with periodontal disease in the MRM dataset. But as indicated by a large negative interaction term, this difference was far more pronounced in the PHEU than PHIV group, with a modest 1.2-fold elevation for the PHIV group versus a 3.6-fold elevation in the PHEU group. This nearly four-fold difference for the simple effect of periodontal disease in the PHEU subgroup was significant even when controlling for false-discovery rate <0.1 in this particular contrast (FDR of 0.057); this controls only for the multiplicity of metabolites being testing, whereas the F-test controls for the multiple between-group comparisons being made, and the FDR-adjusted F-test controls for both factors. Of note, cadaverine is a polyamine likely resulting from prokaryotic amino acid degradation that has been identified as a potential biomarker for periodontitis (Barnes et al. 2009; Kuboniwa et al. 2016). Additional amino acid catabolites, particularly phenylacetate, that may also be of bacterial origin have recently been associated with periodontal disease measures in a large sample set (Liebsch et al. 2019). We did not identify phenylacetate among metabolites with significant differences in this study. For methionine, periodontal disease was associated with increased levels of methionine in the PHEU group (1.7-fold) but slightly decreased levels in the PHIV group (1.1-fold). For histidine this situation was reversed, with periodontal disease associated with decreased levels of histidine in the PHEU group (1.7-fold) and increased levels in the PHIV group (1.4-fold), suggesting the activation of different metabolic pathways. Arginine and cytosine levels were mildly increased levels in periodontal disease groups (1.3-fold and 1.2-fold, respectively), with consistent direction of change for both the PHIV and PHEU groups. The results for these amino acids and amino acid derivatives are consistent with earlier reports, but we would underscore that due to the multiple hypotheses being tested, differences with nonsignificant FDR-adjusted F-statistics reported in Table 2 should be regarded with some degree of caution. Potential oxidation of metabolites (e.g. methionine and cysteine) may contribute to observed variance in abundance measurements. Our confidence in the observed fold-changes was however bolstered by them generally being in agreement with the fold-changes observed when using a targeted extraction approach to obtain levels for these metabolites from the DIA and DDA datasets (Table 2 and Supplemental Table 4).
Table 2.
Top five metabolite associations with PHIV and periodontal disease measured by MRM-based quantitation and targeted extraction from DIA and DDA-based datasets, ranked by p-value for the F-test in the MRM dataset.
| PHIV/PHEU | moderate/without periodontitis | Interactionc | F-testd | |||||
|---|---|---|---|---|---|---|---|---|
| log2 ratioa | p-value | log2 ratiob | p-value | log2 ratio | p-value | p-value | FDRe | |
| MRM | ||||||||
| Cadaverine | 0.04 | 0.92 | 1.07 | 0.005 | −1.54 | 0.05 | 0.01 | 0.51 |
| Methionine | 0.20 | 0.35 | 0.32 | 0.10 | −0.90 | 0.03 | 0.04 | 0.92 |
| Histidine | 0.22 | 0.41 | −0.17 | 0.50 | 1.27 | 0.02 | 0.08 | 0.92 |
| Arginine | −0.20 | 0.32 | 0.36 | 0.05 | 0.59 | 0.13 | 0.08 | 0.92 |
| Cytosine | 0.12 | 0.46 | 0.31 | 0.04 | 0.37 | 0.23 | 0.10 | 0.92 |
| DIA | ||||||||
| Cadaverine | −0.12 | 0.75 | 0.99 | 0.01 | −0.69 | 0.35 | 0.06 | 0.89 |
| Methionine | 0.06 | 0.84 | 0.18 | 0.53 | −1.44 | 0.01 | 0.09 | 0.89 |
| Histidine | 0.01 | 0.98 | −0.25 | 0.33 | 1.33 | 0.01 | 0.06 | 0.89 |
| Arginine | −0.26 | 0.33 | 0.15 | 0.55 | 0.43 | 0.38 | 0.56 | 0.92 |
| Cytosine | 0.06 | 0.78 | −0.03 | 0.88 | −0.58 | 0.14 | 0.52 | 0.92 |
| DDA | ||||||||
| Cadaverine | −0.04 | 0.85 | 0.43 | 0.06 | 0.00 | 0.99 | 0.29 | 0.89 |
| Methionine | 0.16 | 0.58 | 0.09 | 0.73 | −1.16 | 0.04 | 0.20 | 0.89 |
| Histidine | 0.06 | 0.83 | −0.15 | 0.53 | 1.17 | 0.03 | 0.13 | 0.89 |
| Arginine | −0.18 | 0.58 | 0.58 | 0.06 | 0.97 | 0.13 | 0.11 | 0.89 |
| Cytosine | 0.41 | 0.22 | 0.26 | 0.40 | 0.67 | 0.30 | 0.35 | 0.89 |
: Using the notation HP, Hp, hP, and hp to represent average log2 levels for the four subgroups as in Table 1, the log2 ratio for PHIV/PHEU represents the contrast (HP-hP)/2+(Hp-hp)/2, so is positive when levels are on average higher in PHIV than in PHEU youth.
: The log2 ratio for periodontitis +/− represents the contrast (HP-Hp)/2+(hP-hp)/2, so is positive when levels are on average higher in youth with periodontitis than in youth without periodontitis.
: The interaction term represents the contrast (HP-hP) – (Hp-hp) = (HP-Hp) – (hP-hp), so is positive when the log2 ratio for PHIV/PHEU is greater in Perio+ than in Perio−, or when the log2 ratio for Perio+/Perio− is greater in PHIV than in PHEU youth.) Note that under the null hypothesis of no difference between groups, the standard deviation of the interaction term is twice that of the main effects due to the 2s in the denominator of the main effects. Note also that coefficients for simple effects such as Hp-Hp can be recovered from the appropriate main effect (a or b) +/− half the interaction term.
: An empirical Bayes-moderated F-statistic was used as an omnibus test for the two main effects and their interaction.
: False discovery rates (FDRs) were calculated using the Benjamini and Hochberg method (Benjamini, Hochberg 1995).
Table 3.
Top ten metabolite features in the DDA-discovery dataset (positive and negative ionization mode), ranked by p-value from the F-test. The columns for the PHIV/PHEU and periodontitis (+/−) comparisons and their interaction are as described in the legend for Table 2.
| Positive ionization mode | PHIV/PHEU | moderate/without periodontitis | Interaction | F-test | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| m/z | R.T. | Description | log2 ratio | p-value | log2 ratio | p-value | log2 ratio | p-value | p-value | FDR |
| 382.18 | 3.68 | −0.83 | 0.004 | 0.56 | 0.045 | −1.34 | 0.007 | 0.0004 | 0.74 | |
| 676.70 | 6.63 | 1.20 | 0.001 | −0.76 | 0.038 | −1.74 | 0.008 | 0.0008 | 0.74 | |
| 530.95 | 8.25 | RGPYPPGPLAPPQPF (SMR3B) | −0.90 | 0.013 | 0.90 | 0.014 | 1.71 | 0.008 | 0.0022 | 0.74 |
| 451.22 | 2.54 | −0.65 | 0.081 | 1.27 | 0.002 | −1.01 | 0.128 | 0.0025 | 0.74 | |
| 935.97 | 5.69 | SPPGKPQGPPQQEGNKPQ (PRB4) | 1.05 | 0.000 | −0.54 | 0.060 | −0.40 | 0.416 | 0.0025 | 0.74 |
| 288.20 | 3.86 | D-Leucyl-N~5~-(diaminomethylene)-L-ornithine | −0.89 | 0.009 | 1.06 | 0.003 | −0.35 | 0.545 | 0.0027 | 0.74 |
| 265.08 | 5.93 | −0.99 | 0.027 | 1.20 | 0.010 | −1.50 | 0.059 | 0.0031 | 0.74 | |
| 429.25 | 2.55 | −0.76 | 0.028 | 0.99 | 0.007 | −1.02 | 0.094 | 0.0033 | 0.74 | |
| 565.23 | 4.06 | −0.05 | 0.880 | 0.63 | 0.094 | −2.26 | 0.001 | 0.0034 | 0.74 | |
| 434.74 | 6.86 | RIGRFGY (STAT) | −1.01 | 0.006 | 1.09 | 0.004 | 0.22 | 0.720 | 0.0036 | 0.74 |
| Negative ionization mode | ||||||||||
| m/z | R.T. | Description | log2 ratio | p-value | log2 ratio | p-value | log2 ratio | p-value | p-value | FDR |
| 329.15 | 7.4 | 0.26 | 0.665 | −0.65 | 0.226 | −5.09 | 0.00001 | 0.0002 | 0.19 | |
| 727.30 | 6.45 | −1.78 | 0.002 | 1.44 | 0.004 | 0.80 | 0.384 | 0.0014 | 0.34 | |
| 330.13 | 5.16 | −1.27 | 0.003 | 0.88 | 0.023 | −1.17 | 0.110 | 0.0014 | 0.34 | |
| 684.30 | 7.74 | −1.94 | 0.001 | 1.29 | 0.013 | 0.42 | 0.664 | 0.0022 | 0.34 | |
| 297.04 | 10.37 | 3.97 | 0.001 | −1.40 | 0.157 | −3.88 | 0.044 | 0.0023 | 0.34 | |
| 290.09 | 2.98 | 2.37 | 0.001 | 0.89 | 0.141 | 0.07 | 0.948 | 0.0026 | 0.34 | |
| 442.12 | 7.09 | 0.82 | 0.108 | −0.61 | 0.183 | −3.29 | 0.001 | 0.0029 | 0.34 | |
| 405.18 | 6.11 | −0.96 | 0.007 | 0.95 | 0.003 | 0.55 | 0.354 | 0.0031 | 0.34 | |
| 917.46 | 5.86 | 1.29 | 0.003 | −0.69 | 0.070 | −1.54 | 0.038 | 0.0052 | 0.45 | |
| 556.24 | 7.81 | −1.89 | 0.001 | 0.48 | 0.297 | 0.23 | 0.794 | 0.0052 | 0.45 | |
Discovery of unknown metabolite associations using data-dependent acquisition profiling.
As anticipated from the total ion chromatograms, the complexity of the salivary metabolome exceeded the 52 metabolites targeted by our MRM panel (Figure 1b). Unlike MRM methods that utilize predefined transition lists, DDA allows for automated measurements over a predefined mass range (i.e. m/z 50-1000) to collect precursor and fragment ion data based on real-time decisions without user intervention. A priori knowledge about the composition of a sample is unnecessary, since identities are determined via software-based algorithms that utilize retention time, accurate precursor masses, isotopic distributions and MS/MS fragmentation spectra post acquisition. In our dataset, we detected 3317 distinct metabolite features in positive and 874 in negative ionization mode after processing (Supplemental Table 5). Next, we used empirical Bayes statistics to test for differences in metabolite levels associated with PHEU/PHIV status, periodontal disease status, and their interaction (Figure 2). Results for the top 10 metabolites in the positive and negative ionization modes, again ranked based on their p-value from the F-test and again subject to the caution regarding multiple hypothesis testing, are summarized in Table 3.
Figure 2.
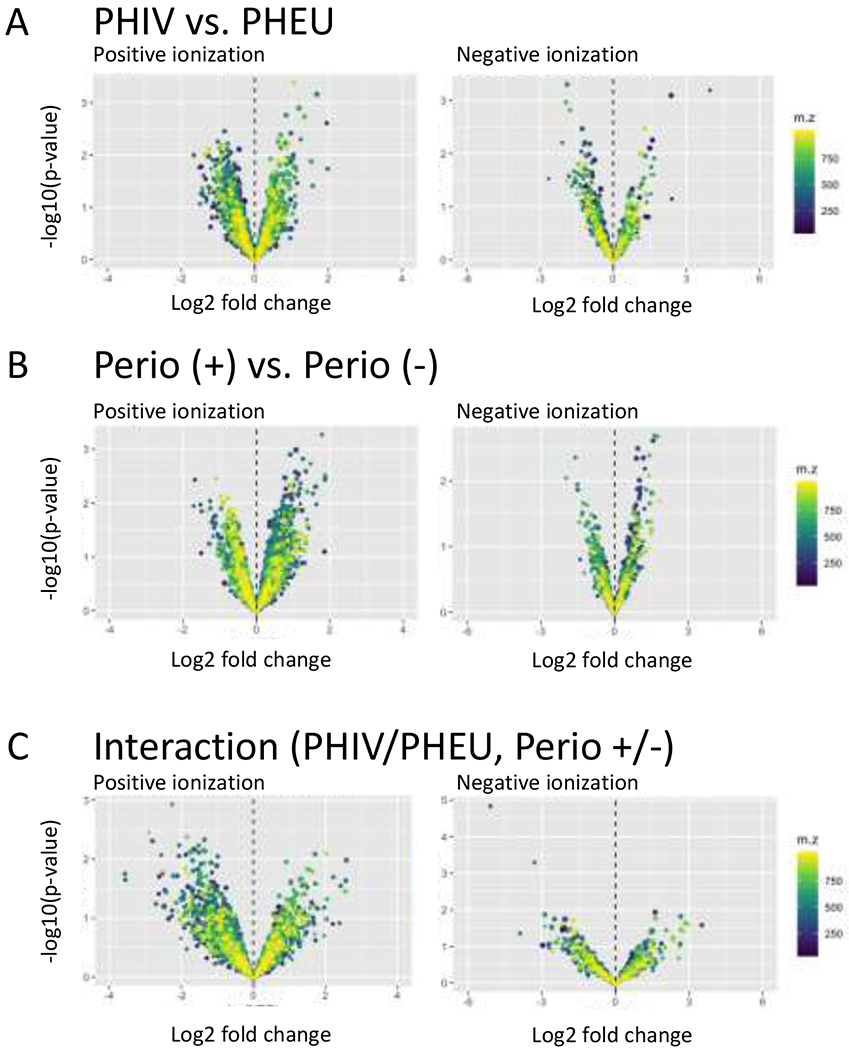
Global untargeted metabolomics profiling using DDA-MS. Volcano plots display log2 ratios against the −log10 p-values for main effects (A) PHIV vs PHEU and (B) moderate vs. no periodontal disease, and (C) their interaction, with separate plots for metabolites acquired in positive (left) and negative (right) ionization mode. See the footnotes for Table 2 for a precise description of these contrasts.
Initially, we identified metabolite features using our in-house accurate mass metabolite spectral database (SCIEX) containing reference spectra for over 500 compounds. For the remaining unknown components of interest, elemental compositions were predicted based on accurate mass measurements and MS/MS fragmentation patterns compared with those of compounds registered in publicly available metabolomics databases. Disappointingly, this approach led only to the identification of few metabolites, which reiterates a common current limitation in discovery metabolomics. Manual interrogation revealed that for many examined metabolites features the fragmentation patterns resembled MS2 spectra observed in peptide analysis. We therefore submitted corresponding peak lists to the PEAKS proteomics software package for peptide de novo sequencing which resulted in the identification of proteolytic cleavage products of known salivary proteins. Encouraged by this finding, we submitted the entire DDA dataset through the PEAKS identification workflow that included database searching that resulted in the identification of 564 endogenous peptides in the metabolite extract of which the majority could be mapped back to sequences derived from salivary precursor proteins including histatin, statherin and several proline-rich proteins. PEAKS analysis also revealed that the charge state of many metabolite features was greater than 1, emphasizing the importance of independent charge state determination/correction. Altered abundances of these peptide fragments could reflect abundance changes of their respective precursor proteins and/or alterations of the underlying proteolytic processing mechanisms by which these peptides are formed. Salivary proteins are known to undergo extensive proteolytic processing even in healthy individuals (Hardt et al. 2005; Helmerhorst, Oppenheim 2007) and little is known about the mechanisms by which HIV infection and/or HAART, which typically includes protease inhibitors, could influence this complex proteolytic network.
Conclusions
We developed a dual targeted and discovery LC-MS/MS strategy to characterize changes in the composition of the salivary metabolome. While previous salivary metabolite profiling studies employed single acquisition modes (Kageyama et al. 2015; Mueller et al. 2014; Sugimoto et al. 2010; Tsuruoka et al. 2013; Wang et al. 2014; Wei et al. 2011; Zheng et al. 2012), our multi-acquisition strategy allowed us to expand beyond a predefined set of targeted metabolites and discover unanticipated molecules that are potentially associated with disease. Our data showed that compounds linked to amino acid/protein/peptide metabolism such as the biogenic amine cadaverine were potentially associated with periodontitis, and potentially interacting with PHIV/PHEU status. These results suggest that the metabolism of oral bacteria may be influenced by HIV infection or by HAART (as all but one of the PHIV youth but none of the PHEU youth were on HAART regimens). We did not attempt to further stratify the PHIV groups by duration of combination of drugs in their HAART regimen due to the limited sample sizes. As the results of this study were based on a relatively small number of individuals, these potential associations should be considered preliminary and need to be validated in future studies using larger patient cohorts. Given the inclusion of protease inhibitors in many HAART regimens, our result stresses the importance to study the effect of HAART on salivary protein metabolism by integrated proteomics/peptidomics.
Supplementary Material
Acknowledgements
We thank the children and families for their participation in PHACS, and the individuals and institutions involved in the conduct of PHACS.
The following institutions, clinical site investigators and staff participated in conducting PHACS AMP and AMP Up in 2018, in alphabetical order: Ann & Robert H. Lurie Children’s Hospital of Chicago: Ellen Chadwick, Margaret Ann Sanders, Kathleen Malee, Yoonsun Pyun; Baylor College of Medicine: William Shearer, Mary Paul, Chivon McMullen-Jackson, Mandi Speer, Lynnette Harris; Bronx Lebanon Hospital Center: Murli Purswani, Mahboobullah Mirza Baig, Alma Villegas; Children’s Diagnostic & Treatment Center: Lisa Gaye-Robinson, Sandra Navarro, Patricia Garvie; Boston Children’s Hospital: Sandra K. Burchett, Michelle E. Anderson, Adam R. Cassidy; Jacobi Medical Center: Andrew Wiznia, Marlene Burey, Ray Shaw, Raphaelle Auguste; Rutgers - New Jersey Medical School: Arry Dieudonne, Linda Bettica, Juliette Johnson, Karen Surowiec; St. Christopher’s Hospital for Children: Janet S. Chen, Maria Garcia Bulkley, Taesha White, Mitzie Grant; St. Jude Children’s Research Hospital: Katherine Knapp, Kim Allison, Megan Wilkins, Jamie Russell-Bell; San Juan Hospital/Department of Pediatrics: Midnela Acevedo-Flores, Heida Rios, Vivian Olivera; Tulane University School of Medicine: Margarita Silio, Medea Gabriel, Patricia Sirois; University of California, San Diego: Stephen A. Spector, Megan Loughran, Veronica Figueroa, Sharon Nichols; University of Colorado Denver Health Sciences Center: Elizabeth McFarland, Carrie Chambers, Emily Barr, Mary Glidden; University of Miami: Gwendolyn Scott, Grace Alvarez, Juan Caffroni, Anai Cuadra.
The study was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development with co-funding from the National Institute on Drug Abuse, the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, the National Institute on Deafness and Other Communication Disorders, the National Institute of Dental and Craniofacial Research, the National Cancer Institute, the National Institute on Alcohol Abuse and Alcoholism, the Office of AIDS Research, and the National Heart, Lung, and Blood Institute through cooperative agreements with the Harvard T.H. Chan School of Public Health (HD052102) (Principal Investigator: George R Seage III; Program Director: Liz Salomon) and the Tulane University School of Medicine (HD052104) (Principal Investigator: Russell Van Dyke; Co-Principal Investigator: Ellen Chadwick; Project Director: Patrick Davis). Data management services were provided by Frontier Science and Technology Research Foundation (PI: Suzanne Siminski), and regulatory services and logistical support were provided by Westat, Inc (PI: Julie Davidson). The Forsyth Center for Salivary Diagnostics was funded by the Massachusetts Life Science Center.
The conclusions and opinions expressed in this article are those of the authors and do not necessarily reflect those of the National Institutes of Health or U.S. Department of Health and Human Services.
Abbreviations
- AAP
American Academy of Periodontology
- CDC
Centers for Disease Control and Prevention
- CV
coefficient of variation
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- TOF
time of flight
- PHIV
perinatally HIV-infected
- PHEU
perinatally HIV-exposed, uninfected
- PHACS
Pediatric HIV/AIDS Cohort Study
- HAART
highly-active antiretroviral therapy
- PI
protease inhibitor
- HIV
human immunodeficiency virus
- IQR
interquartile range
Footnotes
Conflict-of-interest:
The authors declare no conflicts of interests.
Compliance with Ethical Standards
All procedures involving human participants in this study were performed in accordance with the ethical standards of the IRBs at the clinical sites and the Harvard T.H. Chan School of Public Health and with the 1964 Helsinki declaration and its later amendments.
Data availability.
Raw metabolomics data reported in this article are available via the Metabolights repository (https://www.ebi.ac.uk/metabolights/MTBLS1012) under study identifier MTBLS1012. Requests for analysis data sets can be made to the PHACS network by following article-related data request instructions on the PHACS website: https://phacsstudy.org/ under the Quick Link “Data requests for published articles” or by request to the PHACS Data and Operations Center, at phacs_datarequest@fstrf.org.
Supplementary Data.
Supplementary data related to this article is available.
References
- Álvarez-Sánchez B, Priego-Capote F, Luque de Castro MD (2012). Study of sample preparation for metabolomic profiling of human saliva by liquid chromatography-time of flight/mass spectrometry. Journal of chromatography A 1248, 178–181 doi: 10.1016/j.chroma.2012.05.029 [DOI] [PubMed] [Google Scholar]
- Barnes VM, et al. (2014). Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE 9, e105181 doi: 10.1371/journal.pone.0105181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes VM, et al. (2009). Acceleration of purine degradation by periodontal diseases. Journal of Dental Research 88, 851–855 doi: 10.1177/0022034509341967 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B Methodological 57, 289–300 doi: 10.2307/2346101 [DOI] [Google Scholar]
- Cuevas-Córdoba B, Santiago-García J (2014). Saliva: A Fluid of Study for OMICS. Omics 18, 87–97 doi: 10.1089/omi.2013.0064 [DOI] [PubMed] [Google Scholar]
- Dame ZT, et al. (2015). The human saliva metabolome. Metabolomics, 1–20 doi: 10.1007/s11306-015-0840-5 [DOI] [Google Scholar]
- Ghannoum MA, et al. (2013). Metabolomics reveals differential levels of oral metabolites in HIV-infected patients: toward novel diagnostic targets. Omics 17, 5–15 doi: 10.1089/omi.2011.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt M, et al. (2005). Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry 44, 2885–2899 doi: 10.1021/bi048176r [DOI] [PubMed] [Google Scholar]
- Haug K, et al. (2013). MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Research 41, D781–6 doi: 10.1093/nar/gks1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst EJ, Oppenheim FG (2007). Saliva: a dynamic proteome. Journal of Dental Research 86, 680–693 [DOI] [PubMed] [Google Scholar]
- Howell RB, Jandinski JJ, Palumbo P, Shey Z, Houpt MI (1996). Oral soft tissue manifestations and CD4 lymphocyte counts in HIV-infected children. Pediatric dentistry 18, 117–120 [PubMed] [Google Scholar]
- Kageyama G, et al. (2015). Metabolomics analysis of saliva from patients with primary Sjögren's syndrome. Clinical and Experimental Immunology 182, 149–153 doi: 10.1111/cei.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Sakanaka A, Hashino E, Bamba T, Fukusaki E, Amano A (2016). Prediction of Periodontal Inflammation via Metabolic Profiling of Saliva. J Dent Res 95, 1381–1386 doi: 10.1177/0022034516661142 [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–3 doi: 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebsch C, et al. (2019). The Saliva Metabolome in Association to Oral Health Status. J Dent Res 98, 642–651 doi: 10.1177/0022034519842853 [DOI] [PubMed] [Google Scholar]
- Lloyd A (1996). HIV infection and AIDS. Papua and New Guinea medical journal 39, 174–180 [PubMed] [Google Scholar]
- Ma B, et al. (2003). PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid communications in mass spectrometry : RCM 17, 2337–2342 doi: 10.1002/rcm.1196 [DOI] [PubMed] [Google Scholar]
- Moscicki A-B, et al. (2016). The Burden of Oral Disease among Perinatally HIV-Infected and HIV-Exposed Uninfected Youth. PLoS ONE 11, e0156459 doi: 10.1371/journal.pone.0156459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller DC, Piller M, Niessner R, Scherer M, Scherer G (2014). Untargeted metabolomic profiling in saliva of smokers and nonsmokers by a validated GC-TOF-MS method. Journal of Proteome Research 13, 1602–1613 doi: 10.1021/pr401099r [DOI] [PubMed] [Google Scholar]
- Navazesh M (1993). Methods for collecting saliva. Annals of the New York Academy of Sciences 694, 72–77 [DOI] [PubMed] [Google Scholar]
- Neyraud E, et al. (2020). Longitudinal analysis of the salivary metabolome of breast-fed and formula-fed infants over the first year of life. Metabolomics 16, 37 doi: 10.1007/s11306-020-01661-7 [DOI] [PubMed] [Google Scholar]
- Peltenburg NC, et al. (2018). Persistent metabolic changes in HIV-infected patients during the first year of combination antiretroviral therapy. Scientific Reports 8, 16947 doi: 10.1038/s41598-018-35271-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK (2016). Robust Hyperparameter Estimation Protects Against Hypervariable Genes and Improves Power to Detect Differential Expression. The Annals of Applied Statistics 10, 946–963 doi: 10.1214/16-AOAS920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J (2007). Control of HIV/AIDS and AIDS-related conditions in Africa with special reference to periodontal diseases. Journal of the International Academy of Periodontology 9, 2–12 [PubMed] [Google Scholar]
- Ritchie ME, et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47–e47 doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano F, et al. (2018). Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J Periodontol 89, 1452–1460 doi: 10.1002/JPER.18-0097 [DOI] [PubMed] [Google Scholar]
- Ryder MI, Nittayananta W, Coogan M, Greenspan D, Greenspan JS (2012). Periodontal disease in HIV/AIDS. Periodontology 2000 60, 78–97 doi: 10.1111/j.1600-0757.2012.00445.x [DOI] [PubMed] [Google Scholar]
- Ryder MI, Shiboski C, Yao TJ, Moscicki AB (2020). Current trends and new developments in HIV research and periodontal diseases. Periodontol 2000 82, 65–77 doi: 10.1111/prd.12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder MI, Yao T-J, Russell JS, Moscicki A-B, Shiboski CH, Study PHAC (2017). Prevalence of periodontal diseases in a multicenter cohort of perinatally HIV-infected and HIV-exposed and uninfected youth. Journal of clinical periodontology 44, 2–12 doi: 10.1111/jcpe.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siberry GK, et al. (2011). CD4+ lymphocyte-based immunologic outcomes of perinatally HIV-infected children during antiretroviral therapy interruption. J Acquir Immune Defic Syndr 57, 223–9 doi: 10.1097/QAI.0b013e318218e068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr JR, et al. (2018). Oral microbiota in youth with perinatally acquired HIV infection. Microbiome 6, 100 doi: 10.1186/s40168-018-0484-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M (2010). Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95 doi: 10.1007/s11306-009-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G (2012). An accelerated workflow for untargeted metabolomics using the METLIN database. Nature Biotechnology 30, 826–828 doi: 10.1038/nbt.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka M, et al. (2013). Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. ELECTROPHORESIS 34, 2865–2872 doi: 10.1002/elps.201300019 [DOI] [PubMed] [Google Scholar]
- Van Dyke RB, et al. (2011). Antiretroviral treatment of US children with perinatally acquired HIV infection: temporal changes in therapy between 1991 and 2009 and predictors of immunologic and virologic outcomes. Journal of Acquired Immune Deficiency Syndromes 57, 165–173 doi: 10.1097/QAI.0b013e318215c7b1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseliu N, et al. (2005). Longitudinal study of the prevalence and prognostic implications of oral manifestations in romanian children infected with human immunodeficiency virus type 1. The Pediatric infectious disease journal 24, 1067–1071 [DOI] [PubMed] [Google Scholar]
- Wang Q, Gao P, Wang X, Duan Y (2014). The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Scientific Reports 4, 6802 doi: 10.1038/srep06802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, et al. (2011). Salivary metabolite signatures of oral cancer and leukoplakia. International journal of cancer 129, 2207–2217 doi: 10.1002/ijc.25881 [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. (2009). HMDB: a knowledgebase for the human metabolome. Nucleic Acids Research 37, D603–10 doi: 10.1093/nar/gkn810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Sun H, Wang X (2012). Saliva Metabolomics Opens Door to Biomarker Discovery, Disease Diagnosis, and Treatment. Appl Biochem Biotechnol 168, 1718–1727 doi: 10.1007/s12010-012-9891-5 [DOI] [PubMed] [Google Scholar]
- Zheng J, Dixon RA, Li L (2012). Development of isotope labeling LC-MS for human salivary metabolomics and application to profiling metabolome changes associated with mild cognitive impairment. Analytical Chemistry 84, 10802–10811 doi: 10.1021/ac3028307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


