Abstract
Tetrathionate hydrolase (4THase) plays an important role in dissimilatory sulfur oxidation in the acidophilic iron‐ and sulfur‐oxidizing bacterium Acidithiobacillus ferrooxidans. The structure of recombinant 4THase from A. ferrooxidans (Af‐Tth) was determined by X‐ray crystallography to a resolution of 1.95 Å. Af‐Tth is a homodimer, and its monomer structure exhibits an eight‐bladed β‐propeller motif. Two insertion loops participate in dimerization, and one loop forms a cavity with the β‐propeller region. We observed unexplained electron densities in this cavity of the substrate‐soaked structure. The anomalous difference map generated using diffraction data collected at a wavelength of 1.9 Å indicated the presence of polymerized sulfur atoms. Asp325, a highly conserved residue among 4THases, was located near the polymerized sulfur atoms. 4THase activity was completely abolished in the site‐specific Af‐Tth D325N variant, suggesting that Asp325 plays a crucial role in the first step of tetrathionate hydrolysis. Considering that the Af‐Tth reaction occurs only under acidic pH, Asp325 acts as an acid for the tetrathionate hydrolysis reaction. The polymerized sulfur atoms in the active site cavity may represent the intermediate product in the subsequent step.
Keywords: Acidithiobacillus ferrooxidans, hydrolase, protein tertiary structure, site‐directed mutagenesis, sulfur oxidation, tetrathionic acid
Short abstract
1. INTRODUCTION
Tetrathionate hydrolase (4THase), which catalyzes tetrathionate hydrolysis, is a unique enzyme distributed in acidophilic sulfur oxidizers. 4THases have been isolated only from acidophilic sulfur‐oxidizing bacteria such as Acidithiobacillus caldus, 1 Acidithiobacillus thiooxidans, 2 , 3 Acidithiobacillus ferrooxidans, 4 Acidiphilium acidophilum, 5 and the thermoacidophilic archaeon Acidianus ambivalens. 6 It plays an important role together with thiosulfate:quinone oxidoreductase in the S4I pathway. 7 , 8 The gene encoding 4THase in A. ferrooxidans has been identified and termed Af‐tth. 4 Later, 4THase genes from A. caldus (Ac‐tetH) and Ad. ambivalens (Ad‐tth1) were experimentally identified. 6 , 9 The deduced amino acid sequences of the three genes were similar, and they encode proteins of the pyrroloquinoline quinone (PQQ)‐containing protein family (Pfam family 10 : PQQ_2). Recombinant Af‐Tth was obtained heterologously from Escherichia coli and successfully refolded in an active form under an acidic condition without PQQ or any other cofactors. 11 Conversely, quinoid compounds could be detected in Ac‐TetH–expressing bacteria. 9 The localization was different for Af‐Tth (outer membrane) 4 and Ac‐TetH (periplasm). 1 Recently, specific gene disruption systems have been developed for both Acidithiobacillus species. tth‐ and tetH‐deficient mutants cannot use tetrathionate as a sole growth substrate, suggesting that Af‐Tth and Ac‐TetH were necessary for tetrathionate metabolism in these species. 12 , 13 Although some different properties were reported between Af‐Tth and Ac‐TetH, they both functioned as tetrathionate‐metabolizing enzymes physiologically.
The catalytic reaction of tetrathionate hydrolysis has been proposed for the purified enzyme from A. ferrooxidans. Beard et al. 14 suggested that 4THase from the bacterium catalyzes the following reaction:
| (1) |
One molecule of tetrathionate is hydrolyzed by Af‐Tth to generate disulfane monosulfonic acid (DSMSA) and sulfuric acid (sulfate) in reaction (1). DSMSA has been reported as a key intermediate in the biological oxidation of pyrite. 15 Because DSMSA is highly reactive, several abiotic reactions might spontaneously occur. For instance, four molecules of DSMSA could be elongated to form elemental sulfur (S8) and sulfurous acid (sulfite). Consequently, polythionates, S8, and sulfite are the final products. 16 , 17 Meulenberg et al. 18 suggested that one molecule of tetrathionate was hydrolyzed to thiosulfate, sulfur, and sulfate in equimolar amounts by the enzyme according to the following equation:
| (2) |
Concerning the Af‐Tth reaction, we detected turbidity in the reaction mixture because of the production of S8 and the same level of thiosulfate production accompanying tetrathionate consumption using the ion‐pair HPLC method. 4 This result supported reaction (2). As sulfur atoms (S0) are unstable, reaction (2) should be repeated eight times to produce S8 as a stable end‐product according to the following equation:
| (3) |
However, determining the specific reaction is difficult because tetrathionate hydrolysis in these Acidithiobacillus species generates multiple products with chemical interactions among thiosulfate, sulfane sulfur chemicals, polythionate, sulfite, and possibly S8 under high sulfate concentrations and low pH. Therefore, the precise tetrathionate hydrolysis reaction and reaction mechanism remain unknown. Because Af‐Tth exhibits a unique primary structure, the catalytic reaction is unprecedented. Cysteine residues in the catalytic center play an important role in many other enzymes involved in dissimilatory sulfur oxidation such as sulfide:quinone oxidoreductase, 19 sulfur oxygenase reductase, 20 and thiosulfate dehydrogenase. 21 Although cysteine residues are normally conserved in each enzyme among different sulfur‐oxidizing microorganisms, only one cysteine residue (Cys301) can be detected in the primary structure of Af‐Tth, and it is not conserved among 4THases from Acidithiobacillus spp. 22 Furthermore, the site‐specific variant (Af‐Tth C301A) exhibited similar activity as wild‐type Af‐Tth. 22 This result suggests that Af‐Tth has a novel cysteine‐independent reaction mechanism. Therefore, clarifying the precise overall reaction mechanism in Af‐Tth is of interest.
We previously obtained crystals of recombinant Af‐Tth. 23 In this study, we have reported the X‐ray crystallographic results of Af‐Tth. A site‐specific Af‐Tth variant constructed on the basis of the structural information lost its enzymatic activity. Based on the structure–function relationship, we propose the first step of the tetrathionate hydrolysis reaction mechanism of Af‐Tth.
2. RESULTS
2.1. Structure determination of Af‐Tth
Recombinant Af‐Tth was synthesized in E. coli as inclusion bodies in an inactive form. The active enzyme was refolded from inclusion bodies under acidic conditions and purified to homogeneity as a homodimer via gel‐permeation column chromatography (GPC) at pH 4.0. 22 The crystallization of Af‐Tth was previously described in detail. 23 The diffraction dataset was collected at a resolution of 1.95 Å. Resolution diffraction data (at 2.8 Å) for selenomethionyl Af‐Tth was collected at a wavelength of 0.9785 Å for the initial phasing by selenium single‐wavelength anomalous diffraction (Se‐SAD) analysis. The Af‐Tth crystals belonged to space group P32 with unit‐cell parameters a = b = 91.0 Å and c = 232 Å, and it contained six Af‐Tth chains (three dimers and chains A–C, D–B, and F–E) in the asymmetric unit. Electron density analysis after phase improvement allowed us to model all chains excluding some residues at the N‐ and C‐termini and in some loop regions (disordered residues are stated in the caption of Figure S1). Six chains superimposed on each other with root‐mean‐square deviations (RMSDs) of 0.40–0.88 Å for the corresponding Cα atoms at 423–452. The final model, including 2,708 residues, 30 sulfate ions, 3 β‐alanines, 2 glycines, and 697 water molecules, was refined to a crystallographic R‐factor of 17.8% (free R‐factor of 21.9%). Sulfate ions, β‐alanine, and glycine molecules were derived from the crystallization solution. Of 30 sulfate ions, six and five ions existed at the dimerization interface and dimer–dimer interfaces, respectively. In chains B, D, E, and F, there was a strong positive difference electron density (>5σ) at the center of three Sδ atoms in the side chains of Met172, Met238, and Met279. We assigned the electron density as water molecules (highlighted by the red arrow in Figure 2b). These water molecules were closer to the Sδ atom in Met172 (O–S distance: 2.10–2.27 Å) than the other two Sδ atoms (O–S distance: 2.37–2.58 Å). The former distance appeared to be too short to be considered a hydrogen bond. However, water molecules converged at this position with adequate temperature factors (16.2–25.6 Å2), and there were no excess electron densities, positive or negative, after crystallographic refinement. Data collection, processing, and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
| Native | Se‐SAD | D325N | Substrate‐soaked | ||
|---|---|---|---|---|---|
| Data collection | |||||
| Cell constants (Å) | a = b = 91.0, c = 232 | a = b = 92.4, c = 234 | a = b = 94.3, c = 236 | a = b = 95.1, c = 235 | a = b = 95.5, c = 235 |
| Wavelength (Å) | 1.0000 | 0.9785 | 1.0000 | 1.0000 | 1.9000 |
| Resolution (Å) | 42.4–1.95 (2.02–1.95) | 30.7–2.80 (2.90–2.80) | 43.8–2.80 (2.90–2.80) | 47.9–2.30 (2.38–2.30) | 47.7–2.50 (2.59–2.50) |
| Unique reflections | 156,136 (15,660) | 54,412 (5397) | 57,563 (5788) | 105,170 (10,564) | 82,717 (8259) |
| Redundancy | 3.8 (3.8) | 6.4 (6.3) | 5.1 (4.7) | 5.1 (5.1) | 10.1(9.6) |
| I/σ (I) | 10.3 (2.9) | 20.1 (3.7) | 16.5 (2.7) | 14.7 (2.8) | 16.3 (2.7) |
| R meas a | 0.109 (0.563) | 0.138 (0.778) | 0.085 (0.632) | 0.091 (0.678) | 0.129 (0.810) |
| Completeness (%) | 99.7 (99.9) | 100 (100) | 99.5 (99.7) | 99.6 (99.2) | 99.8 (99.7) |
| CC 1/2 b | 0.995 (0.875) | 0.996 (0.886) | 0.997 (0.776) | 0.998 (0.842) | 0.998 (0.848) |
| Wilson B (Å b ) | 25.8 | 48.4 | 62.2 | 40.7 | 37.8 |
| Phasing | |||||
| Figure of merit | 0.42 | ||||
| BAYES‐CC | 40 | ||||
| Refinement | |||||
| Resolution (Å) | 42.4–1.95 (2.02–1.95) | 43.8–2.80 (2.90–2.80) | 47.9–2.30 (2.38–2.30) | ||
| Used reflections | 156,129 (15,661) | 57,466 (5782) | 105,166 (10,564) | ||
| R work c | 0.178 (0.226) | 0.225 (0.320) | 0.179 (0.234) | ||
| R free d | 0.219 (0.291) | 0.277 (0.423) | 0.239 (0.340) | ||
| RMSD | |||||
| Bond length (Å) | 0.007 | 0.003 | 0.014 | ||
| Bond angle (°) | 0.97 | 0.74 | 1.64 | ||
| Ramachandran plot | |||||
| Favored (%) | 97.8 | 96.2 | 96.6 | ||
| Allowed (%) | 2.0 | 3.5 | 3.0 | ||
| Outliers (%) | 0.2 | 0.3 | 0.4 | ||
| No. of atoms | |||||
| Macromolecules | 20,437 | 19,882 | 20,767 | ||
| Ligands | 168 | 45 | 124 | ||
| Solvent | 697 | 10 | 535 | ||
| Mean B value (Å b ) | |||||
| Macromolecules | 25.8 | 63.0 | 42.1 | ||
| Ligands | 41.2 | 72.1 | 57.9 | ||
| Solvent | 25.3 | 53.2 | 40.6 | ||
| PDB code | 6L8A | 7CQY | |||
Note: Statistics for the highest‐resolution shell are presented in parentheses.
R meas is the multiplicity‐weighted R merge. R merge = ∑ |I hkl − <I hkl>|/∑ I hkl, <I hkl> is the mean value of I hkl.
CC 1/2 values were calculated by splitting the data randomly in half.
R work = ∑||F o| − |F c||/∑ |F o|.
R free was calculated from the test set (5% of the total data).
2.2. Crystal structure of Af‐Tth
The monomer structure was rich in β‐sheets, which included 33 β‐strands, and exhibited an eight‐bladed β‐propeller motif, with each blade (I–VIII) consisting of four antiparallel β‐strands, excluding blade I (five antiparallel β‐strands; Figures 1a and S1). The ring closure, also called a “Velcro strap,” 24 was formed with both termini of the four antiparallel beta‐strands (β2, and β31–33) in the propeller domain. The approximate dimension of the β‐propeller region was 50 Å × 50 Å × 30 Å. A structural similarity search using the DALI server 25 indicated that the outer membrane protein factor BamB from the β‐barrel assembly machinery complex exhibited high Z scores (35.2; Protein Data Bank [PDB] ID 4XGA 26 ). Superimposition of the Af‐Tth structure on the BamB structure gave an RMSD of 2.05 Å for 303 corresponding Cα atoms (22.4% sequence identity). Both Af‐Tth and BamB localized at the periplasmic side of the outer membrane, but there were no relationships between their functions.
FIGURE 1.
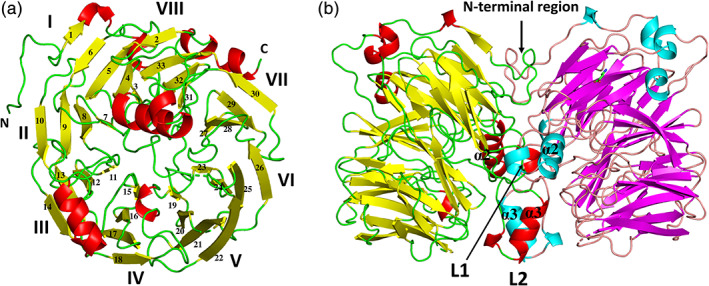
Overall structure of recombinant tetrathionate hydrolase from A. ferrooxidans. Monomer (a) and dimer (b) structures. Each monomer of chains B and D in the dimer is colored yellow and magenta (β‐strands), respectively, red and cyan (α‐ and 310 helices), respectively, and green and wheat (loops), respectively. Monomer (a) is the view from the right side of (b). The monomer structure exhibits an eight‐bladed (I–VIII) β‐propeller motif. L1 and L2 are the insertion loops of the eight‐bladed β‐propeller motif
Four α‐helices and five 310 helices are located outside the β‐propeller region (Figure 2a). The N‐terminal region before blade I and two insertion loops, L1 (including α2 helix and one 310 helix) between blade I and VIII, and L2 (including α3 helix and one 310 helix) in blade II, participate in dimerization (the area of the dimer interface is 2,139–2,459 Å2; Figure 1b).
FIGURE 2.
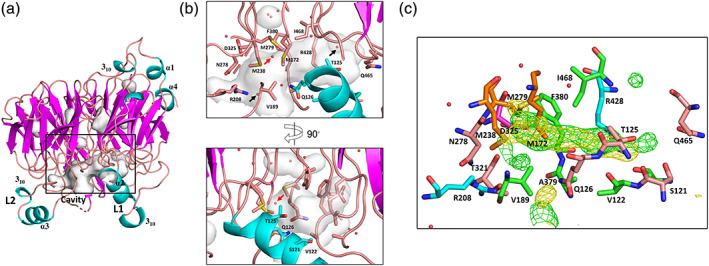
Active site cavity of tetrathionate hydrolase from A. ferrooxidans. (a) The side view of the wild‐type monomer (chain F). (b) A close‐up view of the rectangular box in (a). Cavities were displayed using the Cavities & Pockets (Culled) command of PyMOL. (c) Electron densities in the cavity of the substrate‐soaked structure (chain C). Side chains of residues lining the active site cavity are indicated as a stick model. The water molecules and the three, surrounding methionine residues (M172, M238, and M279) in the cavity are marked with red and black arrows, respectively. In (c), methionines, arginines, aspartic acid, hydrophobic residues (Val, Ala, Phe, Ile), and other residues (Ser, Thr, Asn, Gln) are colored orange, cyan, magenta, green, and wheat, respectively. The green and yellow contour shows the F o − F c omit map (3.0σ, λ = 1.0 Å) and anomalous difference map (3.0σ, λ = 1.9 Å), respectively
In each monomer, the L1 loop formed a cavity in the β‐propeller region (Figure 2b). Hydrophobic residues (Val122, Val189, Ala379, Phe380, Ala424, and Ile468) surrounded this cavity in each monomer. Three methionine residues, namely Met172, Met238, and Met279, were closest to the cavity. Some ordered water molecules were present at both edges of the cavities (two water molecules are highlighted by the black arrows in Figure 2b). Although cysteine residues are indispensable in the reactions of many enzymes of dissimilatory sulfur oxidation metabolism, we previously reported that the sole cysteine residue (Cys301) of Af‐Tth is not involved in the hydrolysis reaction. 22 Cys301 was more than 25 Å from this cavity.
2.3. Substrate‐soaked structure of Af‐Tth
The X‐ray diffraction datasets with two different wavelengths (1.0 and 1.9 Å) from the same crystal of Af‐Tth soaked in precipitant solution with the substrate tetrathionate were collected at resolutions of 2.3 and 2.5 Å (Table 1). The substrate‐soaked structure at a resolution of 2.3 Å was refined to a crystallographic R‐factor of 17.9% (free R‐factor of 23.9%). The final model included 2,752 residues, 18 sulfate ions, 4 β‐alanines, 2 glycines, and 535 water molecules.
The F o − F c omit maps revealed a large electron density in the cavity between the L1 loop and β‐propeller motif. This electron density stretched approximately 10 Å from the center of the three Sδ atoms of Met172, Met238, and Met279 (Figure 2c, S2). The anomalous difference map using the diffraction data collected at 1.9 Å revealed significant signals at the same location of unexplained electron densities in the F o − F c omit maps. These signals were probably attributable to sulfur atoms because the anomalous difference maps clearly identified signals from Met172, Met238, and Met279. In addition to hydrophobic residues (Val122, Val189, Ala379, Phe380, Ala424, and Ile468), the side chain atoms of Ser121, Thr125, Gln126, Arg208, Asn278, Thr321, Asp325, Arg428, and Gln465 were also situated within 4 Å of these electron densities. Although the quality of the electron densities did not permit exact model building, these densities, which were observed only in the substrate‐soaked data, may be derived from the polymerized sulfur atoms.
2.4. Mutation analyses of Af‐Tth
Asp325 was the only acidic amino acid (Asp or Glu) located within 4 Å of these large electron densities. The Asp residue was conserved among 4THases from Acidithiobacillus spp. and the thermoacidophilic archaeon Ad. ambivalens (Figure S1). To clarify the role of the only acidic residue located near the presumed product molecules in Af‐Tth, a site‐specific variant enzyme was engineered in which Asp325 was replaced by asparagine. The Af‐Tth D325N variant showed similar properties to the wild‐type in the refolding procedures, SDS‐PAGE analysis, and GPC. The apparent molecular mass of the D325N variant was 92.8 ± 0.9 kDa, which was similar to that of the wild‐type enzyme (92.5 ± 0.8 kDa) according to GPC. The D325N variant crystallized in the same crystal form as the wild‐type, and the diffraction dataset was collected at a resolution of 2.8 Å (Table 1). In the D325N crystal, disorder was noted for the same loop regions disordered in the wild‐type. The final model of the D325N variant, including 2,630 residues, 9 sulfate ions, and 10 water molecules, was refined to a crystallographic R‐factor of 22.5% (free R‐factor of 27.7%). Monomer structures of the D325N variant superimposed on those of the wild‐type with RMSDs of 0.36–0.77 Å for the corresponding Cα atoms at 403–457. These RMSDs indicate that the D325N variant has a similar tertiary structure as the wild‐type. The side chain conformation at Asp/Asn325 had also few conserved regions. Conversely, the D325N variant displayed no activity (Figure 3), whereas the activity of the Af‐Tth wild‐type was 2.5 ± 0.18 U·mg−1. These results suggested that the Asp325 residue was necessary for tetrathionate hydrolysis activity.
FIGURE 3.
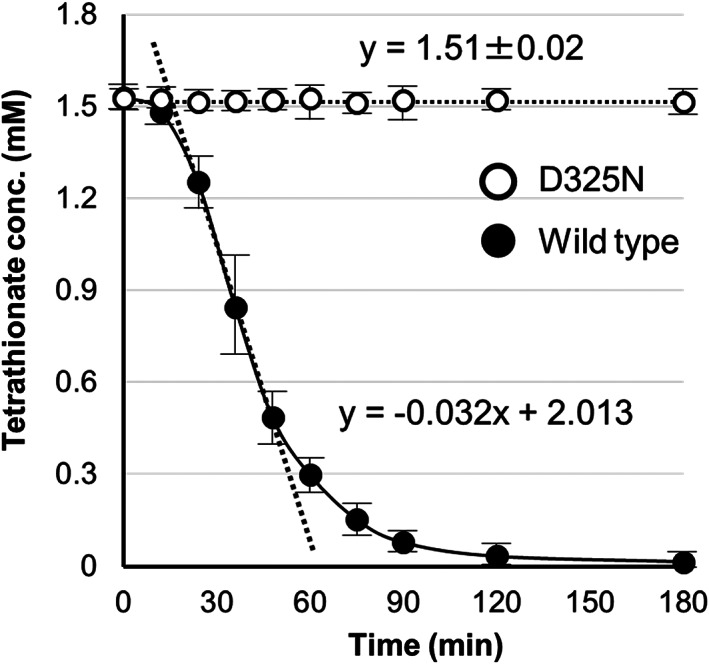
Activity measurement. The activities of wild‐type and D325N tetrathionate hydrolase from Acidithiobacillus ferrooxidans are indicated by closed and open circles, respectively. The activities were determined by monitoring the tetrathionate concentration in each reaction mixture
3. DISCUSSION
Af‐Tth was initially reported to encode a novel outer membrane protein of unknown function. 27 Although archaeal 4THase tertiary structures have been predicted, 6 , 28 this is the first report of the structure of a 4THase determined by X‐ray crystallography.
The localization of 4THases differs among Acidithiobacillus spp. Considering the low optimal pH and solubility of the enzyme, 4THases from A. ferrooxidans, 29 A. thiooxidans, 2 and A. caldus 1 were suggested to be periplasmic proteins. However, we detected activity in the total membrane fraction of tetrathionate‐grown cells of A. ferrooxidans type strain ATCC23270, and found that the enzyme was easily solubilized in the presence of 0.4 M ammonium sulfate. 4 Thus, we suggested that Af‐Tth was an outer membrane‐associated protein. The activity of purified Af‐Tth was significantly enhanced in the presence of sulfate ions. 4 Contrarily, Ad‐TTH1 did not appear to require sulfate ions for its catalytic activity. 6 To understand this difference based on tertiary structure information, we constructed a homology model of Ad‐TTH1 using the Af‐Tth structure as a template on the SWISS‐MODEL server. A previously reported homology model of Ad‐TTH1 was constructed using the structure of quinoprotein alcohol dehydrogenase from Pseudomonas putida (PDB ID 1KV9 30 ) as a template. The new homology model of Ad‐TTH1 based on the Af‐Tth structure had better statistics than the previous model; specifically, the QMEAN scores were of the new and models were −2.01 and −6.96, respectively, and their sequence identities were 40.5 and 16.9%, respectively. The Af‐Tth crystal structure exhibited a more positive electrostatic surface potential than the Ad‐TTH1 model structure (Figure 4). The positively charged surface of Af‐Tth can bind sulfate anions, as demonstrated by the observation that some sulfate ions from crystallization reagents are located on its surface. Such a structural feature may explain the difference of activation by sulfate ions between Af‐Tth and Ad‐TTH1.
FIGURE 4.
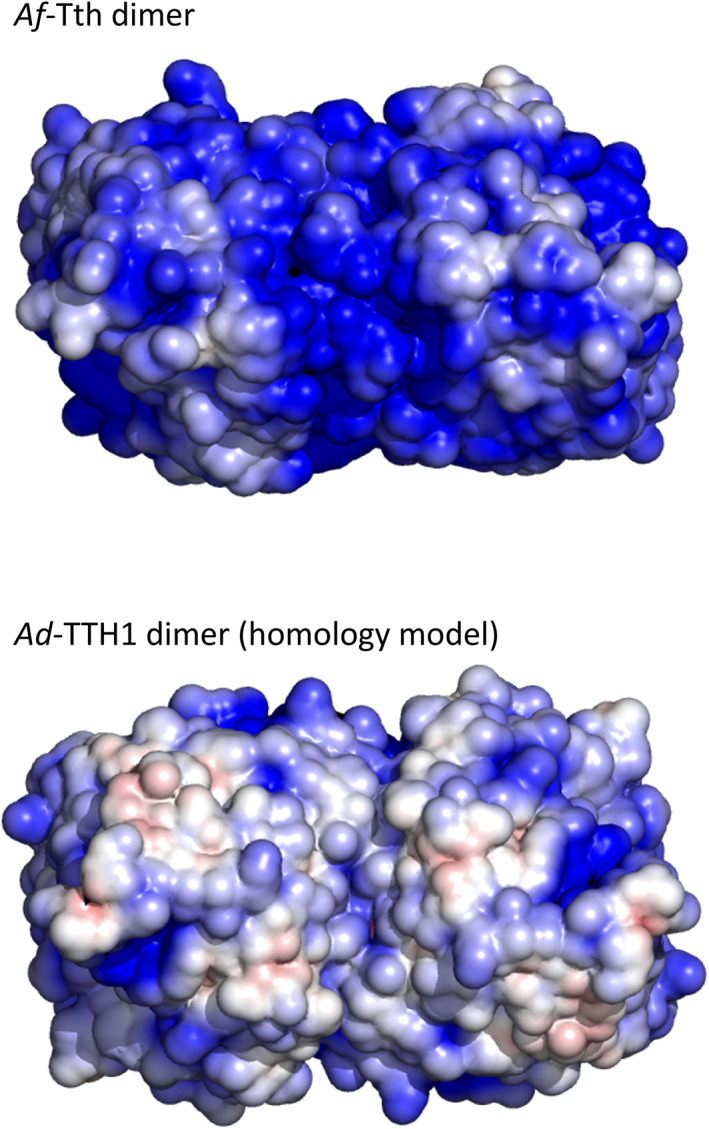
Electrostatic surface potentials of tetrathionate hydrolase dimers at pH 2.0. Van der Waals surfaces of the tetrathionate hydrolase from A. ferrooxidans (Af‐Tth, upper figure) and tetrathionate hydrolase from Ad. ambivalens (Ad‐TTH1) homology models (lower figure) are colored according to the local electrostatic potential as calculated using APBS 41 from −4kT (red) to +4kT (blue)
Af‐Tth exhibits a cysteine‐independent reaction mechanism without protein‐bound intermediates. Although researchers previously speculated that tetrathionate might be hydrolyzed to DSMSA and sulfate, 14 the reaction mechanism has not been clarified. Our structural and mutation analyses of Af‐Tth revealed that Asp325 played a crucial role in the first step of tetrathionate hydrolysis. In general, an aspartic acid residue can behave as an acid or base in catalytic reactions by enzymes. If Asp325 in Af‐Tth acts as an acid, then an Sαatom of the tetrathionate molecule can be protonated by Asp325 (Figure 5). Because of the protonated Sαatom, the terminal sulfur atom of the tetrathionate molecule is easily hit by the nucleophilic attack of a water molecule. The sulfur–sulfur bond between the terminal sulfur atom and protonated S(alpha) atoms would be broken in an SN2 reaction, thereby generating sulfate and DSMSA. Conversely, if Asp325 acts as a base, then the terminal sulfite of a tetrathionate molecule could be attacked by a nucleophilic carboxylate of Asp325, and then the S‐S bond between the S(alpha) atom and sulfite–Asp325 is cleaved in an SN2 reaction. The sulfonated Asp325 residue (R—C(=O)—O—SO3 −) is attacked by a water molecule. However, Af‐Tth exhibited maximal activity at a pH of 3.0, and its activity could not be detected at a pH exceeding 6.0 (Figure S3). The calculated pK a values using the Depth server 31 of Asp325 were 4.54 (chain A), 4.22 (chain B), 4.57 (chain C), 5.05 (chain D), 4.66 (chain E), and 5.27 (chain F). Considering that the reaction occurs under only acidic pH, Asp325 acts as an acid in the reaction, and then DSMSA and sulfate are generated via tetrathionate hydrolysis. These results were consistent with reaction (1) in Section 1.
FIGURE 5.
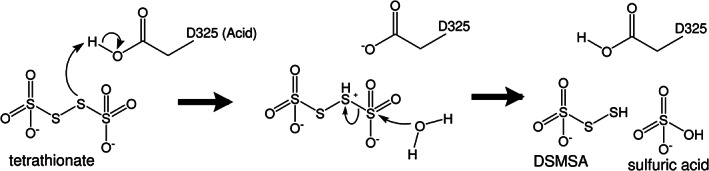
Proposed catalytic mechanism of tetrathionate hydrolysis by tetrathionate hydrolase from A. ferrooxidans. Asp325 works as a general acid under neutral or acidic conditions
Sauvé et al. 32 clarified the hydrolysis mechanism of a sulfur–sulfur bond in the reaction of SoxB as a thiosulfohydrolase. Two manganese ions (Mn2+) in the active site of SoxB are chelated by six histidine and two aspartate residues. The two Mn2+ ions and the arginine residue rigidly stabilize the cysteine‐conjugated sulfonate substrate. The nucleophilic hydroxyl ion then attacks the sulfone sulfur atom of the substrate in the SN2 reaction to break the sulfur–sulfur bond. The reaction mechanism of SoxB is thus considerably different from our proposed hydrolysis mechanism for Af‐Tth.
The next step of the Af‐Tth catalytic reaction is proton abstraction from DSMSA, followed by the release of the sulfur (S0) atom:
| (A) |
Reaction (2) is obtained from combining reaction (1) and reaction (A). Furthermore, the Af‐Tth reaction appears to be a multi‐step reaction. Released sulfur (S0) atoms oligomerize and finally cyclize as S8. Our present results do not provide a definitive answer to elucidate the next step. However, unexplained electron densities at the cavity between the L1 loop and β‐propeller motif appear to correspond to oligomeric sulfur atoms, which may be the intermediate product before the final product (S8). The hydrophobic environment of this cavity, including Val122, Ala131, Ala379, Phe380, and Ile468, is suitable for the storage of hydrophobic sulfur atoms. Three methionines, namely Met172, Met238, and Met279, which are located at the starting point of the large electron densities, may contribute to the release of the sulfur (S0) atom from DSMSA. The three methionine residues are located approximately 10 Å from Asp325, which is the catalytic residue for the first step of tetrathionate hydrolysis. Such a structural environment allows for a smooth transition from the first step (DSMSA production from hydrolysis at Asp325) to the next step (the release of sulfur [S0] atom from the DSMSA by three methionine residues). We assigned a water molecule to a spherical electron density at the center of three Sδ atoms of the methionine residues in the native and D325N structures. Conversely, we confirmed significant anomalous electron densities at this position in some chains of the substrate‐soaked structure (Figure 2c, Figures S2d and S2e), indicating that they may have been derived from sulfur (S0) atom.
Although the complete reaction mechanism of Af‐Tth should be further clarified, we revealed the tertiary structure of this unique enzyme and the hydrolysis reaction mechanism of tetrathionate in the active site in this study. Further studies to investigate the reaction mechanism including the three methionine residues are currently underway.
4. MATERIALS AND METHODS
4.1. Recombinant Af‐tth expression, refolding, and purification
The pET4TH and Af‐tth expression vector was constructed and introduced into E. coli BL21 Star (DE3) cells to obtain recombinant Af‐Tth, as previously described in detail. 4 The recombinant cells harboring pET4TH were inoculated into modified Terrific Broth supplemented with 50 μg·ml−1 ampicillin at 37°C to an OD660 of 1.0. The expression of Af‐tth in pET4TH was induced by the addition of 1 mM isopropyl 1‐thio‐β‐d‐galactopyranoside (IPTG) to the culture, followed by incubation at 20°C for 36 hr. After the incubation, cells were harvested and disrupted by sonication on ice (the total ‘on’ period was 15 min in cycles of 30 s ‘on’ and 30 s “off”). The insoluble fraction was collected by centrifugation at 10,000 × g for 10 min, and the supernatant was removed. The pellet was washed three times with 10 mM Tris–HCl buffer (pH 8.0) containing 1 mM EDTA. To collect the inclusion bodies, the pellet was washed with 100 mM potassium phosphate buffer (pH 7.0) containing 4% vol/vol Triton X‐100. The inclusion bodies were washed again three times with sterilized distilled water to remove the detergent. To obtain soluble and active Af‐Tth, the inclusion bodies were treated using a previously described acidic refolding protoco. 11 After refolding, Af‐Tth was highly purified via hydrophobic column chromatography (TOYOPEARL Butyl‐600M, Tosoh, Tokyo, Japan) and GPC using a TSKgel G3000SW (Tosoh) column equilibrated with 10 mM β‐alanine buffer (pH 4.0) containing 0.4 M ammonium sulfate. The detailed purification procedure was reported previously. 23 The purity of the Af‐Tth solution was confirmed by SDS‐PAGE, and the protein concentration of purified Af‐Tth was determined by the Bradford method using bovine serum albumin as a standard. Selenomethionine‐labeled (SeMet) Af‐Tth was obtained according to the method previously described by Guerrero et al. 33
4.2. Construction of the Af‐Tth D325N variant
A point mutation was introduced using the QuickChange Site‐Directed Mutagenesis kit (Stratagene, La Jolla, CA). Two complementary oligonucleotide primers were designed: Primer D325N_F, 5′‐GCCAGTAATACTGGTATTGGCAACGTATCTCCGG CGGC‐3′; and Primer D325N_R, 5′‐GCCGCCGGAGATACGTTGCCAATACCAGTAT TACTGGC‐3′. The codon for Asp325 (GAC) was replaced by an asparagine codon (underlined). Experiments were performed as advised by the manufacturer. Introduction of the point mutation and the absence of unintended mutations were confirmed by DNA sequencing. The gene expression, refolding, and purification of the D325N variant were performed using the same procedures described for wild‐type Af‐Tth.
4.3. Catalytic activity measurement
Af‐Tth activity was measured using a modified cyanolysis assay. 4 In the standard procedure, the reaction mixture (total volume of 3 ml) contained 1.5 mM K2S4O6, 200 mM K2SO4, 100 mM β‐alanine nitrate buffer (pH 3.0), and the enzyme. The reaction was initiated by adding 150 μL of enzyme solution. Aliquots (150 μL) were collected into thin‐wall tubes on ice at appropriate periods, followed by heating to 98°C for 5 min to stop the reaction. Because S8 was produced by the reaction, the samples were centrifuged at 10,000 × g for 1 min to remove insoluble particles. Supernatant (100 μL) in each tube was subjected to the cyanolysis assay to determine tetrathionate concentrations. One unit of activity was defined as the amount of enzyme required for the hydrolysis of 1 μmol of tetrathionate per minute.
4.4. Crystallization, data collection, phasing, and refinement
The detailed crystallization procedure has been reported previously. 23 Crystals of wild‐type, D325N variant, and SeMet Af‐Tth were grown at 20°C in hanging drops using 10 mg/ml protein solution (in 10 mM β‐alanine buffer [pH 4.0] containing 0.4 M ammonium sulfate) over the precipitant solution (20 mM glycine buffer [pH 10.0] containing 50 mM sodium chloride and 33% [vol/vol] PEG 1000). The substrate‐soaked crystals were prepared by soaking Af‐Tth crystals into the precipitant solution including 70 mM tetrathionate for 90 s. Crystals were flash‐frozen in liquid nitrogen directly, and then diffraction data were collected under cryogenic conditions (100 K) at beamlines BL38B1 and BL41XU at SPring‐8 (Harima, Japan) and beamline BL‐17A at Photon Factory (Tsukuba, Japan). The wavelengths used for diffraction data collection were 0.9785 Å for Se‐SAD phasing and 1.0000 Å for native and D325N data. X‐rays with two different wavelengths (1.0000 and 1.9000 Å) were used to target different positions on the same substrate‐soaked crystal. For the later dataset, redundant data were collected to accurately determine the differences between the Friedel pairs. Diffraction datasets were integrated using the XDS program 34 and scaled using the Aimless program 35 in CCP4 software. 36 The Se‐SAD phasing and phase improvement were performed by AutoSol 37 in the program PHENIX. 38 The crystals belonged to space group P32 with unit‐cell parameters a = b = 92 Å and c = 232 Å, and six Af‐Tth chains were present in the asymmetric unit. The auto model building was performed by the Buccaneer program in CCP4 software using 1.95 Å native data and the initial structure at 2.8 Å resolution from AutoSol. Refinement was performed using phenix.refine in the program PHENIX with manual model modification using the program Coot. 39 Ramachandran analysis was calculated using the MolProbity web server. 40 Figures of structural models and electron density maps were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC). Data collection, processing, and refinement statistics were summarized using phenix.table in PHENIX as presented in Table 1.
DATA DEPOSITION REQUIREMENTS
The final coordinates and structural data for the native and D325N structures were deposited in the Protein Data Bank (PDB) with the PDB IDs 6L8A and 7CQY, respectively.
AUTHOR CONTRIBUTIONS
Tadayoshi Kanao: Conceptualization; funding acquisition; investigation; methodology; project administration; resources; supervision; visualization; writing‐original draft. Naruki Hase: Investigation. Hisayuki Nakayama: Investigation. Kyoya Yoshida: Investigation. Kazumi Nishiura: Investigation. Megumi Kosaka: Investigation. Kazuo Kamimura: Supervision. Yu Hirano: Investigation; software. Taro Tamada: Conceptualization; funding acquisition; investigation; methodology; project administration; software; supervision; validation; visualization; writing‐original draft.
CONFLICT OF INTEREST
All authors declare that they have no conflicts of interest with respect to the publication of this article.
Supporting information
Figure S1 Sequence alignment among three 4THases from Acidithiobacillus species
Figure S2. Electron densities in the cavity of the substrate‐soaked data
Figure S3. pH profile of Af‐Tth activity
ACKNOWLEDGMENTS
We thank the beamline scientists for data collection with synchrotron radiation at SPring‐8 (proposal nos. 2010B1994, 2010B2050, 2012A1812, 2014B2000, 2015A1041, 2015B2041, 2016A1001, 2018A1003, and 2019A1005) and Photon Factory (proposal nos. 2013G122, 2015G035, 2017G033, and 2019G104). This work was partly supported by JSPS KAKENHI Grant Numbers 22580375, 26450482, and 17K08169 (to TK), and by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from Japan Agency for Medical Research and Development (to TT).
Kanao T, Hase N, Nakayama H, et al. Reaction mechanism of tetrathionate hydrolysis based on the crystal structure of tetrathionate hydrolase from Acidithiobacillus ferrooxidans . Protein Science. 2021;30:328–338. 10.1002/pro.3984
Funding information Japan Society for the Promotion of Science, Grant/Award Numbers: KAKENHI 17K08169, KAKENHI 22580375, KAKENHI 26450482
REFERENCES
- 1. Bugaytsova Z, Lindström EB. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus . Eur J Biochem. 2004;271:272–280. [DOI] [PubMed] [Google Scholar]
- 2. Tano T, Kitaguchi H, Harada M, Nagasawa T, Sugio T. Purification and some properties of a tetrathionate decomposing enzyme from Thiobacillus thiooxidans . Biosci Biotechnol Biochem. 1996;60:224–227. [DOI] [PubMed] [Google Scholar]
- 3. Kanao T, Onishi M, Kajitani Y, et al. Characterization of tetrathionate hydrolase from the marine acidophilic sulfur‐oxidizing bacterium, Acidithiobacillus thiooxidans strain SH. Biosci Biotechnol Biochem. 2018;82:152–160. [DOI] [PubMed] [Google Scholar]
- 4. Kanao T, Kamimura K, Sugio T. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans . J Biotechnol. 2007;132:16–22. [DOI] [PubMed] [Google Scholar]
- 5. de Jong GAH, Hazeu W, Bos P, Kuenen JG. Isolation of the tetrathionate hydrolase from Thiobacillus acidophilus . Eur J Biochem. 1997;243:678–683. [DOI] [PubMed] [Google Scholar]
- 6. Protze J, Müller F, Lauber K, et al. An extracellular tetrathionate hydrolase from the thermoacidophilic archaeon Acidianus ambivalens with an activity optimum at pH 1. Front Microbiol. 2011;2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev. 2009;33:999–1043. [DOI] [PubMed] [Google Scholar]
- 8. Wang R, Lin JQ, Liu XM, et al. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front Microbiol. 2018;9:3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rzhepishevska OI, Valdés J, Marcinkeviciene L, et al. Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl Environ Microbiol. 2017;73:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El‐Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kanao T, Matsumoto C, Shiraga K, Yoshida K, Takada J, Kamimura K. Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol Lett. 2010;309:43–47. [DOI] [PubMed] [Google Scholar]
- 12. van Zyl LJ, van Munster JM, Rawlings DE. Construction of arsB and tetH mutants of the sulfur‐oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl Environ Microbiol. 2008;74:5686–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu Y, Liu X, Wang H, Li X, Lin J. Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans . J Bacteriol. 2014;196:2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beard S, Paradela A, Albar JP, Jerez CA. Growth of Acidithiobacillus ferrooxidans ATCC 23270 in thiosulfate under oxygen‐limiting conditions generates extracellular sulfur globules by means of a secreted tetrathionate hydrolase. Front Microbiol. 2011;2:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schippers A, Jozsa P‐G, Sand W. Sulfur chemistry in bacterial leaching of pyrite. Appl Environ Microbiol. 1996;62:3424–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pronk JT, Meulenberg R, Hazeu W, Bos P, Kuenen JG. Oxidation of reduced inorganic sulphur compounds by acidophilic thiobacilli. FEMS Microbiol Rev. 1990;75:293–306. [Google Scholar]
- 17. Wentzien S, Sand W, Albertsen A, Steudel R. Thiosulfate and tetrathionate degradation as well as biofilm generation by Thiobacillus intermedius and Thiobacillus versutus studied by microcalorimetry, HPLC, and ion‐pair chromatography. Arch Microbiol. 1994;161:116–125. [Google Scholar]
- 18. Meulenberg R, Pronk JT, Frank J, Hazeu W, Bos P, Kuenen JG. Purification and partial characterization of a thermostable trithionate hydrolase from the acidophilic sulphur oxidizer Thiobacillus acidophilus . Eur J Biochem. 1992;209:367–374. [DOI] [PubMed] [Google Scholar]
- 19. Cherney MM, Zhang Y, Solomonson M, Weiner JH, James MN. Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: Insights into sulfidotrophic respiration and detoxification. J Mol Biol. 2010;398:292–305. [DOI] [PubMed] [Google Scholar]
- 20. Veith A, Urich T, Seyfarth K, Protze J, Frazão C, Kletzin A. Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens . Front Microbiol. 2011;2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Denkmann K, Grein F, Zigann R, et al. Thiosulfate dehydrogenase: A widespread unusual acidophilic c‐type cytochrome. Environ Microbiol. 2012;14:2673–2688. [DOI] [PubMed] [Google Scholar]
- 22. Kanao T, Nakayama H, Kato M, Kamimura K. The sole cysteine residue (Cys301) of tetrathionate hydrolase from Acidithiobacillus ferrooxidans does not play a role in enzyme activity. Biosci Biotechnol Biochem. 2014;78:2030–2035. [DOI] [PubMed] [Google Scholar]
- 23. Kanao T, Kosaka M, Yoshida K, et al. Crystallization and preliminary X‐ray diffraction analysis of tetrathionate hydrolase from Acidithiobacillus ferrooxidans . Acta Cryst. 2013;F69:692–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fülöp V, Jones DT. β propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. [DOI] [PubMed] [Google Scholar]
- 25. Holm L. DALI and the persistence of protein shape. Protein Sci. 2020;29:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Z, Zhan LH, Hou HF, et al. Structural basis for the interaction of BamB with the POTRA3‐4 domains of BamA. Acta Cryst. 2016;D72:236–244. [DOI] [PubMed] [Google Scholar]
- 27. Buonfiglio V, Polidoro M, Soyer F, Valenti P, Shively J. A novel gene encoding a sulfur‐regulated outer membrane protein in Thiobacillus ferrooxidans . J Biotechnol. 1999;72:85–93. [DOI] [PubMed] [Google Scholar]
- 28. Krupovic M, Peixeiro N, Bettstetter M, Rachel R, Prangishvili D. Archaeal tetrathionate hydrolase goes viral: Secretion of a sulfur metabolism enzyme in the form of virus‐like particles. Appl Environ Microbiol. 2012;78:5463–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Jong GAH, Hazeu W, Bos P, Kuenen JG. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans . Microbiology. 1997;143:499–504. [DOI] [PubMed] [Google Scholar]
- 30. Chen ZW, Matsushita K, Yamashita T, et al. Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5. Structure. 2002;10:837–849. [DOI] [PubMed] [Google Scholar]
- 31. Tan KP, Nguyen TB, Patel S, Varadarajan R, Madhusudhan MS. Depth: A web server to compute depth, cavity sizes, detect potential small‐molecule ligand‐binding cavities and predict the pKa of ionizable residues in proteins. Nucleic Acids Res. 2013;41:W314–W321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sauvé V, Roversi P, Leath KJ, et al. Mechanism for the hydrolysis of a sulfur‐sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J Biol Chem. 2009;284:21707–21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guerrero SA, Hecht H‐J, Hofmann B, Biebl H, Singh M. Production of selenomethionine‐labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl Microbiol Biotechnol. 2001;56:718–723. [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W. XDS. Acta Cryst. 2010;D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Cryst. 2013;D69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Cryst. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terwilliger TC, Adams PD, Read RJ, et al. Decision‐making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Cryst. 2009;D65:582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liebschner D, Afonine PV, Baker ML, et al. Macromolecular structure determination using X‐rays, neutrons and electrons: Recent deelopments in PHENIX. Acta Cryst. 2019;D75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lovell SC, Davis IW, Arendall WB III, et al. Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins. 2003;50:437–450. [DOI] [PubMed] [Google Scholar]
- 41. Baker NA, Sept D, Joseph S, Holst MJ, Mc Cammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequence alignment among three 4THases from Acidithiobacillus species
Figure S2. Electron densities in the cavity of the substrate‐soaked data
Figure S3. pH profile of Af‐Tth activity


