Abstract
Background
Currently, PCR assay is a golden standard for diagnosis of Covid-19. However, it needs nasopharyngeal swabs, expensive instruments and expertise. It even causes PCR errors.
Methods
We validated the antibody assay (Roche) in 36 followed patients and 1879 controls (medical staffs).
Results
Of 1879 medical staffs, only two (0.11%) were positive by Cut off Index (COI; 1.0) (mean ± SD, 0.094 ± 0.047). Thirty six patients were composed of three groups; Group A,4 from Diamond Princess cruise ship, Group B, 2 infected in Africa, and Group C, 30 infected in Japan. PCR assays were conducted at outside laboratories before and repeated in house after hospitalized. Of 36 at admission, positive antibody was seen in 4/4 from the ship, 0/2 from Africa, and 5/30 from Japan. Two from Africa showed the increase of COI and became positive on days 8 and 13. Thirty Japanese was divided in two groups, e.g., 23 showed dynamic increase of COI up to 84.4 within 3 days while active virus replication present (Group C). In remaining 7 (7/30, 23%) (Group C'), no rise of antibody nor positive in house PCR assays, indicative of false positive results of PCR at the beginning.
Conclusion
This antibody testing has a wide dynamic ranges of COI and, thus, could be utilized in the early infection phase. This may also compliment and even help to avoid possible PCR errors. Therefore, this can serve as a powerful diagnostic tool, needed in the frontline of the clinic and hospitals.
Keywords: Covid-19, Early diagnosis, Antibody test, PCR assay
Introduction
The number of the individuals infected by Covid-19 virus may reach over 10 million at the end of June, 2020.1 , 2 The diagnostic tool for this infection includes PCR assay as the only measure employed worldwide.3 However, PCR assay is time consuming, expensive, cumbersome, and need some expertise.4 In particular, PCR error causes tremendous confusions. The preventive measures were required for people involved including the family, office workers, hospital staff and close contacts, if PCR assay were reported positive.
However, PCR assay is still a golden standard to deicide the prevention and choice of treatment modality.5 , 6 Other modalities of the antigen–antibody assay is about to be developed.7 However, the knowledge about these antigen and antibody assay is still limited.
In this communication, we report the developed antibody assay that we employed may become a very powerful diagnostic tool not only for patients in the convalescent phase but in the early phase of this infection. This can even be easily available at clinics and hospitals. Comparison was made to the golden standard PCR assays in serial swab samples and the time course of the development of the antibodies in our Japanese patients along with patients from outside (Diamond Princess cruise ship and Africa).
Methods
Patients and controls
Group A. 4 patients from Diamond Princess
On February 11th, 2020, we were asked from the government to see patients from the Diamond Princess. We are 150 km west of Tokyo, in the city of Kofu, where 400,000 people are living.
No infected patients by Covid-19 virus had existed till then. However, we are the 600 bed medical center of the district, with emergency helicopters and intensive care units. We are prepared to have all the patients in critical conditions. We received the patient on February 11th, one couple of Americans, husband and wife. At the beginning, husband walked in but in a few days, needed ventilator. However, no improvement of blood gas was seen. On February 19th, our emergency team did a bronchoscope drainage to take the build-up plaque out of the major bronchial tree. After that, blood gas dramatically improved. In the meantime, the infected wife developed massive brain hemorrhage. Decompensating surgery was done for this wife and they both went back to the homeland safely in the end of April, after 2.5 months hospital stay. If they passed away, they could be the 1st American.
The other two from Diamond Princess were one Chinese and one Japanese passenger who went home uneventfully by the end of February.
During the hospital stays of four patients, we took 11 blood samples and 19 nasopharyngeal swabs.
Group B. 2 patients from Africa
In March, one couple (pregnant wife and husband) back from Africa were transferred from other hospital. They were obviously infected in Africa. Their clinical course was uneventful and discharged in 2 weeks. During their hospital stay, we took 12 serial blood samples and 15 nasopharyngeal swabs for PCR assay.
Group C and C'. 30 from Japan, Yamanashi
After the experience on 6 aforementioned patients from outside with the very stressful daily practices and some relief for whole hospital staff, we hoped this will not happen in Japan. However, number of infected patients incrementally increased in our country from the beginning of the April and reached the height on April 12th 7. Furthermore, we are now facing the “2nd wave” by the end of October, 2020.
Since then, 30 patients from our district were sent to us with the diagnosis of Covid-19 infection by PCR assay performed outside laboratories. During the hospital stay of 30 Japanese patients, we took 213 serial blood samples and 276 nasopharyngeal swabs for PCR assay.
Controls
By the end of October, 1879 blood samples were taken from 1879 medical staffs from all divisions of hospitals including infection care and emergency units. None of them had any signs and symptoms. Informed consent was obtained.
Sample collection for the antibody test (Roche) and PCR assays
At outside laboratories, PCR assays were conducted, diagnosed as having Covid-19 infection, and sent to us in all 36 patients except eight. The information on PCR assays, namely positive or negative results and cycle thresholds (Ct) values, were sent from outside laboratories. In the eight cases (#13, #17, #23, #24, #25, #27, #28, #29, Group C), the PCR assay for the diagnosis of Covid-19 infection was performed by in house laboratory at our hospital and immediately hospitalized.
As soon as they were hospitalized, we took the swab and blood and tested for Covid-19 nucleic acid RNA and the antibody. Collected samples of nasopharyngeal swab after admission was used for in house PCR testing. This swab was taken by our medical staff with fully equipped personal protective equipment (PPE).
Altogether, 310 nasopharyngeal swabs (average 8.6 per patient) and 236 serial blood samples (average 6.6 per patient) were collected from 36 patients.
Antibody test and COI (cut off index)
To screen the presence of antibody against SARS-CoV-2, serum were tested by using the Elecsys Anti SARS CoV 2 (S300) RUO (Roche Diagnostics, Basal, Switzerland) on cobas 8000 automated platform. Recombinant protein representing the nucleocapsid (N) antigen was used for the determination of antibodies. This assay utilizes the electrochemiluminescence immunoassay (ECLIA) principle.8
In brief, biotinylated recombinant antigen and ruthenium-labeled recombinant antigen form a sandwich complex. After addition of streptavidin-coated microparticles, sandwich complexes are magnetically captured onto the surface of the electrode, then induces chemiluminescent emission.
Samples with a Cut off Index (COI; signal sample/cut-off) <1.0 were considered as negative, those with a COI≥1.0 were considered as reactive (positive).
Viral nucleic acid extraction
We collected nasopharyngeal swabs with cotton swabs and universal transport media (Copan, Murrieta, CA). Total nucleic acid was automatically isolated using the MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA) on automated machine KingFisher Duo Prime as previously described.9
Briefly, we added 200 μL of universal transport media, 5 μL of Proteinase K, 265 μL Binding Solution, 10 μL Total Nucleic Acid Binding Beads, 0.5 mL Wash Buffer, and 1 mL or 0.5 mL of 80% Ethanol to each well of a Deep-well 96-well plate. 70 μL of Elution solution was added to Elution Strip. Total nucleic acids were stored at −80 °C.
Reverse transcription PCR (RT-PCR)
The protocol was designed by the National Institute of Infectious Diseases (NIID), Japan.10 To detect SARS-CoV-2, we performed one-step RT-PCR according to the NIID protocol (version 2.7).
The reaction mixture comprised 5 μL of 4 × TaqMan Fast Virus 1-Step Master Mix, 1.0 μL of 10 μM forward primer (5′-AAATTTTGGGGACCAGGAAC-3), 1.4 μL of 10 μM reverse primer (5′-TGGCAGCTGTGTAGGTCAAC-3′), 0.8 μL of 5 μM probe (5′-FAM-ATGTCGCGCATTGGCATGGA-TAMRA-3′), 6.8 μL of nuclease-free water, and 5 μL of sample in a 20 μL total volume. For the internal positive control, the human ribonuclease P 30 subunit (RPP30) gene was used (Integrated DNA Technologies, Coralville, IA).11
The RT-qPCR assays were conducted on a StepOnePlus Real-Time PCR Systems machine (Thermo Fisher Scientific) with the following cycling conditions: 50 °C for 5 min for reverse transcription, 95 °C for 20 s, and 45 cycles of 95 °C for 3 s and 60 °C for 30 s. The threshold line was set at 0.2. The Ct value was assigned to each PCR reaction and the amplification curve was visually assessed.
Study protocol
This study protocol was approved by the Institutional Review Board of our hospital.
Results
Four patients from Diamond Princess (Group A)
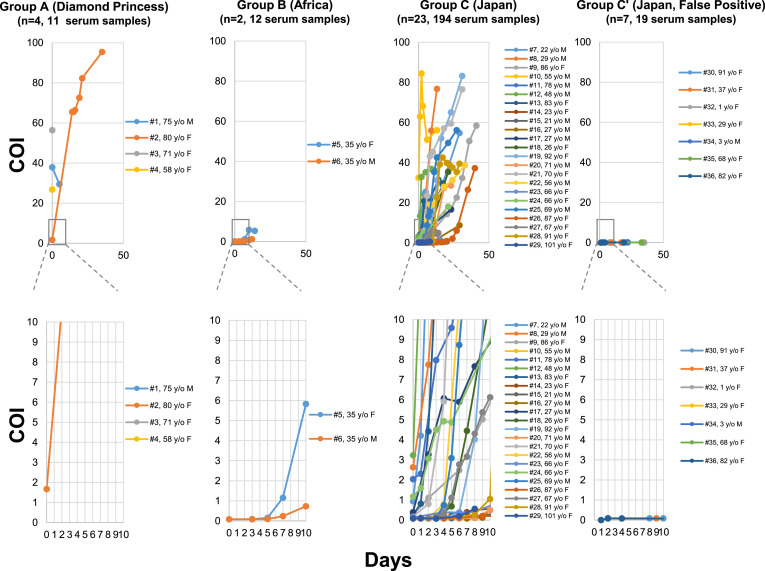
In all four patients, PCR assays were conducted at, and turned out positive and directly transferred from the Diamond Princess cruise ship. Exact dates of infection were not known, but all were positive for the antibody on admission with rise of Cut off Index (COI) of antibody (Fig. 1 ). Initial COI of four cases were 37.9, 1.67, 56.4, and 26.8, in cases #1, #2, #3, and #4, respectively (Fig. 1, Group A). In case #2 which showed the lowest COI, we tested the change of COI in 12 serial samples from day one to day 40. The COI markedly increased to 95.4 on day 40 (Fig. 1, Group A).
Figure 1.
Antibody response in 36 patients. Altogether 236 serum samples from immediately after hospitalization were collected from 36 patients (#1 to #36). Thirty six patients were divided into 4 groups. Group A; 4 patients from Diamond Princess cruise ship, Group B; 2 patients from Africa, Group C; 23 Japanese patients who were diagnosed PCR positive at outside laboratories before transferred to our hospital and re-confirmed by our in house PCR assay, Group C’; 7 Japanese patients diagnosed positive at outside laboratory with Ct (threshold cycle values 36 to 41), but could not be re-confirmed by our laboratory after hospitalization, Serial COI (Cut off Index) was shown from day 0 to day 50 in the upper panel and COI from day 0 to day 10 (expanded) in the lower panel. Abbreviation, y/o, years old; M, male; F, female.
Two patients infected in Africa (Group B)
A couple of pregnant wife (#5) and husband (#6) living in Africa came back to Japan for preparing delivery (Fig. 1, Group B). They were transferred to our hospital because we have neonatal intensive care facility. PCR assays were repeatedly positive, but the antibody response was not reactive on admission (0.0782 and 0.0815), and sluggish in increase of COI but turned positive on day eight (COI: 1.15) and on day 14 (COI: 1.15), respectively (Fig. 1, Group B).
Twenty three Japanese patients (Group C)
Aforementioned six patients were infected outside Japan. From the end of March, 2020, 30 patients from our area were admitted to our hospital.
Of the 30 patients, 23 (#7 to #29) showed very brisk antibody response after admission (Fig. 1, Group C and Table 1 ). We serially checked the change of COI in 194 blood samples from the 23 patients (Group C). Although 18 patients with “negative” tests at admission by pre-defined criteria of COI above 1.0, steady increase of COI were noted in all cases afterwards (Fig. 1, Group C and Table 1).
Table 1.
Serial testings of antibody COI (cut off index) and Ct (cycle threshold) values of PCR assay in 10 days after hospital admission.
| Group | Pt | Method | PCR performed at outside laboratory prior to admission |
Days after admission |
Day of “negative” in two serial PCR assays | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ct value | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||||
| A | #1 | PCR (Ct) | 35 | D21 | ||||||||||
| Ab (COI) | ||||||||||||||
| #2 | PCR (Ct) | 36 | Positive (19) | D13 | ||||||||||
| Ab (COI) | 1.67 | |||||||||||||
| #3 | PCR (Ct) | 29 | Negative | D11 | ||||||||||
| Ab (COI) | 56.4 | |||||||||||||
| #4 | PCR (Ct) | 29 | Negative | Negative | D8 | |||||||||
| Ab (COI) | 26.8 | |||||||||||||
| B | #5 | PCR (Ct) | 21 | Positive (30) | Negative | Positive (36) | D15 | |||||||
| Ab (COI) | 0.08 | 0.08 | 0.15 | 1.15 | ||||||||||
| #6 | PCR (Ct) | 14 | Positive (24) | Negative | Positive (37) | D13 | ||||||||
| Ab (COI) | 0.08 | 0.08 | 0.09 | 0.23 | ||||||||||
| C | #7 | PCR (Ct) | 16 | Positive (19) | Positive (37) | Positive (35) | D15 | |||||||
| Ab (COI) | 0.93 | 4.21 | 15.3 | 24.2 | 25.4 | |||||||||
| #8 | PCR (Ct) | 33 | Positive (33) | Negative | D14 | |||||||||
| Ab (COI) | 2.63 | 7.75 | 22.7 | 55.9 | ||||||||||
| #9 | PCR (Ct) | 22 | Positive (32) | Positive (33) | Positive (43) | Positive (35) | D45 | |||||||
| Ab (COI) | 0.26 | 1.1 | 2.49 | 5.02 | ||||||||||
| #10 | PCR (Ct) | 27 | Positive (>40) | Positive (>40) | Positive (40) | Negative | Positive (41) | Negative | Positive (>40) | D14 | ||||
| Ab (COI) | 32.4 | 62.9 | 84.4 | 68.2 | 51.4 | |||||||||
| #11 | PCR (Ct) | 28 | Positive (28) | Positive (31) | Negative | Positive (37) | D19 | |||||||
| Ab (COI) | 2.04 | 2.31 | 7.97 | 9.59 | 13.3 | |||||||||
| #12 | PCR (Ct) | 19 | Positive (36) | Positive (42) | Positive (33) | Positive (33) | Negative | D13 | ||||||
| Ab (COI) | 3.23 | 13.3 | 32.8 | 35.2 | 36.8 | |||||||||
| #13 | PCR (Ct) | 25a | Positive (25) | Positive (25) | Positive (31) | Positive (28) | Positive (24) | Positive (29) | Positive (35) | Positive (33) | Positive (43) | Positive (35) | D24 | |
| Ab (COI) | 0.25 | 0.82 | 4.42 | 20.8 | 18.5 | 18.2 | 21.6 | |||||||
| #14 | PCR (Ct) | 23 | Positive (30) | Positive (27) | Positive (32) | Positive (37) | Positive (37) | Negative | NA | |||||
| Ab (COI) | 0.08 | 0.08 | 0.09 | 0.13 | 0.16 | 0.22 | 0.29 | 0.39 | 0.50 | |||||
| #15 | PCR (Ct) | 24 | Positive (18) | Positive (16) | Positive (18) | Positive (25) | Positive (28) | NA | ||||||
| Ab (COI) | 0.07 | 0.08 | 0.09 | 0.09 | ||||||||||
| #16 | PCR (Ct) | 18 | Positive (22) | Positive (19) | Positive (29) | Positive (35) | Positive (43) | NA | ||||||
| Ab (COI) | 0.10 | 0.09 | 0.09 | 0.08 | 0.12 | |||||||||
| #17 | PCR (Ct) | 25a | Positive (34) | Positive (39) | Positive (35) | Positive (37) | Negative | NA | ||||||
| Ab (COI) | 0.39 | 3.27 | 6.07 | 5.89 | 7.66 | |||||||||
| #18 | PCR (Ct) | 21 | Positive (25) | Positive (20) | Positive (28) | Positive (38) | NA | |||||||
| Ab (COI) | 0.10 | 0.11 | 0.13 | 0.68 | 4.45 | |||||||||
| #19 | PCR (Ct) | 12 | Positive (16) | Positive (17) | Positive (21) | Positive (19) | Positive (25) | Positive (31) | Positive (30) | Positive (23) | Positive (36) | NA | ||
| Ab (COI) | 0.08 | 0.08 | 0.10 | 0.37 | 4.02 | |||||||||
| #20 | PCR (Ct) | 18 | Positive (17) | Positive (20) | Positive (21) | Positive (21) | Positive (18) | Positive (21) | Positive (19) | Positive (25) | Positive (22) | Positive (29) | NA | |
| Ab (COI) | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.10 | ||||||||
| #21 | PCR (Ct) | 20 | Positive (27) | Positive (31) | Positive (35) | Positive (34) | Positive (31) | Positive (33) | Positive (32) | Positive (35) | Positive (40) | NA | ||
| Ab (COI) | 0.16 | 0.81 | 5.92 | 23.40 | 43.10 | |||||||||
| #22 | PCR (Ct) | 23 | Positive (28) | Positive (17) | Positive (27) | Positive (21) | Positive (26) | Positive (34) | Positive (34) | NA | ||||
| Ab (COI) | 0.08 | 0.09 | 0.80 | 10.90 | 19.10 | |||||||||
| #23 | PCR (Ct) | 23a | Positive (27) | Positive (31) | Positive (30) | Positive (34) | Negative | Positive (39) | Positive (38) | NA | ||||
| Ab (COI) | 0.13 | 0.39 | 0.37 | 0.41 | ||||||||||
| #24 | PCR (Ct) | 32a | Positive (22) | Positive (27) | Positive (35) | Positive (35) | Negative | NA | ||||||
| Ab (COI) | 1.17 | 1.59 | 3.06 | 4.50 | 4.93 | 4.87 | ||||||||
| #25 | PCR (Ct) | 19a | Positive (23) | Positive (29) | Positive (33) | Positive (29) | Positive (37) | Positive (37) | Positive (34) | Positive (38) | Negative | Positive (40) | D22 | |
| Ab (COI) | 0.08 | 0.08 | 0.08 | 0.15 | 0.73 | 3.09 | 8.73 | 13.20 | 15.50 | |||||
| #26 | PCR (Ct) | 16 | Positive (19) | Positive (22) | Positive (21) | Positive (29) | Positive (33) | Positive (31) | Positive (32) | Positive (36) | Positive (38) | Positive (36) | D21 | |
| Ab (COI) | 0.08 | 0.08 | 0.08 | 0.12 | ||||||||||
| #27 | PCR (Ct) | 20a | Positive (23) | Positive (27) | Positive (24) | Positive (27) | Positive (28) | Positive (28) | Positive (32) | Positive (31) | Positive (30) | NA | ||
| Ab (COI) | 0.09 | 0.08 | 0.10 | 0.18 | 0.32 | 1.11 | 2.77 | 3.16 | 4.31 | 5.37 | ||||
| #28 | PCR (Ct) | 19a | Positive (16) | Positive (24) | Positive (23) | Positive (24) | Positive (29) | Positive (29) | Positive (31) | Positive (26) | D29 | |||
| Ab (COI) | 0.07 | 0.08 | 0.08 | 0.16 | 0.28 | |||||||||
| #29 | PCR (Ct) | 16a | Positive (16) | Positive (19) | Positive (20) | Positive (16) | Positive (17) | Positive (25) | Positive (22) | Positive (22) | NA | |||
| Ab (COI) | 0.08 | 0.08 | 0.07 | 0.19 | 0.56 | |||||||||
| C' | #30 | PCR (Ct) | 40 | Negative | Negative | Negative | Negative | PCR repeatedly negative | ||||||
| Ab (COI) | 0.08 | 0.09 | 0.08 | |||||||||||
| #31 | PCR (Ct) | 39 | Negative | Negative | PCR repeatedly negative | |||||||||
| Ab (COI) | 0.08 | 0.08 | ||||||||||||
| #32 | PCR (Ct) | 41 | Negative | Negative | Negative | PCR repeatedly negative | ||||||||
| Ab (COI) | 0.08 | |||||||||||||
| #33 | PCR (Ct) | 37 | Negative | Negative | PCR repeatedly negative | |||||||||
| Ab (COI) | 0.09 | |||||||||||||
| #34 | PCR (Ct) | 39 | Negative | Negative | PCR repeatedly negative | |||||||||
| Ab (COI) | 0.08 | |||||||||||||
| #35 | PCR (Ct) | 37 | Negative | Negative | Negative | Negative | Negative | Negative | Negative | PCR repeatedly negative | ||||
| Ab (COI) | 0.08 | 0.08 | ||||||||||||
| #36 | PCR (Ct) | 36 | Negative | Negative | Negative | PCR repeatedly negative | ||||||||
| Ab (COI) | 0.08 | 0.08 | ||||||||||||
In Case #13, 17, 23, 24, 25, 27, 28 and 29: PCR assay prior to admission was performed at our out-patient clinic by in-house assay.
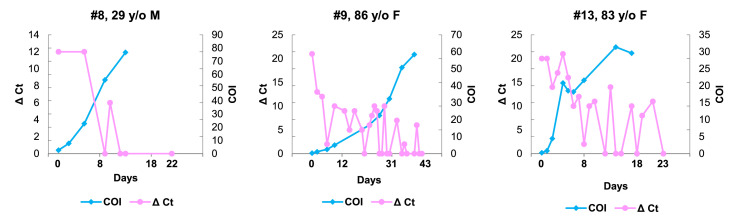
For these 23 patients during the hospital stays, we took nasopharyngeal swabs 248 times and performed PCR assays (average 10.8 times per patient, ranges 4 to 27). Of particular interest, 26 (96%) patients showed steady increase of antibody level while PCR assay demonstrated active virus replication within 10 days (Fig. 2 and Table 1).
Figure 2.
Three representative Japanese patients (#8, #9 and #13) demonstrating early and rapid rise of COI of the antibody from the time of admission while PCR assays repeatedly revealed active virus replication. Delta Ct (Δ Ct) are arbitrarily set to show the changes of virus load by subtraction of Ct values from 45. Abbreviation, y/o, years old; M, male; F, female.
Seven Japanese patients (Group C')
In contrast to the 23 patients with brisk antibody response, seven Japanese patients (#30 to #36) stayed negative for the antibody with very low COI (0.074–0.096) throughout and never became positive (Fig. 1, Group C' and Table 1). All these patients were admitted to our hospital with positive PCR assay results at outside laboratories. After admission, we conducted in house PCR assays for the seven patients 28 times (range, 2 to 7 times). None of them yielded positive results. Thus, in these cases, no evidence of Covid-19 infection was obtained.
The Ct values of PCR assays in these seven cases which we obtained from the outside laboratories were 40, 39, 41, 37, 39, 37 and 36 in cases #30, 31, 32, 33, 34, 35 and 36, respectively (Table 1). These results suggest the antibody test elucidates the presence or absence of Covid-19 infection and compliment the results of PCR assays and salvage the possible errors.
COI and Ct values in 30 Japanese patients
To further ensure possible PCR errors in these 7 cases (Group C′), we compared Ct values of PCR assays and COI of the antibody between 23 cases (Group C) and 7 (Group C'). We obtained the information on Ct values of PCR assays from outside laboratories where the presence of Covid-19 infection was first diagnosed by their PCR assays.
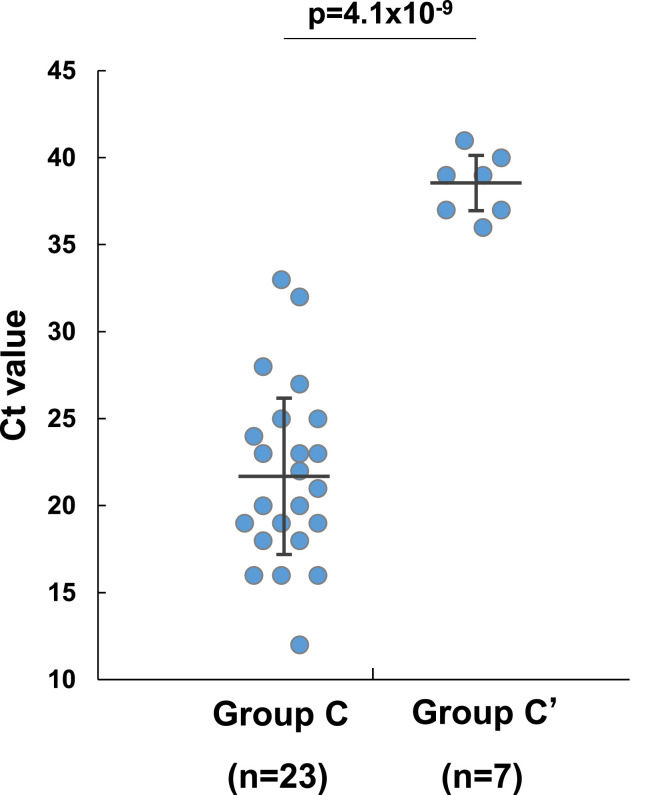
Of the 30 Japanese cases (Groups C and C′), in 23 cases (#7 to #29) the Ct values of PCR ranged from 12 to 33 (mean ± SD, 21.7 ± 5.1), suggesting high virus load. All these cases were proven positive by our repeated in house PCR assays. In addition, the newly introduced antibody assay revealed dynamic change of the indices in serum samples (Fig. 2). In contrast, 7 in Group C’ (#30 to #36) which had basically no antibody response, showed Ct values ranging from 36 to 41 (mean ± SD, 38.4 ± 1.8) (Fig. 3 and Table 1). There were statistically significant differences of Ct values between Group C and Group C' (p = 4.1 × 10−9) (Fig. 3).
Figure 3.
Comparison of Ct values of PCR test in Group C (n = 23) and Group C’ (n = 7). There are significant differences between these two groups (p = 4.1 × 10−9). Group C never reached Ct values of 36 whereas Group C′ was always above. Black bar shows mean ± SD.
COI in 1879 controls
We determined the COI in 1879 medical staffs as a control. Of these, COI was less than 1.0 in 1877 (99.89%). Only two (0.11%) medical staffs had over 1.0 COI (1.220 and 1.100 COI, respectively). However, the COI did not show the increase by the second test (1.180 and 1.080, respectively) and PCR tests were negative in these two control.
These results clearly indicate the Ct values of PCR should be carefully defined to give plus minus results because it is the golden standard of Covid-19 infection. Our data clearly indicate that Ct values of PCR above 36 could be indicative false positive results. However, now the antibody assay may salvage or cover the defects of errors intrinsic to PCR assay.
Discussion
Early diagnosis of the virus infection, especially COIVD-19 is urgently needed. We are in frontline of the patients care in a medical center in Japan. Not only the patients care, we also have to prevent our hospital staff from the infection.12 If you include in and out patients altogether, 3000 to 4000 people in our hospital are at the very high risk of the infection at any time.
We have conducted in house PCR assay in our hospital more than 2000 cases in the past 2 months.4 , 11 , 13 , 14 We have made a swab taking team with full protection for Covid-19 15. Thus, by combining of swab sampling and PCR assay team working hard 24 h a day, we performed PCR assay for all the patients before hospitalization and tried to maintain our ordinary hospital function. In addition, we performed PCR assays for our staff to protect them from invisible agent.15 Fortunately, we started genome analysis center (GAC) 6 years ago in our hospital and some of the member had been involved in molecular biological studies of viruses.16, 17, 18 Thus, we could quickly set up in house PCR assay systems with self-validations.11 , 15 However, it is still time consuming, expensive and need expertise to run PCR assays in a general hospital which require round the clock results. We need more practical method.
When we first tried to detect antibodies against Covid-19 by immunochromatography method, we anticipated IgM antibody can be diagnostic for early Covid-19 infection like hepatitis A or hepatitis B IgM class antibody. However, our preliminary testing by a chromatography assay only showed simultaneous elevation of the both antibodies at the same time (data not shown). Also, for this newly introduced antibody assay (Roche) against Covid-19 in the current study, we anticipated this can be used mainly for patients in convalescent phase. However, as was shown, the index, COI, of the antibody had a very wide dynamic range from 0.08 to over 80, e.g., nearly 1000 times difference. As a control group, we took blood samples from medical staffs. Their COI was extremely low (0.094) and their ranges very narrow (0.047 SD).
If you see the Cut of Index (COI) of the antibody assay, it gave us the very early sign of the infection. At the time of the admission, COI of the antibody was elevated in almost all the cases (Groups A and C). In a few cases, COI of the antibody is below pre-set level of 1.0, but immediately after their values steady increased and reached above the cut of level within 10 days (Fig. 1, Group B).
The current study contained different groups of patients; 4 from Diamond Princess cruise ship who were obviously infected from the end of January, during Wuhan epidemics (Group A)19 and 23 from Japan who were infected in April (Group C). In these two groups, the antibody response is remarkably brisk to the antigens employed in this assay system, whereas the antibody response was present but took time in the couple of pregnant wife and husband from Africa (Fig. 1, Group B). The time and place of infection are so different, namely virus strains may differ. The targeted epitopes of this assay was only defined as selected targets on nucleocapsid portion of virus.20 Hopefully, epitopes of this assay could be presented in public domain and it is of particular interest to sequence the virus nucleotide changes of the epitopes used in our samples.
In countries like Japan where the prevalence of Covid-19 infection in general population is still very low, this test can be a very powerful tool for early diagnosis of Covid-19 infection. In fact, only 2 of 1879 medical staffs were positive with COI of 1.220 and 1.100, respectively. However, in other geographical areas, the presence of antibody by single testing may not be diagnostic for early phase of the infection. However, COI levels at admission may help to make provisional diagnosis of early, intermediate or late phase of infection. Furthermore, by combining clinical features and “acute” increase of COI, the diagnosis on the phases of Covid-19 infection may become more accurate.
We demonstrated 7 cases of seemingly false positive cases of PCR (Group C'). PCR methods were employed so widely, and the only available diagnostic tool for Covid-19 infection. We believe people, unfortunately, overlooked PCR errors (false positive and false negative). We have conducted PCR assays since 198916 , 21 , 22 and always we were troubled by PCR errors.
At least, people have to realize this method is a semiquantitative assay and not simply giving black and white. Our present results clearly indicate high Ct values of PCR could be a “hint” to suspect false positive results (Fig. 3). If PCR needed 36 or more cycles to see the “band”, we need to confirm whether they are true positive results. However, laboratories are obliged, at least in Japan, to give plus and minus answers by predefined indices. Our data certainly indicate PCR cycles over certain numbers have a risk of “false” positive and we advocate setting the gray zone between black and white. Because false positive results, in particular, of Covid-19 infection may provoke enormous amount of not only the medical but also social ‘confusions’, sometime even infringement of privacy.23 , 24
This communications may provide the information on the utility at the antibody against Covid-19. It can be utilized as a very powerful, convenient tool and give the comfort to the medical staff by this assay. This methods can even be employed at the Emergency Department waiting for 10 min and get the useful result whether they are not having any sign of the infection or the possibility of the ongoing current infection by measuring Cut off Index of the antibody. This method could be employed not only for epidemiological mass survey but also in the frontline hospital and clinics. However, obviously the limitation of this study includes the antibody assay alone cannot be utilized as diagnostic tool in countries where many are already infected and antibody prevalence is much higher than Japan.25, 26, 27 In addition to antibody assay, convenient antigen–antibody assay may be needed as was employed in other virus infection.17 , 18
We are currently testing the amino antigen detection method. At end, if we can have nucleic acid, and antigen and antibody tests, we can get back a little closer to ordinal daily hospital practice.
Declaration of competing interest
None.
Acknowledgements
We thank all of the medical and ancillary hospital staff and the patients for consenting to participate. This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.).
References
- 1.Zhu N., Zhang D., Wang, X. Li, B. Yang, J. Song W. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Coronavirus disease (COVID-2019) situation reports. https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 3.World Health Organization Laboratory testing for coronavirus disease (COVID-19) in suspected human cases. https://wwwwhoint/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117
- 4.Hirotsu Y., Maejima M., Shibusawa, Y. Nagakubo, K. Hosaka, K. Amemiya M. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Sci Rep. 2020;10(1):18899. doi: 10.1038/s41598-020-76043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Royal College of Surgeons of England. https://wwwrcsengacuk/coronavirus/
- 6.CDC Evaluating and Testing Persons for Coronavirus Disease 2019 (COVID-19) https://wwwcdcgov/coronavirus/2019-ncov/hcp/clinical-criteriahtml
- 7.COVID-19 Map Johns Hopkins Coronavirus Resource Center. https://coronavirusjhuedu/maphtml
- 8.Yu H. Comparative studies of magnetic particle-based solid phase fluorogenic and electrochemiluminescent immunoassay. J Immunol Methods. 1998;218(1–2):1–8. doi: 10.1016/s0022-1759(98)00047-7. [DOI] [PubMed] [Google Scholar]
- 9.Hirotsu Y., Maejima M., Shibusawa, K. Amemiya, Y. Nagakubo, K. Hosaka M. Analysis of Covid-19 and non-Covid-19 viruses, including influenza viruses, to determine the influence of intensive preventive measures in Japan. J Clin Virol. 2020;129:104543. doi: 10.1016/j.jcv.2020.104543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirato K., Nao N., Katano, I. Takayama, S. Saito, F. Kato H. Development of Genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73(4):304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 11.Hirotsu Y., Mochizuki H., Omata M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020;284:113926. doi: 10.1016/j.jviromet.2020.113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirotsu Y., Maejima M., Nakajima M., Mochizuki H., Omata M. Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: a patient from the Diamond Princess cruise ship. Infect Control Hosp Epidemiol. 2020;41(9):1105–1106. doi: 10.1017/ice.2020.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirotsu Y., Maejima M., Shibusawa, K. Amemiya, Y. Nagakubo, K. Hosaka M. Analysis of a persistent viral shedding patient infected with SARS-CoV-2 by RT-qPCR, FilmArray Respiratory Panel v2.1, and antigen detection. J Infect Chemother. 27 (2), 2021, 406–409 doi: 10.1016/j.jiac.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirotsu Y., Maejima M., Shibusawa, Y. Nagakubo, K. Hosaka, K. Amemiya M. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int J Infect Dis. 99, 2020, 397–402 doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirotsu Y., Maejima M., Shibusawa, Y. Nagakubo, K. Hosaka, K. Amemiya M. Pooling RT-PCR test of SARS-CoV-2 for large cohort of 'healthy' and infection-suspected patients: a prospective and consecutive study on 1,000 individuals. medRxiv. 2020 doi: 10.1038/s41598-020-76043-z. 2020.05.04.20088146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato N., Yokosuka O., Omata M., Hosoda K., Ohto M. Detection of hepatitis C virus ribonucleic acid in the serum by amplification with polymerase chain reaction. J Clin Invest. 1990;86(5):1764–1767. doi: 10.1172/JCI114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omata M., Ehata T., Yokosuka O., Hosoda K., Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324(24):1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 18.Omata M., Yokosuka O., Takano, N. Kato, K. Hosoda, F. Imazeki S. Resolution of acute hepatitis C after therapy with natural beta interferon. Lancet. 1991;338(8772):914–915. doi: 10.1016/0140-6736(91)91774-o. [DOI] [PubMed] [Google Scholar]
- 19.Huang C., Wang Y., Li, L. Ren, J. Zhao, Y. Hu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ElecsysAnti-SARS-CoV-2. https://wwwfdagov/media/137605/download
- 21.Tada M., Omata M., Ohto M. Analysis of ras gene mutations in human hepatic malignant tumors by polymerase chain reaction and direct sequencing. Canc Res. 1990;15(50):1121–1124. [PubMed] [Google Scholar]
- 22.Tada M., Ohashi M., Shiratori, T. Okudaira, Y. Komatsu, T. Kawabe Y. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology. 1996;110(1):227–231. doi: 10.1053/gast.1996.v110.pm8536861. [DOI] [PubMed] [Google Scholar]
- 23.Devakumar D., Shannon G., Bhopal S.S., Abubakar I. Racism and discrimination in COVID-19 responses. Lancet. 2020;395(10231):1194. doi: 10.1016/S0140-6736(20)30792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rzymski P., Nowicki M. Preventing COVID-19 prejudice in academia. Science. 2020;367(6484):1313. doi: 10.1126/science.abb4870. [DOI] [PubMed] [Google Scholar]
- 25.Pollán M., Pérez-Gómez B., Pastor-Barriuso, J. Oteo, MA. Hernán, M. Pérez-Olmeda R. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396(10250):535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anand S., Montez-Rath M., Han, J. Bozeman, R. Kerschmann, P. Beyer J. Prevalence of SARS-CoV-2 antibodies in a large nationwide sample of patients on dialysis in the USA: a cross-sectional study. Lancet. 2020;396(10259):1335–1344. doi: 10.1016/S0140-6736(20)32009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward H., Atchison C.J., Whitaker, A. Kylie EC, E. Joshua, O. Lucy M. Antibody prevalence for SARS-CoV-2 in England following first peak of the pandemic: REACT2 study in 100,000 adults. medRxiv. 2020 08.12.20173690. [Google Scholar]