Abstract
Fanconi anemia (FA) is a clinically heterogenous and genetically diverse disease with 22 known complementation groups (FA-A to FA-W), resulting from the inability to repair DNA interstrand cross-links. This rare disorder is characterized by congenital defects, bone marrow failure, and cancer predisposition. FANCA is the most commonly mutated gene in FA and a variety of mostly private mutations have been documented, including small and large indels and point and splicing variants. Genotype–phenotype associations in FA are complex, and a relationship between particular FANCA variants and the observed cellular phenotype or illness severity remains unclear. In this study, we describe two siblings with compound heterozygous FANCA variants (c.3788_3790delTCT and c.4199G > A) who both presented with esophageal squamous cell carcinoma at the age of 51. The proband came to medical attention when he developed pancytopenia after a single cycle of low-dose chemotherapy including platinum-based therapy. Other than a minor thumb abnormality, neither patient had prior findings to suggest FA, including normal blood counts and intact fertility. Patient fibroblasts from both siblings display increased chromosomal breakage and hypersensitivity to interstrand cross-linking agents as seen in typical FA. Based on our functional data demonstrating that the c.4199G > A/p.R1400H variant represents a hypomorphic FANCA allele, we conclude that the residual activity of the Fanconi anemia repair pathway accounts for lack of spontaneous bone marrow failure or infertility with the late presentation of malignancy as the initial disease manifestation. This and similar cases of adult-onset esophageal cancer stress the need for chromosome breakage testing in patients with early onset of aerodigestive tract squamous cell carcinomas before platinum-based therapy is initiated.
Keywords: esophageal carcinoma, oropharyngeal squamous cell carcinoma
INTRODUCTION
Fanconi anemia (FA) is a rare genetic disorder that results in chromosomal instability and is characterized by congenital defects, progressive pancytopenia, and cancer predisposition. FA is the most common genetic bone marrow failure syndrome affecting approximately one in 100,000 births and is caused by germline mutations in one of the 22 known FANC genes (FANCA-W) (Wang and Smogorzewska 2015; Niraj et al. 2019). A hallmark of FA is cellular hypersensitivity to DNA-damaging agents, such as diepoxybutane (DEB) and mitomycin C (MMC), because of an inability to remove DNA interstrand cross-links (ICLs) by the Fanconi anemia repair pathway.
Presentation of FA is very heterogeneous. With respect to hematologic function, the majority of patients (90%) exhibit aplastic anemia in the first decade of life by a median age of 7 yr (Rosenberg et al. 2011). Nearly 80% of individuals have at least one or more physical abnormalities, which may include short stature, skeletal defects, microcephaly, microphthalmia, structural defects of the heart or other organ systems, and dermatologic findings such as hypopigmentation and café au lait spots (Fiesco-Roa et al. 2019). Associations with VACTERL birth defects are well reported (Alter and Rosenberg 2013). There is also a high incidence of endocrine dysfunction including growth hormone (GH) deficiency, impaired glucose metabolism, hypothyroidism, hypogonadism, and infertility (Giri et al. 2007; Petryk et al. 2015). Data suggest that both a primary gonadal defect and impairment of the hypothalamus–pituitary axis are responsible for reduced fertility (Petryk et al. 2015).
Individuals with FA are also highly predisposed to malignancies, predominantly acute myelogenous leukemia (AML) and squamous cell carcinomas (SCCs) of the aerodigestive tract. The relative risk for AML for FA patients is increased ∼500-fold and the reported cumulative incidence is 13% by 50 yr of age (Rosenberg et al. 2008; Alter et al. 2010; Tamary et al. 2010). They are particularly susceptible to developing tumors of the head and neck, skin, and gastrointestinal and genitourinary tract. In patients with FA, head and neck squamous cell carcinomas (HNSCCs) are the most common solid tumor with a reported 700-fold increased risk. The HNSCC in FA patients often present earlier and more aggressively (Kutler et al. 2003, 2016).
Genotype–phenotype associations in FA are complex. Some of the clinical heterogeneity of the disease can be linked to the genetically distinct subtypes of FA. Biallelic pathogenic variants in BRCA2/FANCD1 and PALB2/FANCN result in a severe subtype of FA with the majority of patients developing embryonal tumors (Wilms, neuroblastoma, medulloblastoma) and AML in early childhood (Howlett et al. 2002; Alter et al. 2007; Xia et al. 2007). However, even within these subtypes, atypical presentations have been reported, suggesting milder phenotypes when some protein functions are preserved (Byrd et al. 2016; Rickman et al. 2020). A study comparing FA-A and FA-G complementation groups found that FA patients with null variants in FANCA or pathogenic variants in FANCG will manifest with a more severe clinical course, earlier bone marrow failure onset, and hematologic malignancies (Faivre et al. 2000). Similarly, a recent study demonstrated in a cohort of patients with FANCB mutations that less severe disease correlated with increased residual FANCB function (Jung et al. 2020b).
Moreover, the phenomenon of hematopoietic mosaicism is responsible for some of the varied clinical presentations observed in a small subset of FA patients and can be diagnostically challenging (Rickman et al. 2015; Asur et al. 2018). Hematopoietic mosaicism can result in partial or full rescue of chromosomal breakage levels in the peripheral blood and protect against aplastic anemia through restoration of a functional allele (Gregory et al. 2001; Nicoletti et al. 2020). In suspected cases, cultured fibroblasts are required for definitive diagnosis of FA.
Pathogenic variants in FANCA, the most commonly mutated FA gene, are responsible for disease in two-thirds of FA patients (Wang and Smogorzewska 2015; Jung et al. 2020b). The spectrum of pathogenic variants consists of point mutations, small insertions and deletions, splicing mutations, and large deletions (Castella et al. 2011; Flynn et al. 2014; Kimble et al. 2018). The high frequency and diverse spectrum of pathogenic variants in FANCA are owed in part to the large size of the gene (79 kb) with an open reading frame of 4.3 kb consisting of 43 exons. FANCA is susceptible to Alu–Alu nonallelic homologous recombination because of a large number of small interspersed nuclear elements (SINE) within the gene locus, accounting for a large number the identified indels (Flynn et al. 2014; Kimble et al. 2018). Many pathogenic FANCA variants are private mutations; however, common founder variants have been identified in the South African Afrikaners, Spanish Gypsies, and Tunisian populations (Castella et al. 2011; Flynn et al. 2014).
Previous reports have shown that there is not a clear correlation between specific FANCA mutations (i.e., homozygous null variants versus hypomorphic gene product) and the severity of the clinical phenotype with respect to congenital abnormalities and age at which hematologic abnormalities arise (Castella et al. 2011). Hematological progression significantly correlated between siblings harboring the same mutations in a recent study (Jung et al. 2020a). However, siblings may also present with widely dissimilar phenotypes (Faivre et al. 2000), suggesting that other, lesser understood factors, such as environmental exposures or modifying genetic variants, may also play a role in disease manifestation.
Our study further highlights the heterogeneous nature of FA by describing an atypical clinical presentation of two brothers diagnosed with FA in their sixth decade of life. The sibling pair initially presented with esophageal malignancies and were subsequently found to have FA by chromosomal breakage and molecular testing. We show that their atypical and delayed presentation, with lack of hematologic failure, is most likely secondary to the mitigating effects of a rare missense hypomorphic FANCA allele, c.4199G > A/p.R1400H.
RESULTS
Clinical Presentation, Family History, and Treatment Outcomes
The subjects of this study are two brothers diagnosed with Fanconi anemia in adulthood (see Table 1 for clinical summary). The proband was a previously healthy man who presented at the age of 51 after a 9-mo history of progressive dysphagia and a 20-pound weight loss and was found to have invasive SCC of the distal esophagus and proximal stomach. Other than male gender and social alcohol consumption, no additional risk factors such as race, tobacco, chronic gastroesophageal reflux, or Barrett's esophagus were present. Chemotherapy was initiated, and after a single cycle of low-dose carboplatin and paclitaxel, the proband developed pancytopenia (WBC 3.3 × 103/µL, Hgb 9.6 g/dL, platelet 13 × 103/µL) requiring granulocyte-colony stimulating factor (G-CSF) and platelet transfusion. Because of the unusual hypersensitivity to chemotherapy, the proband was referred to a hematologist for further evaluation.
Table 1.
Summary of patient history, physical exam findings, clinical presentation, and diagnosis
| Proband | Affected sibling | |
|---|---|---|
| Age at FA diagnosis | 51 yr | 51 yr |
| PMH | Hyperlipidemia | History of melanoma |
| No hematologic concerns | Hyperlipidemia | |
| Intact fertility | No hematologic concerns | |
| Intact fertility | ||
| Physical exam | Minor thumb anomalies | Minor thumb anomalies |
| Single café au lait spot | ||
| Clinical presentation | Invasive SCC of the distal esophagus and proximal stomach | Invasive well-differentiated distal esophagus SCC (T2 N0) |
| Severe pancytopenia after initial round of chemotherapy | Laryngeal SCC (T2 N0 M0) Invasive moderately differentiated anal SCC | |
| Treatment | Surgical: esophagectomy with gastric pull through | Surgical: esophagectomy with gastric pull through and local resection for anal cancer |
| Chemotherapy | Radiation for laryngeal and anal cancer | |
| DEB/MMC testing | Elevated chromosomal breakage upon clinical testing (2.22 breaks/cell at 0.1 µg/mL DEB) | Elevated chromosomal breakage in research testing (4.48 breaks/cell at 50 nM MMC) |
| Molecular testing | Heterozygous for FANCA variant c.4199G > A | Heterozygous for FANCA variant c.4199G > A |
| Heterozygous for FANCA variant c.3788_3790delTCT | Heterozygous for FANCA variant c.3788_3790delTCT | |
| Family history | Father pancreatic cancer (death at 63) | |
| Paternal uncle melanoma (death at 72) | ||
| Paternal grandfather melanoma (death in 70s) | ||
(FA) Fanconi anemia, (PMH) past medical history, (SCC) squamous cell carcinoma, (DEB) diepoxybutane, (MMC) mitomycin C.
The proband's physical exam was notable for a single café au lait spot on his left flank and minor thumb anomalies including a small left thumb with flexion contraction of the phalangophalangeal joint and a right thumb that was proximally implanted. Bone marrow biopsy revealed hypoplastic marrow (40% cellularity), decreased myeloid and erythroid precursors, megaloblastoid changes, and an inappropriately low megakaryocyte count, which raised concern for FA. Subsequent chromosomal breakage studies with DEB testing revealed elevated breakage (2.22 breaks/cell at a dose of 0.1 µg/mL). The proband was found to be compound heterozygous for FANCA variants, c.3788_3790delTCT and c.4199G > A (Table 2).
Table 2.
Summary of FANCA variants
| Gene | Genomic location | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | Observed effect | dbSNP/dbVar ID | Parent of origin |
|---|---|---|---|---|---|---|---|---|
| FANCA | 16q24.3 |
NG_011706.1:
g.80814_80816delTCT NM_0.000135.4: c.3788_3790delTCT |
NP_000126.2: p.F1263del | Deletion | Pathogenic | Pathogenic | 397507553/41003 | Unknown |
| FANCA | 16q24.3 |
NG_011706.1:g.82715G > A NM_000135.4: c.4199G > A |
NP_000126.2: p.R1400H | Substitution (missense mutation) | Uncertain significance | Likely pathogenic; hypomorphic | 149851163/408175 | Unknown |
Prevention Genetics Fanconi Anemia Nextgen sequencing panel tested full coding regions plus ∼20 bases of noncoding DNA flanking each exon of these genes: BRIP1, FANCM, PALB2, FANCL, FANCA, FANCC, FANCG, FANCE, FANCF, FANCB, FANCI, SLX4, ERCC4, FANCD2 isoform A and B, RAD51C.
Further treatment for the proband was largely surgical including a subsequent esophagectomy with gastric pull-through for esophageal SCC. The proband was found to have a mediastinal mass consistent with tumor reoccurrence within 6 mo. Treatment with capecitabine was initiated but was later changed to oxaliplatin and 5-fluorouracil. These agents were ultimately discontinued because of complications of diarrhea and acute pericarditis. The proband developed disseminated esophageal cancer and died at the age of 52.
A year after the proband's diagnosis of FA, his younger brother presented with a well to moderately differentiated SCC of the upper esophagus on screening endoscopy, also at the age of 51. Targeted FANCA testing was performed and he was found to also carry the familial FANCA variants, c.3788_3790delTCT and c.4199G > A. His past medical history was relevant for melanoma of his left upper extremity, which had been resected 2 yr prior. His physical exam was significant for an underdeveloped thenar eminence, but no hematological abnormalities. He underwent esophagectomy with gastric pull-through for the esophageal cancer. Approximately 18 mo later the patient was diagnosed with synchronous primary laryngeal and anal SCC, which were both treated with radiation (5400 cGy to the pelvis and 6900cGy to the laryngeal and hypopharyngeal regions). The patient then developed reoccurrence of the esophageal cancer and died at the age of 55.
As is typical with FANCA etiology, the family history was negative for FA prior to the proband's diagnosis. However, the father died of pancreatic cancer and the extended family history was notable for a paternal uncle and paternal grandfather who both had melanoma. Neither parent was reported to have abnormal cell counts. Both proband and younger brother had healthy biological children.
Identification of FANCA Variants
Clinical testing by Prevention Genetics was performed on peripheral blood from the proband. A next-generation sequencing panel for Fanconi anemia identified the heterozygous FANCA variants c.3788_3790delTCT and c.4199G > A (Table 2). A CGH array analysis of the FANCA gene revealed no evidence of a genomic deletion. The affected sibling was also found heterozygous for these same two variants by clinical targeted gene sequencing of FANCA at Prevention Genetics the following year. CancerNext-Expanded panel (Ambry Genetics) identified no other pathogenic variants in 43 genes associated with hereditary cancer (Supplemental Table S1). The presence of the two FANCA variants was confirmed by research-based Sanger sequencing on blood of the proband (Supplemental Fig. S1) and complementary DNA (cDNA) from fibroblasts derived from both siblings (Supplemental Fig. S2). The c.3788_3790delTCT variant is a known pathogenic variant that produces an in frame deletion in exon 38 (p.F1263del) and has an allele frequency of 0.0001 (ExAC database) (Levran et al. 2005; Castella et al. 2011). The second variant (c.4199G > A) is a point mutation in exon 42 and is a rare variant (allele frequency of 0.00004, ExAC database) that results in p.R1400H FANCA. It has previously been documented in two FA patients (Ameziane et al. 2008; Kimble et al. 2018). Clinical testing identified the presence of the same FANCA variants in the proband's brother. These findings were confirmed by research-based Sanger sequencing on blood of the proband (Supplemental Fig. S1) and cDNA from fibroblasts derived from both siblings (Supplemental Fig. S2).
Functional Analyses
Cells Derived from the Proband and Brother Display Characteristics Typical of Fanconi Anemia
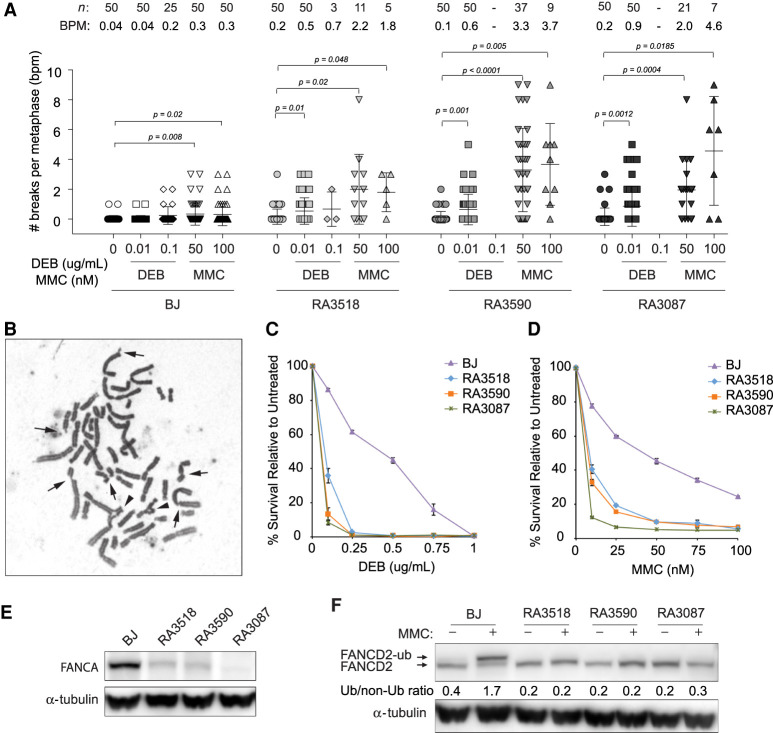
Consistent with a diagnosis of FA, fibroblasts derived from both brothers (proband RA3518, younger sibling RA3590) show increased chromosomal fragility when treated with DNA interstrand cross-linking agents including DEB and MMC (Fig. 1A,B). Chromosomal breakage is elevated after DEB and MMC to levels similar to those observed in fibroblasts from typical FA-A patients (RA3087). RA3087 cells are deficient for FANCA because of homozygous deletion at the FANCA locus (g.88311999_88394999del83001(hg18), delEx9_43) (Kim et al. 2013). Fibroblasts derived from both siblings also show hypersensitivity to these cross-linking agents in cell survival assays (Fig. 1C,D). Cellular sensitivity of RA3518 and RA3590 fibroblast to DEB-treated cells is similar to that seen in RA3087 FA-A patient fibroblast, whereas they are slightly less sensitive to MMC. Analysis of FANCA levels by western blot show that fibroblasts from the siblings (RA3518 and RA3590) have lower levels of endogenous FANCA in comparison to wild-type cells (Fig. 1E).
Figure 1.
Characterization of fibroblasts from FA siblings. (A) Quantification of chromosome breaks of diepoxybutane (DEB)-treated and MMC-treated primary fibroblasts from proband (RA3518) and affected sibling (RA3590) in comparison to BJ wild-type and FANCA-deficient primary fibroblasts (RA3087). Number of metaphases analyzed (n) and breaks per metaphase (BPM) are indicated. Welch's unpaired two-tailed parametric t-test was used to determine statistical significance between untreated and treated cells as indicated. (B) Example of a metaphase spread from the proband's primary fibroblasts following 24-h treatment with 50 nM mitomycin C (MMC). Arrows indicate chromosome breaks and the arrowheads highlight radial figures. (C,D) Cellular sensitivity assays of patients’ E6E7 transformed/hTERT immortalized (EH) fibroblasts in comparison to BJ wild-type and FANCA-null EH fibroblasts after DEB or MMC treatment. Cells were treated in triplicate with increasing drug concentrations. Cells were then counted after 7 d and normalized to untreated control to determine percent survival. Error bars represent standard deviation. (E) Western blot for endogenous FANCA in untreated BJ, affected individuals (RA3518 and RA3590), and RA3087 FANCA-deficient primary fibroblasts. (F) Western blot assessing FANCD2 monoubiquitination in BJ, affected individuals (RA3518 and RA3590), and RA3087 FANCA-deficient primary fibroblasts. Cells were either untreated or cultured with 1 µM MMC for 24 h. Slower migrating band represents monoubiquitinated FANCD2. Relative ratio of monoubiquitinated to nonubiquitinated FANCD2 was measured for each variant.
A key step in the FA repair pathway is monoubiquitination of the FANCI and FANCD2 proteins by FANCL, part of the multiprotein core complex composed of eight FA proteins including FANCA. Fibroblasts from the siblings (RA3518 and RA3590) were found to be deficient for FANCD2 monoubiquitination (Fig. 1F). Taken together these findings confirm that both subjects share a classic FA cellular phenotype, have reduced FANCA levels, and have defective FA pathway function.
Cellular Complementation with Wild-Type FANCA Rescues Sensitivity to ICL-Inducing Agents
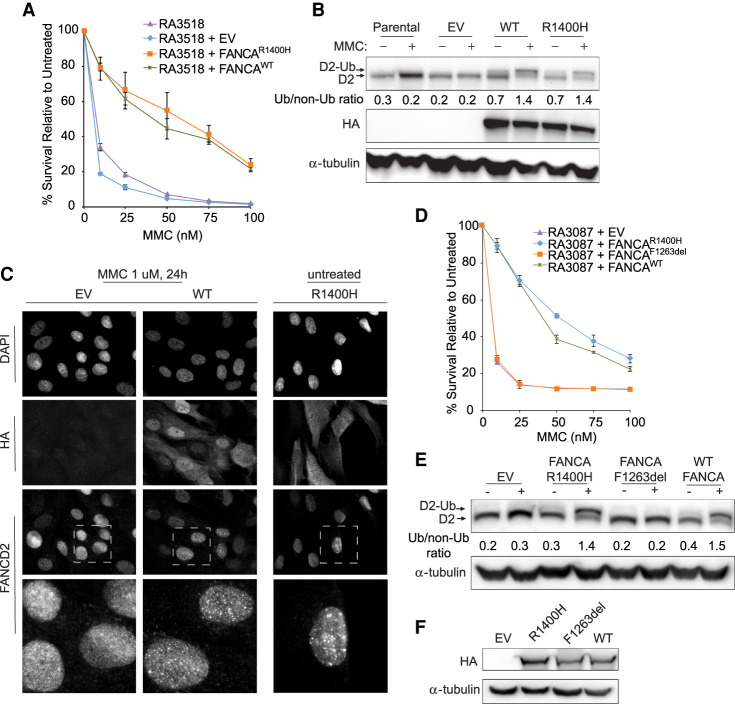
To demonstrate that FANCA deficiency causes the observed cellular defects, cells derived from the proband (RA3518) were complemented with wild-type FANCA cDNA. Overexpression of wild-type FANCA rescued hypersensitivity and FANCD2 monoubiquitination after treatment with MMC (Fig. 2A,B) Surprisingly, overexpression of FANCAc.4199G>A (p.R1400H) also rescued MMC hypersensitivity and FANCD2 monoubiquitination (Fig. 2A,B). Monoubiquitination of FANCD2 is required for its retention on chromatin and participation in the repair of ICLs, so pathway function can also be assayed by immunofluorescence demonstrating FANCD2 foci formation after DNA damage. Overexpression of wild type and R1400H FANCA in RA3518 cells in immunofluorescence studies also rescued FANCD2 foci formation compared to RA3518 cells expressing an empty vector control (Fig. 2C). However, the overexpressed R1400H FANCA protein was found to have cytoplasmic localization compared to the largely nuclear localization of the wild-type protein (Fig. 2C). Nuclear localization of FANCA is critical for proper function and impairment of this process by pathogenic missense variants has been recognized by multiple investigators (Naf et al. 1998; Castella et al. 2011; Kimble et al. 2018). The proband and sibling carry the c.3788_3790delTCT variant that is predicted to express p.F1263del FANCA, a previously reported and well-studied pathogenic mutant (Adachi et al. 2002). Overexpression of F1263del FANCA does not rescue cellular sensitivity of FA-deficient RA3087 patient fibroblasts or FANCD2 monoubiquitination after treatment with MMC (Fig. 2D–F). These data support a hypothesis that p.R1400H FANCA is a hypomorphic allele and p.F1263del is a loss of function allele. Unexpectedly, FANCA R1400H, when overexpressed, provides enough function to restore resistance to cross-linking agents and rescue FANCD2 ubiquitination to wild-type levels.
Figure 2.
Overexpression of FANCA R1400H rescues hypersensitivity to mitomycin C (MMC), FANCD2 ubiquitination, and localization of FANCD2 to sites of damage. (A) MMC cellular sensitivity assay of proband RA3518 fibroblasts with no vector expression (parental), expressing empty vector (EV), or overexpression of FANCA mutant (FANCAR1400H) or wild-type FANCA (FANCAWT). (B) Western blot assessing FANCD2 monoubiquitination and level of HA tagged FANCA expression in parental (no vector) and EV, FANCAR1400H, and FANCAWT expressing RA3518 fibroblasts. Cells were either untreated or cultured with 1 µM MMC for 24 h. (C) Immunofluorescence images with either anti-HA to detect overexpressed FANCA or anti-FANCD2 to detect FANCD2 localization to chromatin after treatment with 1 µM MMC for 24 h in RA3518 fibroblasts expressing EV, or overexpression of wild-type FANCA (FANCAWT) or FANCA mutant (FANCAR1400H). (D) MMC cellular sensitivity assay of FANCA-deficient RA3087 fibroblasts either expressing EV, a FANCA mutant (FANCAR1400H or FANCAF1263del), or wild-type FANCA (FANCAWT). (E) Western blot assessing FANCD2 monoubiquitination in FANCA deficient RA3087 fibroblasts overexpressing p.F1263del FANCA. Cells were either untreated or cultured with 1 µM MMC for 24 h. Relative ratio of monoubiquitinated to nonubiquitinated FANCD2 was measured for each variant. (F) Western blot assessing level of HA-tagged FANCA expression in RA3087 cells.
Genome Engineered Cells Expressing R1400H FANCA from the Endogenous Locus Have Defective ICL Repair
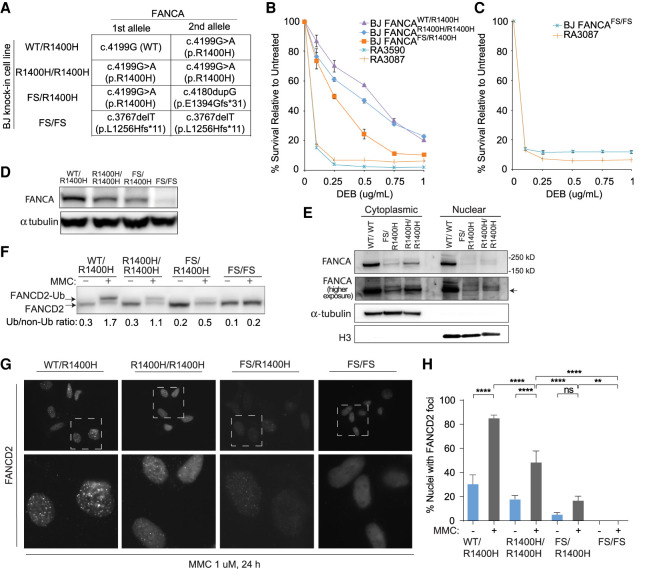
To further support our hypothesis that the R1400H FANCA is responsible for the milder FA phenotype observed in the siblings, the c.4199G > A missense variant was knocked-in at the endogenous FANCA locus of BJ cells to produce R1400H FANCA expressing cells (Fig. 3A; Supplemental Fig. S3). A biallelic c.4199G > A knock-in created a homozygous state. A carboxy-terminal frameshift allele (p.E1394Gfs*31) was engineered in trans with c.4199G > A variant creating a compound heterozygous state. In addition, a cell line with biallelic p.L1256Hfs*11 frameshift (FS) was used as a null FANCA control. BJ cells demonstrated a dose effect with homozygous FANCAR1400H/R1400H-expressing cells having a mild sensitivity to MMC, compound heterozygous FANCAR1400H/FS-expressing cells demonstrating a more moderate sensitivity, and FANCAFS/FS-expressing cells demonstrating sensitivity similar to typical FA-A fibroblasts RA3087 (Fig. 3B,C). Analysis of FANCA levels in BJ fibroblast expressing the FANCA variants by western blot show progressively decreased levels in FANCAR1400H/R1400H and FANCAR1400H/FS-expressing cells and absence of protein in FANCAFS/FS-expressing cells (Fig. 3D). Consistent with the partially cytoplasmic localization of the overexpressed R1400H FANCA protein (Fig. 2C), greater portion of the endogenously expressed R1400H FANCA is present in the cytoplasmic than in the nuclear fraction (Fig. 3E). BJ cells also demonstrated a dose effect with regard to FANCD2 monoubiquitination. A mild decrease in the slower migrating monoubiquitinated FANCD2 band is observed in the homozygous FANCAR1400H/R1400H-expressing cells, a more pronounced decrease in the compound heterozygous FANCAR1400H/FS-expressing cells, and absence in the FANCAFS/FS-expressing cells (Fig. 3F). These results correlated well with the observed defect in FANCD2 foci formation in the BJ fibroblasts expressing the FANCA variants (FANCAR1400H/R1400H < FANCAR1400H/FS < FANCAFS/FS) (Fig. 3G–H). In summary, CRISPR–Cas9 mediated genome engineering of the FANCA c.4199G > A (p.R1400H) variant into an alternate wild-type human fibroblast cell line demonstrates it is a pathogenic hypomorphic allele and that ICL repair is sensitive to the amount of hypomorphic allele available in the cell.
Figure 3.
Genome editing of wild-type fibroblasts to express p.R1400H FANCA endogenously results in decreased FANCA levels and impaired FANCD2 ubiquitination and foci formation upon MMC-induced damage. (A) Genotypes and predicted effects of CRISPR-mediated FANCA variant knock-in clones including heterozygous (WT/R1400H), homozygous (R1400H/R1400H), compound heterozygous frameshift (FS/R1400H), and homozygous frameshift (FS/FS) were generated in BJ fibroblasts. (B,C) DEB cellular sensitivity assays showing survival of patient cell lines in comparison to FANCA variant knock-in clones. (D) Western blot assessing endogenous FANCA expression in untreated BJ fibroblast FANCA clones. (E) Western blot assessing endogenous FANCA upon cellular fractionation of untreated FANCA variant knock-in clones. (F) Western blot assessing FANCD2 monoubiquitination in BJ fibroblast FANCA clones. Cells were either untreated or cultured with 1 µM MMC for 24 h. Relative ratio of monoubiquitinated to nonubiquitinated FANCD2 was measured for each variant. (G) Images of immunofluorescence of FANCD2 foci in BJ fibroblast FANCA clones. Lower panel images are magnified to demonstrate differences in foci formation. Cells were preextracted prior to fixation. (H) Quantification of cells observed to have FANCD2 foci in BJ fibroblast FANCA clones by immunofluorescence. Cells were either untreated or cultured with 1 µM MMC for 24 h. Statistical significance determined by ordinary one-way ANOVA with multiple comparisons. (****) P < 0.0001; (**) 0.0021 < P < 0.0002.
DISCUSSION
Here we have presented a family with two affected brothers both diagnosed with esophageal cancer at the age of 51 yr as their initial presentation of FA. Prior to entering the sixth decade of life, both brothers were relatively healthy. Neither had exhibited any of the characteristic hematologic findings of FA in terms of bone marrow failure or myelodysplasia. Notably, they had very subtle stigmata of FA in the form of minor thumb-related anomalies with otherwise unremarkable physical exams including normal height, weight, and BMI. Additionally, both patients were able to father biological children, in stark contrast to the majority of FA male patients who have hypogonadism and impaired fertility.
This atypical and mild clinical presentation of FA later in life is consistent with the hypothesis that although the p.F1263del allele likely provides no residual FANCA function, the second hypomorphic p.R1400H allele provides partial function. p.R1400H FANCA likely supports enough FA pathway function to protect germ cell development and the bone marrow under non–stress conditions. However, the excessive toxicity experienced by the proband upon exposure to chemotherapy shows that the hematologic compartment remains vulnerable to effects of DNA damage under conditions of stress. Cellular sensitivity assays with DEB and MMC confirm hypersensitivity of both patient-derived cell lines to these agents, which is consistent with classic FA findings.
The c.4199G > A/p.R1400H variant has been previously described in two other individuals with Fanconi anemia, but determining pathogenicity was difficult because, similar to our findings, it was reported that overexpression of the mutant allele resulted in complementation comparable to that obtained with the wild-type cDNA (Ameziane et al. 2008). This suggests that overexpression of the mutant protein is able to compensate for protein instability and/or defects in FANCA protein function. Here the additional findings of the c.4199G > A/ p.R1400H variant in another FA family and our functional data support the variant as a most likely pathogenic allele. We observe increased mislocalization of mutant protein to the cytoplasm, decreased FANCD2 ubiquitination and foci formation, and cellular sensitivity to cross-linking agents. Examining the variant by CRISPR–Cas9-mediated genome editing in an alternate cell line was key to demonstrating the defects of the R1400H mutant, which were further exacerbated when present in trans to a loss-of-function allele indicating a dose effect of a mutant protein.
Although the proband and sibling did not develop bone marrow failure, a classic presentation of FA, the diagnosis of SCC in both siblings demonstrates that the tumor-suppressor function of the FANCA hypomorphic allele is incomplete. This raises the question of whether there are other unidentified factors, either environmental or genetic, that account for increased susceptibility to malignancy. The underlying pathophysiology of SCC is not well-understood, but it is possible that the threshold for FA pathway function is tissue-specific, with the epithelial mucosa being in need of higher levels of function than the bone marrow stem cells over a long period of time. It is remarkable, although not unusual, for Fanconi anemia patients to develop multiple SCCs in their lifetime. In the case described here, the younger sibling developed three mucosal SCCs in the span of 2 yr, emphasizing the need for the fully functional FA pathway of DNA repair in the protection against cancer.
This study highlights the significance of recognizing hypomorphic variants in cases of adult-onset FA. Subtle clinical findings underscore why clinicians must have a low threshold for testing for FA in unusually young patients with SCC as highlighted here and in patients with other cancers including AML, breast cancer, liver, and cholangiocarcinoma (Huck et al. 2006; Stevens et al. 2016; Engel et al. 2019). Malignancy was a major cause of morbidity and mortality for these patients, and FA-specific treatment regimens need to be selected given the extreme hypersensitivity to chemotherapy.
METHODS
Cell Culture and Viral Transfection/Transduction
Fibroblasts were grown in DMEM supplemented with 15% [v/v] FBS, 100 units of penicillin per milliliter and 0.1 mg of streptomycin per milliliter, nonessential amino acids, and 1× GlutaMAX (Invitrogen). Fibroblasts were incubated at 3% oxygen. Cell lines were immortalized with a catalytic subunit of human telomerase (hTERT) and were transformed by HPV E6 and E7 protein, unless otherwise stated in the text. cDNAs were delivered using lentiviral transduction after packaging in HEK293T cells according to manufacturer's protocol (Mirus) and selected in puromycin.
Sequencing
Sanger sequencing was used to confirm presence of variants identified through clinical studies. The variants were amplified from the patients’ genomic DNA by polymerase chain reaction (PCR) and the products were sequenced directly. First-strand cDNA synthesis (Superscript III, Invitrogen), reverse-transcription (RT)-PCR, and Topo cloning (Thermo Fisher Scientific) allowed for the segregation of the variant alleles. PCR and sequencing primers are indicated in Supplemental Table S2.
Chromosomal Breakage and Sensitivity Assays
Analysis of chromosomal breakage was performed following treatment with DNA damaging agents as described previously (Kim et al. 2011). For sensitivity assays, fibroblasts were seeded overnight and treated the following day with DEB and MMC at the indicated concentrations. Cells were grown for 3–4 d, passaged at appropriate ratios, and counted using Z2 Coulter counter (Beckman Coulter) upon reaching 90% confluency.
Cellular Fractionation
Cellular fractions were prepared by suspending cells in fractionation buffer (20 mM HEPES pH 7.4, 2 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA) and incubating on ice for 20 min. Following centrifugation, the supernatant was reserved as the cytoplasmic fraction. The remaining nuclear pellet was washed once in fractionation buffer and resuspended in TBS with 0.1% SDS.
Western Blot and Antibodies
Whole-cell lysates were prepared by lysing in Laemmli buffer (Bio-Rad) and sonication. Samples were boiled and separated on 3%–8% gradient gels (Invitrogen) by SDS-PAGE. Immunoblotting was performed using the following antibodies: FANCA (Bethyl Antibody, A301-980A), FANCD2 (Novus NB100-182), H3 (Abcam, ab1791), and HA (Covance MMS-101R).
Immunofluorescence
Cells were fixed with 3.7% formaldehyde, permeabilized with 0.5% NP-40 in PBS, blocked in 5% [v/v] FBS in PBS, and incubated with indicated antibodies (1:1000) in blocking buffer. Cells were incubated with Alexa Fluor secondary antibodies to visualize FANCD2 foci (488) and HA-FLAG tagged plasmid expression (594). The cells were washed and the coverslips were mounted with DAPI Fluoromount-G (SouthernBiotech). When preextraction was used, cells were treated with 0.5% TritonX-100 in PBS for 5 min at room temperature before fixation with 3.7% formaldehyde.
Plasmids
The wild-type (WT) FANCA cDNA was expressed from a CMV promoter and had an amino-terminal HA-FLAG tag. The microdeletion mutant (c.3788_3790delTCT) and the missense mutant (c.4199G > A) were generated with the QuikChange II XL-Site Directed Mutagenesis kit (Agilent Technologies) using a template of WT FANCA cDNA cloned in pENTR plasmid. Mutant constructs were then moved into a lentiviral vector via LR reaction (Gateway system). Sanger gene sequencing verified presence of variants.
CRISPR–Cas9 Genome Editing
Genome editing was performed by electroporation of Cas9/gRNA ribonucleotide complexes as previously described (Rickman et al. 2020). DirectPCR lysis reagent was utilized per manufacturer's protocol to generate lysates from BJ fibroblast clones and used for PCR screening. PCR was performed using Taq DNA polymerase (QIAGEN) per the manufacturer's protocol (for primers see Supplemental Table S2). Amplified PCR products were purified using ExoSAP-IT protocol (ThermoFisher Scientific) and Sanger sequencing was performed to confirm variants.
ADDITIONAL INFORMATION
Data Deposition and Access
Fibroblasts RA3518 and RA3590 are available from the International Fanconi Anemia Registry. The variant c.4199G > A/p.R1400H is in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with accession number VCV000408175.3. The c.3788_3790delTCT/ p.F1263del has ClinVar accession number VCV000041003.13.
Ethics Statement
Both subjects were enrolled in the International Fanconi Anemia Registry (IFAR), under a protocol approved by the Institutional Review Board of Rockefeller University. Medical information and cells were collected after obtaining informed written consent from the subjects. Additional medical information about the subjects was obtained with written consent from the next of kin after subjects’ passing.
Acknowledgments
We would like to thank the subjects and their families for their participation in this study.
Author Contributions
F.P.L., S.S., K.A.R., P.D.R., R.J.N., S.C.C., and A.S. designed and performed experiments and interpreted data. A.S. is IFAR's PI. J.A.K. enrolled the family in the study. K.B.H. and M.D.D. provided clinical data pertaining to the subjects. S.S., K.A.R., and A.S. wrote the manuscript with input from other authors.
Funding
This work was supported by grants from the National Heart Lung and Blood Institute (R01 HL120922) (A.S.), National Cancer Institute (R01 CA204127) (A.S.), and National Center for Advancing Translational Sciences (UL1 TR001866). S.C.C. is supported by the Intramural Research Program of the National Human Genome Research Institute, NIH. K.A.R. was supported by the Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program and the William Randolph Hearst Foundation Fellowship at the Rockefeller University. P.D.R. is supported by the Anderson Center for Cancer Research Fellowship at The Rockefeller University. A.S. is a Howard Hughes Medical Institute (HHMI) Faculty Scholar.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Detlev Schindler
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adachi D, Oda T, Yagasaki H, Nakasato K, Taniguchi T, D'Andrea AD, Asano S, Yamashita T. 2002. Heterogeneous activation of the Fanconi anemia pathway by patient-derived FANCA mutants. Hum Mol Genet 11: 3125–3134. 10.1093/hmg/11.25.3125 [DOI] [PubMed] [Google Scholar]
- Alter BP, Rosenberg PS. 2013. VACTERL-H association and Fanconi anemia. Mol Syndromol 4: 87–93. 10.1159/000346035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Rosenberg PS, Brody LC. 2007. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 44: 1–9. 10.1136/jmg.2006.043257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS. 2010. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol 150: 179–188. 10.1111/j.1365-2141.2010.08212.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameziane N, Errami A, Leveille F, Fontaine C, de Vries Y, van Spaendonk RM, de Winter JP, Pals G, Joenje H. 2008. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat 29: 159–166. 10.1002/humu.20625 [DOI] [PubMed] [Google Scholar]
- Asur RS, Kimble DC, Lach FP, Jung M, Donovan FX, Kamat A, Noonan RJ, Thomas JW, Park M, Chines P, et al. 2018. Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype. Mol Genet Genomic Med 6: 77–91. 10.1002/mgg3.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd PJ, Stewart GS, Smith A, Eaton C, Taylor AJ, Guy C, Eringyte I, Fooks P, Last JI, Horsley R, et al. 2016. A hypomorphic PALB2 allele gives rise to an unusual form of FA-N associated with lymphoid tumour development. PLoS Genet 12: e1005945 10.1371/journal.pgen.1005945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castella M, Pujol R, Callen E, Trujillo JP, Casado JA, Gille H, Lach FP, Auerbach AD, Schindler D, Benitez J, et al. 2011. Origin, functional role, and clinical impact of Fanconi anemia FANCA mutations. Blood 117: 3759–3769. 10.1182/blood-2010-08-299917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel NW, Schliffke S, Schuller U, Frenzel C, Bokemeyer C, Kubisch C, Lessel D. 2019. Fatal myelotoxicity following palliative chemotherapy with cisplatin and gemcitabine in a patient with stage IV cholangiocarcinoma linked to post mortem diagnosis of Fanconi anemia. Front Oncol 9: 420 10.3389/fonc.2019.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre L, Guardiola P, Lewis C, Dokal I, Ebell W, Zatterale A, Altay C, Poole J, Stones D, Kwee ML, et al. 2000. Association of complementation group and mutation type with clinical outcome in Fanconi anemia. European Fanconi Anemia Research Group. Blood 96: 4064–4070. [PubMed] [Google Scholar]
- Fiesco-Roa MO, Giri N, McReynolds LJ, Best AF, Alter BP. 2019. Genotype–phenotype associations in Fanconi anemia: a literature review. Blood Rev 37: 100589 10.1016/j.blre.2019.100589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn EK, Kamat A, Lach FP, Donovan FX, Kimble DC, Narisu N, Sanborn E, Boulad F, Davies SM, Gillio AP, et al. 2014. Comprehensive analysis of pathogenic deletion variants in Fanconi anemia genes. Hum Mutat 35: 1342–1353. 10.1002/humu.22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri N, Batista DL, Alter BP, Stratakis CA. 2007. Endocrine abnormalities in patients with Fanconi anemia. J Clin Endocrinol Metab 92: 2624–2631. 10.1210/jc.2007-0135 [DOI] [PubMed] [Google Scholar]
- Gregory JJ, Wagner JE, Verlander PC, Levran O, Batish SD, Eide CR, Steffenhagen A, Hirsch B, Auerbach AD. 2001. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci 98: 2532–2537. 10.1073/pnas.051609898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. 10.1126/science.1073834 [DOI] [PubMed] [Google Scholar]
- Huck K, Hanenberg H, Gudowius S, Fenk R, Kalb R, Neveling K, Betz B, Niederacher D, Haas R, Gobel U, et al. 2006. Delayed diagnosis and complications of Fanconi anaemia at advanced age—a paradigm. Br J Haematol 133: 188–197. 10.1111/j.1365-2141.2006.05998.x [DOI] [PubMed] [Google Scholar]
- Jung M, Mehta PA, Jiang CS, Rosti RO, Usleaman G, da Rosa JM C, Lach FP, Goodridge E, Auerbach AD, Davies SM, et al. 2020a. Comparison of the clinical phenotype and haematological course of siblings with Fanconi anaemia. Br J Haematol 10.1111/bjh.17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Ramanagoudr-Bhojappa R, van Twest S, Rosti RO, Murphy V, Tan W, Donovan FX, Lach FP, Kimble DC, Jiang CS, et al. 2020b. Association of clinical severity with FANCB variant type in Fanconi anemia. Blood 135: 1588–1602. 10.1182/blood.2019003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146. 10.1038/ng.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. 2013. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 121: 54–63. 10.1182/blood-2012-07-441212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble DC, Lach FP, Gregg SQ, Donovan FX, Flynn EK, Kamat A, Young A, Vemulapalli M, Thomas JW, Mullikin JC, et al. 2018. A comprehensive approach to identification of pathogenic FANCA variants in Fanconi anemia patients and their families. Hum Mutat 39: 237–254. 10.1002/humu.23366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD. 2003. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101: 1249–1256. 10.1182/blood-2002-07-2170 [DOI] [PubMed] [Google Scholar]
- Kutler DI, Patel KR, Auerbach AD, Kennedy J, Lach FP, Sanborn E, Cohen MA, Kuhel WI, Smogorzewska A. 2016. Natural history and management of Fanconi anemia patients with head and neck cancer: a 10-year follow-up. Laryngoscope 126: 870–879. 10.1002/lary.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Diotti R, Pujara K, Batish SD, Hanenberg H, Auerbach AD. 2005. Spectrum of sequence variations in the FANCA gene: an International Fanconi Anemia Registry (IFAR) study. Hum Mutat 25: 142–149. 10.1002/humu.20125 [DOI] [PubMed] [Google Scholar]
- Naf D, Kupfer GM, Suliman A, Lambert K, D'Andrea AD. 1998. Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol Cell Biol 18: 5952–5960. 10.1128/MCB.18.10.5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti E, Rao G, Bueren JA, Rio P, Navarro S, Surralles J, Choi G, Schwartz JD. 2020. Mosaicism in Fanconi anemia: concise review and evaluation of published cases with focus on clinical course of blood count normalization. Ann Hematol 99: 913–924. 10.1007/s00277-020-03954-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niraj J, Farkkila A, D'Andrea AD. 2019. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol 3: 457–478. 10.1146/annurev-cancerbio-030617-050422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk A, Kanakatti Shankar R, Giri N, Hollenberg AN, Rutter MM, Nathan B, Lodish M, Alter BP, Stratakis CA, Rose SR. 2015. Endocrine disorders in Fanconi anemia: recommendations for screening and treatment. J Clin Endocrinol Metab 100: 803–811. 10.1210/jc.2014-4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O, Chandrasekharappa SC, et al. 2015. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep 12: 35–41. 10.1016/j.celrep.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman KA, Noonan RJ, Lach FP, Sridhar S, Wang AT, Abhyankar A, Huang A, Kelly M, Auerbach AD, Smogorzewska A. 2020. Distinct roles of BRCA2 in replication fork protection in response to hydroxyurea and DNA interstrand cross-links. Genes Dev 34: 21–22. 10.1101/gad.336446.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg PS, Alter BP, Ebell W. 2008. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica 93: 511–517. 10.3324/haemotol.12234 [DOI] [PubMed] [Google Scholar]
- Rosenberg PS, Tamary H, Alter BP. 2011. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A 155a: 1877–1883. 10.1002/ajmg.a.34087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens H, Chyn Chua C, Wallis M, Hew S, Grigg A. 2016. Fanconi anemia in 55-year-old identical twins first presenting as fatal post-chemotherapy pancytopenia. Am J Hematol 91: 1273–1276. 10.1002/ajh.24488 [DOI] [PubMed] [Google Scholar]
- Tamary H, Nishri D, Yacobovich J, Zilber R, Dgany O, Krasnov T, Aviner S, Stepensky P, Ravel-Vilk S, Bitan M, et al. 2010. Frequency and natural history of inherited bone marrow failure syndromes: the Israeli Inherited Bone Marrow Failure Registry. Haematologica 95: 1300–1307. 10.3324/haemotol.2009.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. 2015. SnapShot: Fanconi anemia and associated proteins. Cell 160: 354–354 e351. 10.1016/j.cell.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39: 159–161. 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fibroblasts RA3518 and RA3590 are available from the International Fanconi Anemia Registry. The variant c.4199G > A/p.R1400H is in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) with accession number VCV000408175.3. The c.3788_3790delTCT/ p.F1263del has ClinVar accession number VCV000041003.13.