Abstract
Membrane proteins are involved in numerous vital biological processes, including transport, signal transduction and the enzymes in a variety of metabolic pathways. Integral membrane proteins account for up to 30% of the human proteome and they make up more than half of all currently marketed therapeutic targets. Unfortunately, membrane proteins are inherently recalcitrant to study using the normal toolkit available to scientists, and one is most often left with the challenge of finding inhibitors, activators and specific antibodies using a denatured or detergent solubilized aggregate. The Nanodisc platform circumvents these challenges by providing a self‐assembled system that renders typically insoluble, yet biologically and pharmacologically significant, targets such as receptors, transporters, enzymes, and viral antigens soluble in aqueous media in a native‐like bilayer environment that maintain a target's functional activity. By providing a bilayer surface of defined composition and structure, Nanodiscs have found great utility in the study of cellular signaling complexes that assemble on a membrane surface. Nanodiscs provide a nanometer scale vehicle for the in vivo delivery of amphipathic drugs, therapeutic lipids, tethered nucleic acids, imaging agents and active protein complexes. This means for generating nanoscale lipid bilayers has spawned the successful use of numerous other polymer and peptide amphipathic systems. This review, in celebration of the Anfinsen Award, summarizes some recent results and provides an inroad into the current and historical literature.
Keywords: lipid bilayer, membrane protein, Nanodisc, signaling complex
1. INTRODUCTION
It is an honor to receive the Protein Science Anfinsen Award and provide this article for the journal Protein Science. Dr. Anfinsen 1 was a pioneer in efforts to understand the structure of proteins. Using ribonuclease to show that the folding of a protein was determined by its amino acid sequence, 2 he initiated a vast field of study as to how the three dimensional structure of a protein comes together from a random coil. Together with the assembly of multimeric protein complexes, Nature indeed has revealed the beauty of self‐assembly. Further pioneering work by Fred Richards and others on the elucidation of the ribonuclease crystal structure and the re‐assembly of the cleaved S‐peptide fragment with the A‐protein 3 revealed how complementarity drives the formation of higher order complex entities. As we will see, Nanodiscs, a complex of lipids and an amphipathic helical protein, are a wonderful example of self‐assembly. As Anfinsen used protein modification to understand structure, so modifications of the helical proteins that self‐assemble lipids into discoidal structures have enabled a plethora of Nanodisc applications in the study of membrane proteins and membrane protein/lipid complexes. Fittingly, Anfinsen was also active early in the structure of lipoproteins and their interconversion by enzymatic processes. 4
Our term Nanodiscs refers to a discoidal self‐assembled complex of lipids and a helical protein we have called the membrane scaffold protein (MSP). The current MSP based lipoprotein Nanodiscs are direct descendants of the human high‐density lipoprotein (HDL) particles and the helical MSPs from the primary HDL protein component, apolipoprotein‐A1 (Apo‐AI). Their current wide‐spread use by the biochemical and biophysical community has been aided by modifications to the original Apo‐AI sequence, but the evolution was aided by a vast literature of lipoproteins and atherosclerosis as a human malady. Hence, a brief history is called for.
Human plasma lipoproteins were known to be complexes of lipids, proteins and cholesterol derivatives. 5 The “high density” fraction, defined via ultracentrifugation as a density of 1.063–1.21 g/ml, received study as the so‐called “good cholesterol”. Pioneering work by Brouillette and colleagues noted that the first 43 amino acids of the human Apo‐AI sequence were not required for lipid binding. 6 Continuing with major contributions from Segrest, the subsequent 11‐ or 22‐residue helices, punctuated by glycine or proline residues, were known to provide the major lipid binding domain and to also help control the size of the resulting particle. 6 , 7 , 8 The modularity of the alpha helix content of Apo‐AI led to the hinge domain hypothesis 9 due to the existence of discrete steps in particle sizes. With the extensive work of Jonas and colleagues, 10 , 11 , 12 who honed the detergent dialysis method for forming “reconstituted” HDL particles, it was clear that a large degree of heterogeneity could exist in the size and composition of reconstituted HDLs. 13
Our entry into the lipoprotein field came from two very different directions. The first was from a realization that much of the machinery of the living cell involved organized multi‐component complexes of proteins, lipids and nucleic acids. Protein structure determination, via x‐ray or NMR, could not easily capture these large entities. Hence we began adapting the then revolutionary idea of atomic force microscopy (AFM) to work under aqueous solutions. 14 In looking for something in the ~10–100 nm size range we were struck with the discoidal form of HDL that could be easily generated by the dialysis method developed by our Illinois colleague. 13 When we examined a population of HDL particles assembled from various Apo‐AI to lipid stoichiometries, the heterogeneity was obvious. 15 , 16 While there seemed to be a preference for a certain ratios, the origin of the heterogeneity was not clear, even though fractionating by size exclusion chromatography could yield a much more narrow size distribution. 15 Nevertheless, the AFM experiments worked and part of the laboratory went on to use this tool to pattern and quantitate single bilayers on atomically flat surfaces. 15 , 17
The second direction, that formed our branch point in the study of lipoproteins, came from a problem in need of a solution. Most of my laboratory was heavily involved in understanding the bioinorganic mechanisms of metalloprotein oxygenase, particularly the cytochromes P450. 18 Much of the functional understanding, and first crystal/NMR structure results came from the study of a soluble bacterial P450cam. However, a great relevance of P450s is their key role in human drug metabolism and hormone biosynthesis. Believing Alexander Pope's 1773 poem noting that “The proper study of Mankind is Man” we wanted to study the mammalian P450s. But, horror of horrors, these were all membrane bound! Frustrating were the human proteins isolated from their membranes via detergent. Often these were then studied in detergent, with sometimes a small amount of lipid added or as an aggregate following removal of the solubilizing agent. However, the mechanistic parameters derived from study of these “solubilized” P450s did not match those obtained from the bacterial versions, for example, Reference 20. Was there something fundamentally different, or did the isolation from the normal membrane bilayer environment render the protein either partially or wholly inactive? We needed to put these membrane proteins back into a bilayer environment. While reconstitution into liposomes was well known, these large heterogeneous entities were not stable to the techniques we had become used to with soluble proteins (e.g., stopped‐flow spectrophotometry). Hence the wild idea: What about just adding the detergent solubilized membrane protein into the same mix of detergent solubilized helical Apo‐AI and lipid that was used to assemble Nanodiscs? Would the membrane protein target magically assemble into the discoidal bilayer? Would this be stable and amiable to biophysical studies? The answer, of course, is that indeed, Nature's process of self‐assembly allowed everyone to find their appropriate home.
Our first efforts were with a simple target that had only a short membrane anchor, with the main catalytic portion of the structure in a soluble “ecto” domain. The P450 reductase (a 68 kD flavoprotein) provided this initial test. Indeed it worked, could be imaged by the AFM, was shown to be active in the Apo‐AI supported bilayer and even be structurally characterized by measuring to high precision the height above the bilayer surface. 16 , 19 Subsequent tries with a more complex membrane anchoring topology, provide by a mammalian P450 that is “embedded” into the membrane (Figure 1b), also worked, and again AFM under physiological conditions could provide a model for how these two proteins could come together to effect electron transfer and catalysis. 21 Subsequent work over the past 20 years by many laboratories has shown that the same self‐assembly process works for almost any membrane protein, including multi‐span integral proteins and even complex multi‐component assemblies such as the photosynthetic reaction center, 22 cytochrome oxidases 23 and transporters. 24 , 25 Multiple examples will be described in later sections.
FIGURE 1.
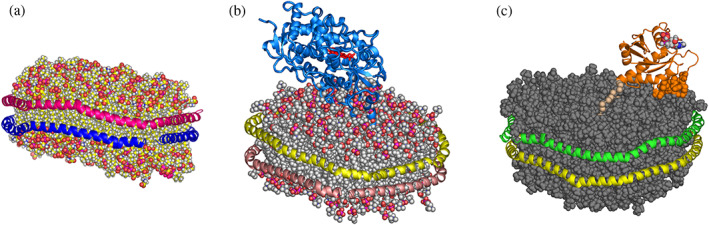
The Nanodisc System. All images are based on long‐term molecular dynamics simulations and illustrate two main uses of Nanodiscs. (a) The self‐assembled discoidal bilayer formed by DMPC lipids and two encircling MSP1D1 membrane scaffold proteins (MSPs; blue and magenta helices). (b) Human cytochrome P450 CYP3A4 as an example of an incorporated membrane protein target in its natural bilayer environment, rendered soluble in aqueous solution by the amphipathic MSP. (c) The use of Nanodiscs as a membrane surface of defined lipid composition. The fully processed oncoprotein KRas4b, with methylated carboxy terminus and covalently attached farnesyl group as it associates with the bilayer (grey surface). The polycationic hyper variable domain (wheat color, Cα atoms shown) and head group residues (helix 4 surface representation) both associate with the anionic bilayer surface. GDP substrate is shown in a Van der Waals atomic surface representation
1.1. The question of size and functionality
In the historical context, we were still concerned with size heterogeneity as we ultimately would like to apply Nanodiscs to three dimensional structural determination. In addition, some uses, such as the reconstitution of large multi‐protein complexes, may need discoidal bilayers larger than ~10 nm. Given the discussion in the HDL literature that one or more internal helices, in addition to the amino terminal “globular domain,” may or may not be involved in lipid protein interactions, we thought to increase the length of the MSP by adding one, two or three additional 22‐mer helices. This corresponds to what is now called MSP1E1, MSP1E2, and MSP1E3, made by adding another helix‐4, helix‐4 plus helix‐5 or helix‐4, helix‐5 and helix‐6 respectively. 26 But a long problem in the literature surfaced—what was the topology of the Apo‐AI protein relative to the lipids? Two models were proposed. Much of the earlier experimental 9 , 27 and theoretical 8 work envisioned a “belt” configuration, yet infrared linear dichroism 28 and molecular dynamics simulations 29 proposed a “picket fence” configuration where the helical components were punctuated by turns and ran up‐down. Ultimately, this was resolved by numerous experimental approaches including solid state NMR, 30 solution NMR, 31 chemical crosslinking and mass spectrometry 32 and molecular dynamics trajectories. 33 As will be discussed, recent high‐resolution cryo‐electron microscopy (cryo‐EM) of Nanodisc embedded membrane proteins clearly show the belt configuration. More recent long‐time molecular dynamics calculations have added motion to the picture of the encircling MSP, capturing the collective dynamical motion of the discoidal bilayer. 34
An indirect quantitation of lipid count in the Nanodisc versus the length of the MSP could also give an indication as to which model was correct, since the width of a “picket fence” helix is clearly smaller than the length of a 22‐mer “belt.” Realizing that the most precise determination of lipid content was via radioactive counting, Denisov et al. measured the number of DPPC lipids in purified, homogeneous MSP1, MSP1E1, MSP1E2 and MSP1E3. 26 , 35 In a belt model, if the lipid count is plotted against the square root of the MSP length, then a straight line should form. This was observed, and clearly indicated the preference for the “belt” configuration. However, the fit to the data did not go through the origin, suggesting that the first part of the MSP sequence was not contributing to the formation of the phospholipid bilayer. Genetic engineering of the MSP synthetic gene that removed 11, 22 and 33 amino acids, size characterization 26 and molecular dynamics simulation 36 showed that deletion of the first 11 amino acids of the truncated Apo‐AI sequence (already missing the residues 1–43 of the globular domain) gave the most stable and homogeneous Nanodiscs. These MSPs were then labeled with D1 (for deletion) as MSP1D1, MSP1E1D1, MSP1E2D1 and MSP1E3D1. While the MSP1D1 and MSP1E3D1, which generate ~9.8 nm and ~12.7 nm discs respectively, are used the most frequently, even larger constructs were made. 37 Also successful, and of important use in recent cryo‐EM investigations, is MSP2N2. 38 This was an evolution from the earlier MSP2, which was simply a fusion of two MSP1 sequences, but did not form homogeneous or stable discs. 39 Removing the extra amino acids from the N‐terminus and generating a fusion of two MSP1D1 sequences with a simple glycine‐serine linker generates nice ~17 nm discoidal bilayers. How big can one go? At some point this will look like a mixture of lipids and amphipathic protein with the aspect ratio of a dinner plate, and fluctuations will result in collapse. Remember, Nature evolved the Apo‐AI sequence to form the netting around the predominant spherical HDL particle.
Over the years, the utility of Nanodiscs has been expanded by including a variety of tags or altered amino acid residues into the MSP sequence. These have been summarized in the more recent reviews. 40 , 41 Briefly, histidine tags are available at both the N‐ and C‐ terminus of MSP and the N‐terminal variants with Factor X or TEV protease sites. FLAG tags are available at both termini. Unique cysteine residues have been incorporated for attaching fluorescent or other labels. Most recently, “Dark” MSP with no tryptophans, “Ultra Dark” which also deletes the tyrosine residues, and “Ultra Bright” with five additional tryptophans per MSP have been generated. 42 These have found use in some forms of assay, such as analytical ultracentrifugation, fluorescence polarization or microscale thermophoresis.
1.2. The current popularity of nanodiscs
The current use and overall utility of Nanodiscs can be summarized in two graphs. Following the discovery that Nanodiscs could be used to self‐assemble essentially any membrane protein in a phospholipid bilayer with maintenance of function, it was initially utilized by only a few laboratories as evidenced from publications, quoted from PubMed by title or key word (Figure 2a).
FIGURE 2.
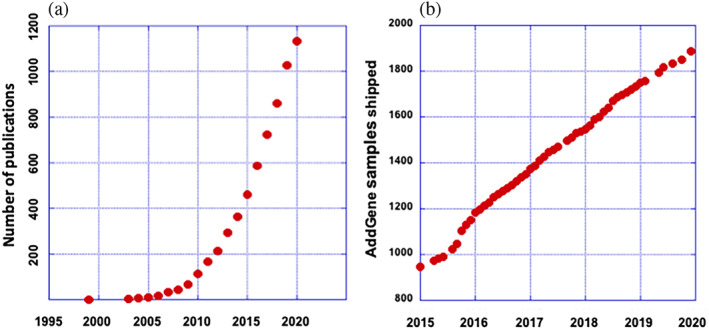
The popularity of Nanodiscs. (a) Indicates the cumulative number of publications that deal with Nanodiscs and their applications. (b) Shows the cumulative number of samples of the membrane scaffold protein (MSP) gene shipped by AddGene to laboratories worldwide. The abscissa indicates the value as of October 1, of the indicated year
Believing that new technologies should be disseminated to the widest academic audience, we provided the synthetic genes for MSPs, optimized for high expression in Escherichia coli, to the scientific community through AddGene. These constructs were widely popular, winning the “Blue Flame” designation from AddGene early on. As evidence of their continued popularity, Figure 2b illustrates that the number of samples provided to the academic community has doubled since 2016. For those not willing to express the MSP in a liter of bacteria, or for commercial entities, Millipore‐Sigma offers a variety of purified and characterized proteins. With increasing use as a controlled membrane surface, in vaccines and in vivo delivery, we can expect the use of these nanometer scale lipid bilayers will only increase. Equally important is the explosion in the use other amphipathic molecules for bilayer stabilization, including full length Apo‐AI, individual oligopeptides and a variety of complex organic polymers. As a general example of amphipathic protein‐lipid assembly, it is amusing to note that even apo‐myoglobin, with five helices, will associate with lipids to form discoidal particles.
1.3. Recent reviews of nanodiscs
There have been numerous reviews of MSPs and the Nanodisc approach, both from our laboratory and others. 40 , 41 , 43 , 44 , 45 Other reviews also describe other amphipathic scaffolds, including zebrafish apolipoproteins, 46 the saposin family of lysosomal lipase activators, 47 and short amphipathic peptides. 48 , 49 Indeed, even α‐synuclein 50 and apo‐myoglobin, with its five helices, can solubilize lipids into 10‐nm structures. 51 The use of amphipathic polymers, first those of styrene maleic acid pioneered by Dafforn and co‐workers, 52 and extended to other block copolymers of defined charge and hydrophobicity 53 , 54 , 55 are also finding increasing use. An extensive comparison of these various means for rendering membrane proteins suitable for biophysical structure determination is beyond the scope of this article. Clearly, however, the choice of approach depends on the end point sought. We focus the remainder of this manuscript on summarizing the latest applications of MSP based Nanodiscs. We use a capital “N” to recognize the former copyright of Nanodisc and to distinguish the use of these MSP proteins, engineered from Apo‐AI, that yield homogeneous and stable nanoscale soluble lipid bilayers.
1.4. How do Nanodiscs assemble?
When the first presentation of the Nanodisc platform was published more than 20 years ago, George Whitesides noted that this was “an excellent example of self‐assembly.” How else to describe hundreds of lipids, long amphipathic MSPs and integral membrane protein targets all coming together into a final functional structure? These core process of lipids and Apo‐AI rearranging, and interacting with enzymes such as LCAT and cholesterol exchange proteins, are at the heart of important transitions in the human lipoproteins that have been studied for many years, 56 aided by an in vitro procedure to form HDL particles with detergent removal, forming so‐called “reconstituted” HDL. 57 The success of incorporating a wide variety of membrane proteins into the resultant discoidal bilayer in their correct configuration further encourages the question as to how the various components come together. How multi‐subunit integral membrane proteins themselves can order correctly is amazing. In each case, the starting condition consists of a detergent solubilizing the lipid, encircling MSP and the individual subunits of the target. Dialysis, or more commonly, the addition of hydrophobic beads, provides a sink for the detergent, through a complex process involving desorption of the detergent from the surfaces of target, MSP and lipid. If there is also a solubilized target, a key question relates to the branch point between correct assembly and the aggregation of components. This leads to a variety of success rates for the incorporation of a membrane protein target into Nanodiscs. Some systems, with the correct stoichiometry of reactants, >90% of the target is functionally incorporated. Others (notably GPCRs) can result in <10% success. Careful optimization of conditions, including detergent choice, amount and exposure time to the hydrophobic beads, temperature and so forth can improve the overall yield. But what is clearly needed is a detailed understanding of the Nanodisc assembly process. Far from being just a philosophical issue, this is the single rate‐limiting step in using the powerful Nanodisc technology to investigate membrane proteins where the quantities are limited or they are sensitive to conditions.
For the relatively simple assembly of “bare” Nanodiscs with the engineered MSPs, early work utilized molecular dynamics to examine the reverse process of detergent disassembly, 58 as well as the collapse of a pre‐arranged MSP with lipids to form a discoidal particle. 59 , 60 This work was extended to examine the conversion of disc to sphere, as is the case for the maturation of circulating HDL particles. 61 However, there has been limited experimental work examining this complex self‐ assembly process. Preliminary solution X‐ray scattering (SAXS) studies after rapid removal of detergent suggests that discs are formed on the minute time scale. One approach has been to examine the structure of the mixture of lipid and scaffold protein at varying concentrations of detergent. Our early work compared experimental SAXS data with calculations to provide a suggested path for assembly. 59 More recent work in our laboratory followed several biophysical parameters, including second derivative optical spectroscopy to monitor the polarity of tyrosine residues, Laurdan fluorescence to suggest when a bilayer‐water interface was forming and the overall rotational correlation time of the particle. 62 By studying both addition and removal of detergent one came to the conclusion that the assembly process was reversible, with the bilayer beginning to form below the critical micellar concentration of the detergent and the association of MSP and lipids starting at even lower detergent levels. Other laboratories, notably that of Arleth, 63 are also beginning to address the details of Nanodiscs assembly. The more complicated process of how a membrane protein can find its correct home in the bilayer, as well as the sequential ordering necessary for the functional incorporation of multi‐subunit proteins and entire assemblies, such as the photosynthetic reaction center or cytochrome oxidase, is only beginning to be addressed.
1.5. Nanodiscs enable control of membrane protein aggregation state
Many complexes and assemblies of multi‐component membrane proteins have been successfully incorporated into Nanodiscs, which are well suited to stabilize the critical protein–protein interactions in a native‐like membrane. An impressive recent example is the multiprotein complex of the mitochondrial calcium uniporter assembled in Nanodiscs containing the cardiolipin necessary for function. Cryo‐EM structures in the presence and absence of Ca2+ were resolved for the uniporter holocomplex, which was formed from MCU and EMRE subunits from the beetle Tribolium castaneum in complex with the human MICU1‐MICU2 heterodimer. 64 , 65 An MCU‐EMRE subcomplex was prepared by coexpressing TcMCU and TcEMRE in mammalian cells, purifying the TcMCU‐EMRE assembly, and reconstituting it into lipid Nanodiscs with MSP1D1 and a lipid mixture (POPC:POPE:cardiolipin 2:2:1). This control over lipid composition of the membrane in Nanodiscs and purification of complex, with well‐defined stoichiometry, enabled acquisition of high resolution data and to discern the mechanism of cation dependent gatekeeping. 64 , 66
The stability of Nanodisc assembled membrane proteins, and lack of protein exchange between Nanodiscs, has provided a useful tool for addressing the question of oligomerization and functional relevance of oligomer formation. A first example was demonstrated early in our laboratory by monitoring the correct assembly of bacteriorhodopsin trimer in Nanodiscs, 67 and then by proof‐of principle experiments comparing transducin activation 68 and arrestin binding to rhodopsin monomers and dimers. 69 As a result, the application of Nanodiscs provided direct evidence of fully active rhodopsin monomers with properties similar to those observed in proteoliposomes, where in this mimetic it is impossible to control the monomer‐dimer equilibrium. In addition, the binding of arrestin was enhanced in the presence of POPG lipids, and conformational changes in visual arrestin upon binding to the phosphorylated rhodopsin monomer were similar to those observed in native membranes. 70
A qualitative understanding of the process of multi‐subunit assembly was realized by the studies of bacterial chemotactic receptors in collaboration with the Hazelbauer laboratory. 71 , 72 The Tar receptor is a trimer of dimers 73 , 74 with separate assays for the formation of dimers and the final oligomeric configuration in the bilayer. This allowed the dissection of the specific roles of dimerization and their roles in activating of chemotaxis kinases and allosteric coupling of the trimer of dimers 75 to the histidine kinase signaling complex. While the extensive protein–protein interface between dimers drove their formation in the correct configuration, the trimer of dimers only forms a small interface at the apical position. Hence, during the assembly process, the sets of dimers are incorporated in multiple orientations relative to the bilayer. The subsequent cryo‐EM structure of E. coli aspartate receptor dimers revealed the presence of two flexible hinges which allow for formation of higher order complexes without steric clash. 76 This example suggests that subunit components with high affinity will form correctly in the resultant Nanodisc, whereas weak or transient protein–protein complexes may incorporate into the bilayer in different overall configurations.
1.6. Cell‐free translation of membrane proteins into Nanodiscs
An alternative means of membrane protein incorporation into Nanodiscs is based on cell‐free in vitro expression of membrane proteins in the presence of Nanodisc components or preformed Nanodiscs. 77 Binding to the preformed Nanodiscs prevents aggregation and precipitation of expressed protein. 78 Co‐translational incorporation of membrane proteins into Nanodiscs significantly improves the yield of folded and fully functional membrane proteins. 79 , 80 This approach was optimized for several proteins expressed with various 13C‐ and 15N‐labeling schemes for solution NMR studies. 81 Cell‐free expression can be also used to bypass solubilization and purification step for unstable enzymes. For instance, Cook et al. 82 successfully used cell‐free expression to assemble and purify to homogeneity the monomeric human UGT2B7 in Nanodiscs using the scaffold protein MSP1D1 with POPC lipids. The enzyme with intact C‐terminal trans‐membrane helix is stable in Nanodiscs and functionally competent, similar to UGT2B7 in Supersomes®. Each Nanodisc is shown to harbor a fully active single UGT2B7 monomer, contrary to the previous suggestions that formation of dimers is needed for activity.
1.7. Biophysical characterization of Nanodiscs
A number of reviews have summarized the plethora of biophysical tools that have been used to characterize Nanodiscs, 40 , 41 which can be only briefly summarized. First, is the case of “bare” Nanodiscs, composed of just lipid and the MSP. The overall shape is that of a circular disc as evidenced by numerous cryo‐EM and TEM structures, 83 , 84 , 85 although depending on packing density of lipids and the models used for interpretation, solution X‐Ray scattering can indicate a more elliptical configuration. 63 , 86 As monitored by both Laurdan fluorescence and differential scanning calorimetry (DSC), the lipid bilayer contained by the encircling MSPs undergoes a thermal phase transition similar to that seen in liposomes, although broader. 87 This phase transition can also be monitored by SAXS where the mean area per head group and packing is quantitated. 88 More recent works have also monitored the phase transition and separated the contributions from the central lipid core and those juxtaposed to the MSP belt. 89 Here the core domain has properties similar to those measured for liposomes with 1–2 layers of more perturbed lipids due to their direct interactions with the scaffold protein. Recent DSC of the very small DMPC Nanodiscs 89 suggests a significantly perturbed bilayer. Long‐time molecular dynamics studies contributed to a picture of fluctuations of the scaffold protein and internal motion of the Nanodisc. 34 , 90 The tumbling motion of the entire Nanodisc has been measured by time resolved fluorescence anisotropy, 91 yielding a rotational correlation time of ~50–70 ns for the MSP1D1 discs, similar to those reported from NMR studies. 92 Interestingly, the overall solution motion of the Nanodisc can be used to monitor the binding of molecules to the Nanodisc, thereby giving a label‐free assay. One excellent example involved discovering a specificity for the association of the signaling protein KRas4b to surfaces of varying ionic lipid composition. 93
As will be described, many investigations use the Nanodisc to provide a surface containing mixed charged lipids. Since one can imagine the difficulty in packing over a 100 charged head groups in ~7–10 nm area, a key question becomes understanding the actual charge on the Nanodisc. To this end, in collaboration Professor Laue, membrane confined electrophoresis was used to determine the actual Debye–Huckel–Henry charge for different lipid mixtures. 94 Not surprisingly, Nanodiscs behave as polyelectrolytes. When the pK of the head group can be altered by the local electrostatic field, such as with POPS, Nanodiscs can be formed with very high percentage of the charged lipid. When quantitating binding events, the true thermodynamic characterization requires analysis of actual charge rather than the number of incorporated lipids. 93
Recent progress in native mass‐spectrometry, in collaboration with Professor M. Gross, enabled the direct counting of lipids in Nanodiscs. 95 , 96 The next step applied this method to studies of membrane proteins and peptides in Nanodiscs, as described in several original papers and reviews. 97 , 98 , 99 An elegant extension of Nanodisc technology for mass‐spectrometry applications via generation of the mutant MSP was designed in order to change total Nanodisc mass and to improve resolution of overlapping species in a single spectrum. 100 An advantage of Nanodiscs is the precise control of lipid composition, which was used in order to probe the effect of specific lipids on dynamic properties of two membrane transporters XylE and LacY. 101 Systematic analysis of HDX data in Nanodiscs of various lipid compositions, combined with molecular dynamics simulations, indicated significant stabilization of the inward‐open conformational state in the presence of PE, as compared to PC headgroups.
Hydrogen‐deuterium exchange experiments performed with MSP1D1 Nanodiscs revealed a picture of scaffold protein dynamics, as compared to the lipid‐free MSP1D1. 102 As expected, MSP1D1 in Nanodiscs had much higher level of protection than lipid free protein on a short time scale, but reached the same level of exchange at 2 hr or longer. The largest dynamic range was found to be at the C‐terminal helices, while the central helices were significantly more stable. This observation is in a good agreement with multiple MD simulations of Nanodiscs, starting from our first publication with only 6 ns long trajectories. 36 Subsequently, a variety of Nanodiscs of different sizes were explored both with all‐atom and coarse‐grain modeling, providing a consistent picture of the dynamic bilayer surrounded by two helical belts having distorted circular or elliptical structures. 34 , 89 , 103 , 104 Direct contact with the scaffold protein usually resulted in more disordered and dynamically constrained lipids at the lipid‐protein interface. The same effect was observed in several NMR studies of Nanodiscs, in both solid‐state 105 and solution. 31
1.8. Nanodiscs as imaging agents
The large surface area of Nanodiscs prompted the idea that they could be used as a vehicle for in vivo imaging. One challenge in magnetic resonance imaging with heavy metal contrast reagents, such as gadolinium (Gd3+), is the relatively short plasma lifetime and overall sensitivity. Nanodiscs are descendent from human HDL particles, which are immunologically neutral and have significant plasma lifetime until cleared by the hepatic scavenger receptors. Additionally, the possibility that multiple sensitizers could be loaded into a single 10 nm particle could offer dramatic increase in sensitivity. Our first efforts with commercially available lipid anchored chelates resulted in precipitation and formation of rouleaux due to crosslinking, whether or not the Gd3+ was incorporated before or after the lipids were self‐assembled into the Nanodisc. Although HDL particles have been suggested for use as a contrast agent platform, 106 Nanodiscs offer the advantages of improved stability and the potential for multi‐mode imaging. Improved Gd3+ chelating agents and lipid anchors developed by Meade and co‐workers 107 were self‐assembled into Nanodiscs together with a fluorescent dye covalently attached to the MSP, producing particles that label cells with high efficiency and provide a dramatic sensitivity increase for magnetic resonance imaging. 108 Further advancing the multi‐factorial aspects of Nanodiscs was realized in collaboration with Shively and co‐workers. Here 64 Cu‐DOTA‐azide click linked to PEGylated lipids were incorporated into Nanodiscs together with an antibody targeting carcinoembryonic antigen (CEA) positive tumors. 109 In addition, the same chemical derivation approach was shown to load therapeutic drugs such as doxorubicin, thus realizing the goal of a three pronged platform of targeting, imaging and treatment. Further examples of Nanodisc utility in positron emission tomography diagnostics were recently reported. 110
1.9. Membrane proteins in nanodiscs
Structural studies of membrane proteins in the phospholipid bilayer of Nanodiscs have made major advances using cryo‐EM and NMR spectroscopy, taking advantage of sample size and lipid composition that mimic the natural membrane environment. These enabled improved resolution through extensive statistics collected with class‐averaged single‐particle images revealing the correct lipid‐protein interactions of the incorporated target. Many such examples were published during recent years, when cryo‐EM reached a new level of resolution with new sophisticated methods of analysis. 111 Because of size limitations of this review we selected only a minor sampling that illustrates various aspects of Nanodisc utility in this field (Figure 3).
FIGURE 3.
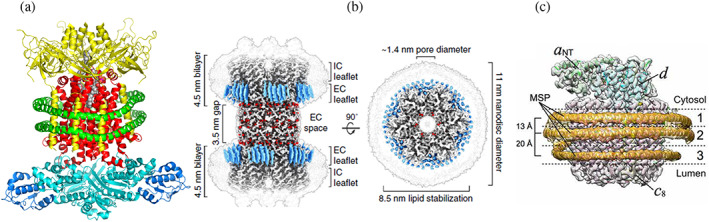
(a) Cryo‐electron microscopy (cryo‐EM) structure of the ABC transporter complex MlaFEDB. Hexamer MLAD (yellow), dimeric transport permease MLAE (red), ATP binding protein (cyan) and transporter‐binding protein MLAB (blue) are enclosed into MSP1D1 Nanodisc (two parallel helices shown in green). 112 (b) Cryo‐EM structure of connexin‐46/50 incorporated in two Nanodiscs (light gray contours). Well resolved lipids are shown in blue, pore with diameter 1.4 nm is shown as the top view on the right. Reproduced with permission from Flores et al. 113 (c) Cryo‐EM structure of the Vo proton channel of yeast vacuolar ATPAse. In this structure three molecules of MSP1E3D1 scaffold stabilize the protein complex by forming rings of different diameter around middle transmembrane part and around c‐ring. Reproduced with permission from Roh et al. 114 Open access article published under the CC BY license
In a recent work, the Lefkowitz group incorporated the M2 muscarinic and β2‐adrenergic receptors in Nanodiscs to study structural and functional properties of their complexes with β‐arrestin. They found that MSP1E3D1 Nanodiscs provided optimal size for interaction of the β‐arrestin C‐domain with the lipid bilayer, as seen by cryo‐EM. 115 Allosteric activation of proto‐oncogene kinase Src was shown to require interaction with the complex of β‐arrestin and phosphorylated at C‐terminal tail GPCR receptors in Nanodiscs, while chimeric M2 receptor with non‐phosphorylated C‐terminus did not activate Src. 116
A spectacular high resolution example is provided by the cryo‐EM structure of the phospholipid ABC transporter, MlaFEDB, in Nanodics formed with MSP1D1 and an E. coli polar lipid extract (PDB 6XBD) that explicitly resolves the encircling MSP (Figure 3a). 112 This transporter was found with two lipid molecules bound as substrates, which may indicate transport of two lipids in each catalytic cycle, or involvement of this transporter in the transport of cardiolipin, which has four acyl chains.
Nanodiscs are used in SARSCoV2 studies, with the cryo‐EM structures of both dimer and tetramer of the 3a ion channel in MSP1E3D1 Nanodiscs were obtained with a novel fold of 3a channel resolved. 117 A co‐ and post‐translational insertase, a large nine‐subunit membrane protein complex from the endoplasmic reticulum (EMC), was incorporated in MSP2N2 Nanodiscs, and the cryo‐EM structure resolved to 3.4 Å in order to decipher the mechanism of substrate insertion. 118 Use of the larger Nanodisc was required to provide sufficient area for interactions with the surface‐oriented helices in addition to the 12 trans‐membrane helices of the seven subunits of the complex. Local thinning of the membrane on one side of EMC was suggested to lower the barrier for the substrate insertion into a hydrophilic conduit formed by three subunits of the complex, with the help of other parts on both sides of the membrane.
Another example of cryo‐EM resolving lipid‐protein interactions is the structure of Cx46/50, a complex of the connexins Cx46 and Cx50, each in Nanodiscs (MSP1E1 scaffold, DMPC lipids) forming a pore between two membranes, mimicking cell‐to‐cell junction with 3.5 nm gap. 113 Connexins form intercellular channels for communication of ions and small molecules between cells, often assembling into arrays, termed gap junctions, which are critical for synapse responses, and signaling. Interactions of connexins with membranes and their assembly strongly depend on the lipid environment, hence molecular mechanisms of these junctions should be studied in direct interactions with membranes. Nanodiscs provide an optimal tool for the single‐particle cry‐EM structural analysis of these interactions, revealing highly ordered lipids shown in blue in Figure 3b.
Very recently a collaboration from several laboratories demonstrated that long chain lipids and expanded hydrophobic surface of Vo‐channel of vacuolar ATPase could assemble with three MSP1E3D1 scaffold proteins and revealed the structure of the resulting nanodisc by Cryo‐EM (Figure 3c). 114 This work provided the membrane environment necessary to reveal the detailed mechanism of proton translocation by the vacuolar ATPase and a high resolution structure of both the proton channel and scaffold protein.
Nanodiscs have also been used for structural studies of membrane proteins in a detergent‐free lipid environment by NMR. 45 , 119 , 120 Although the large sizes of Nanodiscs often put limitations on high resolution NMR spectroscopy in solution, the advantages of this method of membrane protein solubilization can be used by applying new experimental tools to solve this problem. 121 As previously mentioned, 13C‐ and 15N‐enriched proteins for solution NMR studies can be made in cell‐free expression systems 81 or in E. coli 122 with subsequent incorporation into Nanodiscs. Interactions of cytochrome P450 and its redox partners cytochrome b5 and NADPH dependent P450 reductase in Nanodiscs have been documented using NMR. 123 , 124 , 125 , 126 , 127 NMR was also used for screening of the small molecule binding to the membrane enzyme DsbB. 128
Essentially the complete tool‐kit of spectroscopic methods has been applied to reveal the structure and function of membrane proteins self‐assembled into Nanodiscs. In addition to providing a close fit to the native membrane environment, allowing sampling of functionally important conformations, the Nanodisc platform provides an optically transparent medium, is robust to rapid mixing and variable temperature and offers a low viscosity aqueous solution. Uses of resonance spectroscopies, including all forms of paramagnetic resonance, fluorescence energy transfer (FRET), circular dichroism, hydrogen exchange, calorimetry, electrochemistry and high pressure have been applied. An extensive list of citations is provided in our recent reviews. 40 , 41
1.10. Human cytochromes P450: An example of Nanodisc utility
Eukaryotic cytochromes P450 are membrane proteins with a single N‐terminal trans‐membrane helix and a hydrophobic patch embedded in the lipid head groups of the membrane bilayer. This large superfamily has over 300,000 members and is found across all life forms. 116 Fifty‐seven members perform critical roles in human biochemistry, but tend to form aggregates in aqueous solution. In most cases they require significant concentrations of detergent in order to solubilize the oligomers and aggregates to gain even partial activity. As a result, their catalytic properties, measured under various conditions in different solubilizing mixtures, have often been poorly reproducible with a large degree of variability in catalytic parameters such as substrate binding affinity, induced spectral change and product forming rates. 129 , 130 , 131 Another complication arises because of possible heterogeneity of the P450 sample preparation and substrate partitioning into the membrane, which is hard to control. 131 This represents a well‐known problem in all biophysical and biochemical studies of membrane proteins, which carry out their normal function while inserted in the lipid bilayer, or interacting with the membrane surface. For the cytochromes P450 these interactions are even more important, because their redox partners, cytochrome P450 reductase (CPR), adrenodoxin reductase (Adr) or cytochrome b5, are also membrane bound proteins, and their functional interactions with P450 enzymes is dictated by the membrane surface to control height and orientation. 19
As with other membrane proteins, the historic choice for gaining in vitro activity utilized proteoliposomes and mixed micelles consisting of detergent and lipids to solubilize the protein and ascertain function. 43 Both approaches have advantages and disadvantages. Proteoliposomes are good mimics for biological membranes, but hard to use with various spectroscopic methods because of turbidity, and impose diffusional limitations for substrate access due to phase heterogeneity. Since proteins are mobile in the liposome bilayer, control of the stoichiometry of interacting proteins is very difficult. Micelles provide more homogeneous and transparent media, but often destabilize membrane proteins because of the lack of native membrane bilayer environment. Nanodiscs emerged to combine the essential advantages of both systems, making stable, soluble and homogeneous samples of membrane proteins and their functional complexes in bilayers with precisely controlled composition. For P450 enzymes this was initially reported in 2002–2004, when methods were developed to incorporate P450 CYP2B4, 21 CYP6B1, 132 CYP3A4, 133 and CYP73A5 134 in Nanodiscs using self‐assembly from a mixture of target protein, MSP and lipids. From these and further studies, 135 , 136 , 137 , 138 it became clear that P450 enzymes in Nanodiscs are fully functional and significantly more stable with respect to conversion to the inactive P420 form.
Substrate binding and the association of small ligands by P450 enzymes in Nanodiscs can be described by single or double exponential kinetics. 135 , 139 , 140 , 141 The same is true for the autoxidation reaction in the presence or absence of substrates, 135 , 138 , 139 Because of homogeneity of P450 monomers in Nanodiscs, these results can be analyzed in terms of molecular properties with no complications arising from limited access to the active site due to oligomerization or to the competitive binding of detergent molecules at the active site. 129 , 130 In addition, incorporation of cytochromes P450 in Nanodiscs opened the door for various nanoscale sensing techniques, such as localized surface plasmon resonance, 142 , 143 nanoplasmonic cup arrays, 144 and microring resonator arrays 145 yielding highly sensitive detection of interactions with substrates and inhibitors.
Stable and homogeneous monomeric preparations of human CYP3A4 in Nanodiscs allowed for detailed analysis of the homotropic and heterotropic cooperativity of this important drug metabolizing enzyme. 136 , 146 , 147 , 148 , 149 , 150 , 151 An important result from these investigations is that all signature features, such as non‐Michaelis steady‐state kinetics and allosteric effects, are observed to a full extent in a CYP3A4 monomer incorporated in the native‐like lipid bilayer. Thus, this approach helped to identify the remote effector site at the P450‐membrane interface, making possible more detailed understanding of allosteric mechanism in CYP3A4 responsible for drug–drug interactions. 147 , 148 , 150 A similar approach was recently used for analysis of heterotropic interactions of multiple substrates with P450 CYP2J2 in Nanodiscs, 152 , 153 suggesting a generalizable means to understand the mechanisms of cooperativity and drug–drug interactions.
To address a long‐standing question of redox potential coupling to substrate binding in the human P450 enzymes, we used CYP3A4 incorporated into POPC lipid bilayer in Nanodiscs. 137 Careful anaerobic spectro‐electrochemical titrations of CYP3A4, saturated with several different substrates, demonstrated a linear correlation between the fraction of high‐spin heme iron induced by the presence of substrate, and the measured mid‐potential value, validating a universal feature of regulation first reported for the soluble bacterial P450cam 154 , 155 and CYP102. 156 Nanodiscs also provided a convenient tool to measure the effect of electrostatic field from the charged bilayer lipids on the redox potential of CPR. 157 An effect of lipid composition on the activity and redox coupling of CYP3A4 and CPR in Nanodiscs was also demonstrated by the Scrutton group, 158 where they found that specific liver microsomal lipids improved functional properties of cytochrome P450. The importance of the lipid composition for the cytochrome P450 activity was also shown for CYP5A1 in Nanodiscs. 159
Nanodisc incorporation of the human steroidogenic P450 enzymes CYP17A1 and CYP19A1 enabled application of cryoradiolytic reduction and the trapping of unstable intermediates for spectroscopic characterization and mechanistic studies. With CYP19A1, the ferrous dioxygen complex was prepared and its autoxidation to the ferric state documented. 138 , 160 Cryoreduction yielded the peroxo states resulting in the visualization by EPR spectroscopy. 161 , 162 With CYP17A1, we used a combination of biochemical and biophysical studies in order to decipher the mechanism of carbon–carbon bond scission in the formation of androgens, which had remained a subject of debate. Multiple studies using CYP17A1 incorporated in Nanodiscs, together with its redox partners CPR and cytochrome b5, demonstrated that the steady‐state kinetics of the C—C cleavage (lyase) reaction catalyzed by CYP17A1 with 17‐hydroxypregnenolone (17OH‐PREG) and 17‐hydroxyprogesterone {17OH‐PROG) is much more consistent with a peroxo‐ferric intermediate as the main catalytically active intermediate. There is very little involvement of the classic high valent Compound I intermediate that is the main pathway in most P450 oxygenation events. 163 Kinetic solvent isotope effects proved to be a robust probe for the proton transfer events that control the population of iron‐bound oxidants. 164 , 165 Cryotrapping and annealing at increasing temperatures monitors the protonation events and led to the identification of a new hemiacetal intermediate, characterized by resonance Raman spectroscopy, in human P450 CYP17A1. 163 , 166 , 167
The first proof‐of‐principle report on Nanodiscs potential for NMR studies of membrane bound cytochromes P450 was published in 2007, 168 where we expressed and purified 13C,15N‐uniformly enriched CYP3A4 and incorporated in Nanodiscs. Solid state NMR (SS‐NMR) monitored the 13C‐13C 2D chemical shift correlation spectra with 0.5 ppm line width, and the secondary structure distribution within seven amino acids was consistent with the substrate‐free X‐ray structure. CYP3A4 remained fully functional after precipitation. SS‐NMR and solution NMR was applied by Ramamoorthy and collaborators to study interactions of CYP2B4 with redox partners CPR and cytochrome b5 in Nanodiscs formed with amphipathic peptides. 123 , 126 , 169 , 170 , 171 , 172 These efforts demonstrated the importance of generating well‐characterized complexes of P450 with redox partners with known stoichiometry for structural studies.
Another advantage of this platform is that membrane proteins in Nanodiscs can be affixed to as surface without denaturation, enabling a variety of sensing modalities. For example, adsorption of Nanodisc on mica surface, 19 , 21 specific binding of chemically modified lipids (i.e., biotinylated) to the high affinity sites on a streptavidin surface, 173 or interaction via a histidine tag or genetically engineered or chemically modified site on the MSP belt, be can be used to keep a membrane protein in its native bilayer environment but within close proximity to an electrode or sensor surface without altering activity. 143 , 174 , 175 These features are useful for applications of surface methods, such as surface plasmon resonance, 175 , 176 localized plasmon approaches 142 , 143 and single molecule studies. 177 , 178 , 179 , 180
The dynamics of membrane proteins incorporated in Nanodiscs have been studied using hydrogen‐deuterium exchange (HDX). This method provides detailed information on the protein mobility, making possible detection of dynamic changes as a result of substrate or effector binding. 181 In the case of the cytochrome P450s, a substantial difference in the dynamics of the highly specific steroidogenic P450 aromatase CYP19A1 156 and the promiscuous drug metabolizing CYP3A4 182 was noted upon substrate binding. The backbone dynamics of substrate‐specific CYP19A1 is significantly more sensitive to the substrate binding, possibly indicating the hidden dynamic contribution to the high specificity, a hypothesis to be explored further.
Many other enzymes have been incorporated into Nanodiscs for functional studies. Examples include the dimeric heme cyclooxygenase (COX) 183 , 184 the flavoprotein human monoamine oxidase A 185 and methane monooxygenase. 186 DHHC acyltransferase (containing four trans‐membrane helices) was incorporated into the larger MSPE3D1‐POPC Nanodiscs for functional studies to show that significant membrane deformation is enabling hydration of catalytic cysteine at the active site inside the membrane. 187
1.11. Nanodiscs as a controlled surface of defined composition
Another major use of Nanodiscs has been as a vehicle to investigate the role of a membrane surface in the assembly and control of multiprotein complexes. Many cellular reactions act at a membrane surface, including systems where integral membrane proteins carry out catalysis, transport and signal transduction. The membrane surface itself can also play a critical role in bringing multiple protein players to a surface to interact with embedded membrane proteins. Specific phospholipids have been implicated in contributing to overall affinity and the resulting detailed topology necessary for function. This utility of Nanodiscs has been recently reviewed, 188 but an overall summary of the work from our laboratory is included here. Important is the ability to provide a surface of defined size and composition. While lipid specificities and interactions with soluble proteins have been measured using liposomes, 189 a challenge is the mobility of the lipids over the large area of the bilayer that makes determination of stoichiometric interactions difficult. Here the precise homogenous size of the Nanodisc membrane surface and ability to control the lipid, fatty acid, cholesterol and so forth composition is an advantage.
One of the first applications was in the quantitation of the lipid specificity for the recruitment of blood coagulation factors and their interaction with the integral membrane protein tissue factor. Using the Nanodisc system it was shown that the local phospholipid environment modulates the activity of blood clotting. 176 , 190 , 191 Furthermore, the utility of Nanodiscs for solid state NMR studies, and tethering to a sensor surface, enabled a structural understanding of how the carboxyglutamate (GLA) resides on the coagulation factors formed specific a complex with anionic lipids. 192 , 193
A major application of Nanodiscs as a controlled surface has been in the area of signaling. For example, integrins play a central role in the dynamics of cell adhesion and cell–cell interaction and communication, including the control of embryonic development, cell migration, and the immune response. 194 Integrins are membrane‐bound, heterodimeric receptors that are involve interaction with many signaling partners that result in both inside‐out and outside‐in communication between the extracellular matrix and the cytoskeleton. 195 , 196 Talin is a key player in the activation of integrins. Nanodiscs enabled the first documentation of inside‐out signaling by talin via cryo‐EM 195 as well as development of FRET measures that allowed documentation of the lipid specificity of talin binding. 197 , 198
More recently, our laboratory has used Nanodiscs to examine the recruitment of oncoproteins, such as KRas4b, with a membrane surface. Here again, the ability to precisely control the lipid composition enabled the determination of protein affinity with membranes of various lipid compositions. Ras proteins are small GTPases playing a key role in the signaling pathways responsible for cell growth and proliferation. 199 These proteins bind guanine nucleotide and are “off” when GDP is bound and “on” when GTP is at the active site. The signal transduction pathway connects receptor tyrosine kinase integral membrane through Ras activation by guanosine exchange proteins. Nucleotide binding induces a conformational switch that permits the binding and activation of downstream kinases that control cellular function. Nanodiscs have been critical in identifying the lipid specificity in the association of the KRas4b isozyme using a variety of biophysical techniques including FRET and single molecule measurements using AFM. 93 , 200 A structural picture of KRas4b interacting with the Nanodisc membrane has been recently realized. 201
There are several other examples where Nanodiscs have been used to investigate protein binding to membrane surfaces, such as the documentation the association of the 69 kD cytotoxic protein BteA with characterization by combination of NMR, SAXS, X‐ray and fluorescent quenching 202 and the binding of antimicrobial peptides as determined by mass spectrometry. 98 , 203 A particularly interesting use of Nanodiscs was to show the membrane involvement in the activation of the cell adhesion receptor CEACAM1 and the stepwise association of the cytosolic domain to calcium‐calmodulin and then actin. 204
1.12. Nanodiscs in high throughput screening
The robustness of Nanodiscs to hard physical handling has allowed them to be used in a variety of high throughput screening applications. Their stability when on surfaces and, as described earlier, the variety of tags that can be added, has enabled the development of robotic high throughput approaches to drug discovery. One example is the use of Nanodiscs to identify the membrane protein components responsible for the association of the peptide aggregates involved in the initiation of neurodegenerative maladies, such as a‐beta oligomers (ABO) and Alzheimer's disease. 205 , 206 In the ABO hypothesis, the causative process is the binding of ABO to a membrane protein in the post‐synaptic junction that would initiate a damaging signaling event. How to identify the culprit? In collaboration with the Klein laboratory at Northwestern, we solubilized brain membranes and generated a “soluble membrane protein library” where each membrane protein is assembled into a separate Nanodisc. 207 ABO were added and a double pull down using a magnetic bead on the Nanodisc and an ABO antibody. 208 Six potential targets were identified. Subsequent work has focused on the a3‐subunit of the Na/K ATPase. 209 This approach provides an important application of Nanodisc technology for screening not only other toxic oligomers, but a variety of other pharmaceutical targets that are membrane proteins. 210
2. SUMMARY
In this article we have attempted to provide a brief description of the many Nanodisc applications. The Nanodisc platform, consisting of engineered MSPs derived from the human Apo‐AI lipoprotein, has clearly provided an enabling platform for a plethora of investigations focused on understanding membranes and membrane protein structure and function. Perhaps more relevant, has been their use in understanding self‐assembly and molecular recognition events that build on the pioneering work of Christian Anfinsen. Nanodisc successes have encouraged further advances through development of other amphipathic molecules, such as peptides and polymers, that also efficiently self‐assemble lipids and membrane proteins into robust soluble nanoscale entities. Future expansion into larger and more complex interacting components will provide a new look at cellular dynamics and function.
AUTHOR CONTRIBUTIONS
Stephen Sligar: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing‐original draft; writing‐review and editing. Ilia Denisov: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing‐original draft; writing‐review and editing.
ACKNOWLEDGMENTS
Our work has been supported uninterrupted for over four decades through the National Institutes of Health General Medical Sciences. Receipt of a MIRA award GM118145 allowed exploring a breadth of scientific inquiry in different systems where Nanodisc technology could lead to major advances in the understanding of membrane protein structure and function. The research described and referenced herein is the product of an incredibly talented group of graduate students, technicians, post‐doctoral associates and senior research scientists. Our current group includes members that have been with the laboratory for more than 20 years. Finally, the highly collaborative and supportive research environment and colleagues at the University of Illinois Chemistry and Biochemistry departments has encouraged expansion into new arenas of scientific inquiry. All are gratefully acknowledged.
Sligar SG, Denisov IG. Nanodiscs: A toolkit for membrane protein science. Protein Science. 2021;30:297–315. 10.1002/pro.3994
Stephen Sligar is the winner of the 2020 Christian B. Anfinsen Award.
Funding information National Institute of General Medical Sciences, Grant/Award Number: R35GM118145
REFERENCES
- 1. Richards FM. Christian B. Anfinsen (1916‐95). Nature. 1995;376:19. [DOI] [PubMed] [Google Scholar]
- 2. Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. [DOI] [PubMed] [Google Scholar]
- 3. Richards FM. Structure of proteins. Annu Rev Biochem. 1963;32:269–300. [DOI] [PubMed] [Google Scholar]
- 4. Brown RK, Boyle E, Anfinsen CB. The enzymatic transformation of lipoproteins. J Biol Chem. 1953;204:423–434. [PubMed] [Google Scholar]
- 5. Siri‐Tarino PW, Krauss RM. The early years of lipoprotein research: From discovery to clinical application. J Lipid Res. 2016;57:1771–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rogers DP, Brouillette CG, Engler JA, et al. Truncation of the amino terminus of human apolipoprotein A‐I substantially alters only the lipid‐free conformation. Biochemistry. 1997;36:288–300. [DOI] [PubMed] [Google Scholar]
- 7. Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: A review of secondary structure and function. J Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 8. Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: A multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–369. [DOI] [PubMed] [Google Scholar]
- 9. Brouillette CG, Jones JL, Ng TC, Kercret H, Chung BH, Segrest JP. Structural studies of apolipoprotein A‐I/phosphatidylcholine recombinants by high‐field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 1984;23:359–367. [DOI] [PubMed] [Google Scholar]
- 10. Jonas A, Kezdy KE, Wald JH. Defined apolipoprotein A‐I conformations in reconstituted high density lipoprotein discs. J Biol Chem. 1989;264:4818–4824. [PubMed] [Google Scholar]
- 11. Jonas A, Wald JH, Toohill KL, Krul ES, Kezdy KE. Apolipoprotein A‐I structure and lipid properties in homogeneous, reconstituted spherical and discoidal high density lipoproteins. J Biol Chem. 1990;265:22123–22129. [PubMed] [Google Scholar]
- 12. Wald JH, Krul ES, Jonas A. Structure of apolipoprotein A‐I in three homogeneous, reconstituted high density lipoprotein particles. J Biol Chem. 1990;265:20037–20043. [PubMed] [Google Scholar]
- 13. Jonas A, von Eckardstein A, Kézdy KE, Steinmetz A, Assmann G. Structural and functional properties of reconstituted high density lipoprotein discs prepared with six apolipoprotein A‐I variants. J Lipid Res. 1991;32:97–106. [PubMed] [Google Scholar]
- 14. Ohnishi S, Hara M, Furuno T, Okada T, Sasabe H. Direct visualization of polypeptide shell of ferritin molecule by atomic force microscopy. Biophys J. 1993;65:573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carlson JW, Jonas A, Sligar SG. Imaging and manipulation of high‐density lipoproteins. Biophys J. 1997;73:1184–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayburt TH, Carlson JW, Sligar SG. Reconstitution and imaging of a membrane protein in a nanometer‐size phospholipid bilayer. J Struct Biol. 1998;123:37–44. [DOI] [PubMed] [Google Scholar]
- 17. Bayburt T, Carlson J, Godfrey B, Shank‐Retzlaff M, Sligar SG. Structure, behavior, and manipulation of nanoscale biological assemblies. Handb Nanostruct Mater Nanotechnol. 2000;5:637–710. [Google Scholar]
- 18. Denisov IG, Sligar SG. Cytochrome P450 enzymes In: Kadish KM, Smith KM, Guilard R, editors. Handbook of porphyrin science. Volume 5, Singapore: World Scientific, 2010; p. 165–201. [Google Scholar]
- 19. Bayburt TH, Carlson JW, Sligar SG. Single molecule height measurements on a membrane protein in Nanometer‐scale phospholipid bilayer disks. Langmuir. 2000;16:5993–5997. [Google Scholar]
- 20. Guengerich FP. Oxidation‐reduction properties of rat liver cytochromes P‐450 and NADPH‐cytochrome p‐450 reductase related to catalysis in reconstituted systems. Biochemistry. 1983;22:2811–2820. [DOI] [PubMed] [Google Scholar]
- 21. Bayburt TH, Sligar SG. Single‐molecule height measurements on microsomal cytochrome P450 in nanometer‐scale phospholipid bilayer disks. Proc Natl Acad Sci USA. 2002;99:6725–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crisafi E, Pandit A. Disentangling protein and lipid interactions that control a molecular switch in photosynthetic light harvesting. Biochim Biophysm Acta Biomembranes. 2017;1859:40–47. [DOI] [PubMed] [Google Scholar]
- 23. Xu L, Ojemyr LN, Bergstrand J, Brzezinski P, Widengren J. Protonation dynamics on lipid nanodiscs: Influence of the membrane surface area and external buffers. Biophys J. 2016;110:1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie TK, Kwon H, Atkins WM. Conformational analysis of human ATP‐binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J Biol Chem. 2011;286:39489–39496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bao H, Dalal K, Cytrynbaum E, Duong F. Sequential action of MalE and maltose allows coupling ATP hydrolysis to translocation in the MalFGK2 transporter. J Biol Chem. 2015;290:25452–25460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self‐assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. [DOI] [PubMed] [Google Scholar]
- 27. Wlodawer A, Segrest JP, Chung BH, Chiovetti R Jr, Weinstein JN. High‐density lipoprotein recombinants: Evidence for a bicycle tire micelle structure obtained by neutron scattering and electron microscopy. FEBS Lett. 1979;104:231–235. [DOI] [PubMed] [Google Scholar]
- 28. Wald JH, Coormaghtigh E, De Meutter J, Ruysschaert JM, Jonas A. Investigation of the lipid domains and apolipoprotein orientation in reconstituted high density lipoproteins by fluorescence and IR methods. J Biol Chem. 1990;265:20044–20050. [PubMed] [Google Scholar]
- 29. Phillips JC, Wriggers W, Li Z, Jonas A, Schulten K. Predicting the structure of apolipoprotein A‐I in reconstituted high‐density lipoprotein disks. Biophys J. 1997;73:2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y, Kijac AZ, Sligar SG, Rienstra CM. Structural analysis of nanoscale self‐assembled discoidal lipid bilayers by solid‐state NMR spectroscopy. Biophys J. 2006;91:3819–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bibow S, Polyhach Y, Eichmann C, et al. Solution structure of discoidal high‐density lipoprotein particles with a shortened apolipoprotein A‐I. Nat Struct Mol Biol. 2017;24:187–193. [DOI] [PubMed] [Google Scholar]
- 32. He Y, Song HD, Anantharamaiah GM, et al. Apolipoprotein A1 forms 5/5 and 5/4 antiparallel dimers in human high‐density lipoprotein. Mol Cell Proteomics. 2019;18:854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pourmousa M, Pastor RW. Molecular dynamics simulations of lipid nanodiscs. Biochim Biophys Acta Biomembranes. 2018;1860:2094–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schachter I, Allolio C, Khelashvili G, Harries D. Confinement in nanodiscs anisotropically modifies lipid bilayer elastic properties. J Phys Chem B. 2020;124:7166–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ritchie TK, Grinkova YV, Bayburt TH, et al. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys J. 2005;88:548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grinkova YV, Denisov IG, Sligar SG. Engineering extended membrane scaffold proteins for self‐assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel. 2010;23:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arkhipova V, Guskov A, Slotboom DJ. Structural ensemble of a glutamate transporter homologue in lipid nanodisc environment. Nat Commun. 2020;11:998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bayburt TH, Grinkova YV, Sligar SG. Self‐assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 40. Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Denisov IG, Sligar SG. Nanodiscs in membrane biochemistry and biophysics. Chem Rev. 2017;117:4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLean MA, Denisov IG, Grinkova YV, Sligar SG. Dark, ultra‐dark and ultra‐bright nanodiscs for membrane protein investigations. Anal Biochem. 2020;607:113860/113861–113860/113867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popot J‐L. Membrane proteins in aqueous solutions. From detergents to amphipols. Cham, Switzerland: Springer International Publishing, 2018. [Google Scholar]
- 44. Nasr ML, Wagner G. Covalently circularized nanodiscs; challenges and applications. Curr Opin Struct Biol. 2018;51:129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puthenveetil R, Nguyen K, Vinogradova O. Nanodiscs and solution NMR: Preparation, application and challenges. Nanotechnol Rev. 2017;6:111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee Y, Warne T, Nehme R, et al. Molecular basis of β‐arrestin coupling to formoterol‐bound β1‐adrenoceptor. Nature. 2020;583:862–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frauenfeld J, Loving R, Armache JP, et al. A saposin‐lipoprotein nanoparticle system for membrane proteins. Nat Methods. 2016;13:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A. Real‐time monitoring of lipid exchange via fusion of peptide based lipid‐nanodiscs. Chem Mater. 2018;30:3204–3207. [Google Scholar]
- 49. Carlson ML, Young JW, Zhao Z, et al. The Peptidisc, a simple method for stabilizing membrane proteins in detergent‐free solution. Elife. 2018;7:e34085/34081–e34085/34023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eichmann C, Bibow S, Riek R. α‐Synuclein lipoprotein nanoparticles. Nanotechnol Rev. 2017;6:105–110. [Google Scholar]
- 51. Lee JW, Kim H. Fragmentation of dimyristoylphosphatidylcholine vesicles by apomyoglobin. Arch Biochem Biophys. 1992;297:354–361. [DOI] [PubMed] [Google Scholar]
- 52. Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 2009;131:7484–7485. [DOI] [PubMed] [Google Scholar]
- 53. Ravula T, Hardin NZ, Ramamoorthy A. Polymer nanodiscs: Advantages and limitations. Chem Phys Lipids. 2019;219:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Di Mauro GM, Hardin NZ, Ramamoorthy A. Lipid‐nanodiscs formed by paramagnetic metal chelated polymer for fast NMR data acquisition. Biochim Biophys Acta Biomembranes. 1862;2020:183332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Danielczak B, Meister A, Keller S. Influence of Mg2+ and Ca2+ on nanodisc formation by diisobutylene/maleic acid (DIBMA) copolymer. Chem Phys Lipids. 2019;221:30–38. [DOI] [PubMed] [Google Scholar]
- 56. van der Vorst EPC. High‐density lipoproteins and apolipoprotein A1. Subcell Biochem. 2020;94:399–420. [DOI] [PubMed] [Google Scholar]
- 57. Jonas A. Reconstitution of high‐density lipoproteins. Methods Enzymol. 1986;128:553–582. [DOI] [PubMed] [Google Scholar]
- 58. Shih AY, Freddolino PL, Sligar SG, Schulten K. Disassembly of nanodiscs with cholate. Nano Lett. 2007;7:1692–1696. [DOI] [PubMed] [Google Scholar]
- 59. Shih AY, Arkhipov A, Freddolino PL, Sligar SG, Schulten K. Assembly of lipids and proteins into lipoprotein particles. J Phys Chem B. 2007;111:11095–11104. [DOI] [PubMed] [Google Scholar]
- 60. Shih AY, Freddolino PL, Arkhipov A, Sligar SG, Schulten K. Molecular modeling of the structural properties and formation of high‐density lipoprotein particles. Curr Top Membr. 2008;60:313–342. [Google Scholar]
- 61. Shih AY, Sligar SG, Schulten K. Maturation of high‐density lipoproteins. J Roy Soc Interface. 2009;6:863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Camp T, Sligar SG. Nanodisc self‐assembly is thermodynamically reversible and controllable. Soft Matter. 2020;16:5615–5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Skar‐Gislinge N, Johansen NT, Hoiberg‐Nielsen R, Arleth L. Comprehensive study of the self‐assembly of phospholipid nanodiscs: What determines their shape and stoichiometry? Langmuir. 2018;34:12569–12582. [DOI] [PubMed] [Google Scholar]
- 64. Wang C, Jacewicz A, Delgado BD, Baradaran R, Long SB. Structures reveal gatekeeping of the mitochondrial Ca(2+) uniporter by MICU1‐MICU2. Elife. 2020;9:e59991/1–e59991/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Y, Han Y, She J, et al. Structural insights into the Ca(2+)‐dependent gating of the human mitochondrial calcium uniporter. Elife. 2020;9:e60513/1–e60513/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang C, Baradaran R, Long SB. Structure and reconstitution of a MCU‐EMRE mitochondrial Ca(2+) uniporter complex. J Mol Biol. 2020;432(60):5632–5648. 10.1016/j.jmb.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. [DOI] [PubMed] [Google Scholar]
- 68. Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. [DOI] [PubMed] [Google Scholar]
- 69. Bayburt TH, Vishnivetskiy SA, McLean MA, et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin‐1 binding. J Biol Chem. 2011;286:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim M, Vishnivetskiy SA, Van Eps N, et al. Conformation of receptor‐bound visual arrestin. Proc Natl Acad Sci USA. 2012;109:18407–18412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li M, Khursigara CM, Subramaniam S, Hazelbauer GL. Chemotaxis kinase CheA is activated by three neighbouring chemoreceptor dimers as effectively as by receptor clusters. Mol Microbiol. 2011;79:677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water‐soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. [DOI] [PubMed] [Google Scholar]
- 75. Li M, Hazelbauer GL. Selective allosteric coupling in core chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2014;111:15940–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Akkaladevi N, Bunyak F, Stalla D, White TA, Hazelbauer GL. Flexible hinges in bacterial chemoreceptors. J Bacteriol. 2018;200:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Roos C, Kai L, Proverbio D, et al. Co‐translational association of cell‐free expressed membrane proteins with supplied lipid bilayers. Mol Membr Biol. 2013;30:75–89. [DOI] [PubMed] [Google Scholar]
- 78. Periasamy A, Shadiac N, Amalraj A, et al. Cell‐free protein synthesis of membrane (1,3)‐β‐D‐glucan (curdlan) synthase: Co‐translational insertion in liposomes and reconstitution in nanodiscs. Biochim Biophys Acta. 2013;1828:743–757. [DOI] [PubMed] [Google Scholar]
- 79. Lyukmanova EN, Shenkarev ZO, Khabibullina NF, et al. Lipid‐protein nanodiscs for cell‐free production of integral membrane proteins in a soluble and folded state: Comparison with detergent micelles, bicelles and liposomes. Biochim Biophys Acta. 2012;1818:349–358. [DOI] [PubMed] [Google Scholar]
- 80. Proverbio D, Roos C, Beyermann M, Orban E, Dotsch V, Bernhard F. Functional properties of cell‐free expressed human endothelin A and endothelin B receptors in artificial membrane environments. Biochim Biophys Acta. 2013;1828:2182–2192. [DOI] [PubMed] [Google Scholar]
- 81. Henrich E, Löhr F, Mezhyrova J, Laguerre A, Bernhard F, Dötsch V. Synthetic biology‐based solution NMR studies on membrane proteins in lipid environments. Methods Enzymol. 2019;614:143–185. [DOI] [PubMed] [Google Scholar]
- 82. Cook I, Asenjo AB, Sosa H, Leyh TS. The human UGT2B7 nanodisc. Drug Metab Dispos. 2020;48:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Grushin K, Miller J, Dalm D, Stoilova‐McPhie S. Factor VIII organisation on nanodiscs with different lipid composition. Thromb Haemost. 2015;113:741–749. [DOI] [PubMed] [Google Scholar]
- 84. Dai A, Ye F, Taylor DW, Hu G, Ginsberg MH, Taylor KA. The structure of a full‐length membrane‐embedded integrin bound to a physiological ligand. J Biol Chem. 2015;290:27168–27175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hernandez‐Rocamora VM, Garcia‐Montanes C, Rivas G, Llorca O. Reconstitution of the Escherichia coli cell division ZipA‐FtsZ complexes in nanodiscs as revealed by electron microscopy. J Struct Biol. 2012;180:531–538. [DOI] [PubMed] [Google Scholar]
- 86. Skar‐Gislinge N, Simonsen JB, Mortensen K, et al. Elliptical structure of phospholipid bilayer nanodiscs encapsulated by scaffold proteins: Casting the roles of the lipids and the protein. J Am Chem Soc. 2010;132:13713–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2003;556:260–264. [DOI] [PubMed] [Google Scholar]
- 88. Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bengtsen T, Holm VL, Kjølbye LR, et al. Structure and dynamics of a nanodisc by integrating NMR, SAXS and SANS experiments with molecular dynamics simulations. Elife. 2020;9:e56518/1–e56518/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Qi Y, Lee J, Klauda JB, Im W. CHARMM‐GUI nanodisc builder for modeling and simulation of various nanodisc systems. J Comput Chem. 2019;40:893–899. [DOI] [PubMed] [Google Scholar]
- 91. Camp T, McLean M, Kato M, Cheruzel L, Sligar S. The hydrodynamic motion of nanodiscs. Chem Phys Lipids. 2019;220:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high‐resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gregory MC, McLean MA, Sligar SG. Interaction of KRas4b with anionic membranes: A special role for PIP2. Biochem Biophys Res Commun. 2017;487:351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Her C, Filoti DI, McLean MA, et al. The charge properties of phospholipid nanodiscs. Biophys J. 2016;11:989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marty MT, Zhang H, Cui W, Blankenship RE, Gross ML, Sligar SG. Native mass spectrometry characterization of intact nanodisc lipoprotein complexes. Anal Chem. 2012;84:8957–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Marty MT, Zhang H, Cui W, Gross ML, Sligar SG. Interpretation and deconvolution of nanodisc native mass spectra. J Am Soc Mass Spectrom. 2014;25:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Keener JE, Zambrano DE, Zhang G, et al. Chemical additives enable native mass spectrometry measurement of membrane protein oligomeric state within intact nanodiscs. J Am Chem Soc. 2019;141:1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Walker LR, Marzluff EM, Townsend JA, Resager WC, Marty MT. Native mass spectrometry of antimicrobial peptides in lipid nanodiscs elucidates complex assembly. Anal Chem. 2019;91:9284–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sahin C, Reid DJ, Marty MT, Landreh M. Scratching the surface: Native mass spectrometry of peripheral membrane protein complexes. Biochem Soc Trans. 2020;48:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Reid DJ, Keener JE, Wheeler AP, et al. Engineering nanodisc scaffold proteins for native mass spectrometry. Analyt Chem. 2017;89:11189–11192. [DOI] [PubMed] [Google Scholar]
- 101. Martens C, Shekhar M, Lau AM, Tajkhorshid E, Politis A. Integrating hydrogen‐deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid‐modulated conformational changes in membrane proteins. Nat Protocol. 2019;14:3183–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Morgan CR, Hebling CM, Rand KD, Stafford DW, Jorgenson JW, Engen JR. Conformational transitions in the membrane scaffold protein of phospholipid bilayer nanodiscs. Mol Cell Proteomics. 2011;10:M111 010876, 010811 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Debnath A, Schafer LV. Structure and dynamics of phospholipid nanodiscs from all‐atom and coarse‐grained simulations. J Phys Chem B. 2015;119:6991–7002. [DOI] [PubMed] [Google Scholar]
- 104. Siuda I, Tieleman DP. Molecular models of nanodiscs. J Chem Theory Comput. 2015;11:4923–4932. [DOI] [PubMed] [Google Scholar]
- 105. Kijac A, Shih AY, Nieuwkoop AJ, Schulten K, Sligar SG, Rienstra CM. Lipid‐protein correlations in nanoscale phospholipid bilayers determined by solid‐state nuclear magnetic resonance. Biochemistry. 2010;49:9190–9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cormode DP, Frias JC, Ma Y, et al. HDL as a contrast agent for medical imaging. Clin Lipidol. 2009;4:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Carney CE, MacRenaris KW, Mastarone DJ, Kasjanski DR, Hung AH, Meade TJ. Cell labeling via membrane‐anchored lipophilic MR contrast agents. Bioconjug Chem. 2014;25:945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Carney CE, Lenov IL, Baker CJ, et al. Nanodiscs as a modular platform for multimodal MR‐optical imaging. Bioconjug Chem. 2015;26:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wong P, Li L, Chea J, et al. Antibody targeted PET imaging of 64Cu‐DOTA‐anti‐CEA PEGylated lipid nanodiscs in CEA positive tumors. Bioconjug Chem. 2020;31(3):743–753. 10.1021/acs.bioconjchem.9b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huda P, Binderup T, Pedersen MC, et al. PET/CT based in vivo evaluation of 64Cu labelled nanodiscs in tumor bearing mice. PLoS One. 2015;10:e0129310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kalienkova V, Alvadia C, Clerico Mosina V, Paulino C. Single‐particle cryo‐EM of membrane proteins in lipid nanodiscs. Methods Mol Biol. 2020;2127:245–273. [DOI] [PubMed] [Google Scholar]
- 112. Coudray N, Isom GL, MacRae MR, Saiduddin MN, Bhabha G, Ekiert DC. Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. To be published. 2020. http://www.rcsb.org/structure/6XBD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Flores JA, Haddad BG, Dolan KA, et al. Connexin‐46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 Å. Nat Commun. 2020;11:4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Roh SH, Shekhar M, Pintilie G, et al. Cryo‐EM and MD infer water‐mediated proton transport and autoinhibition mechanisms of V(o) complex. Sci Adv. 2020;6(41):eabb9605/1–eabb9605/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Staus DP, Hu H, Robertson MJ, et al. Structure of the M2 muscarinic receptor‐β‐arrestin complex in a lipid nanodisc. Nature. 2020;579:297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nelson DR. Cytochrome P450 diversity in the tree of life. Biochim Biophys Acta Proteins Proteom. 2018;1866:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kern DM, Sorum B, Hoel CM, et al. Cryo‐EM structure of the SARS‐CoV‐2 3a ion channel in lipid nanodiscs. bioRxiv. 2020;2020.06.17.156554:1–32. [Google Scholar]
- 118. Pleiner T, Tomaleri GP, Januszyk K, Inglis AJ, Hazu M, Voorhees RM. Structural basis for membrane insertion by the human ER membrane protein complex. Science. 2020;369:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bibow S. Opportunities and challenges of backbone, sidechain, and RDC experiments to study membrane protein dynamics in a detergent‐free lipid environment using solution state NMR. Front Mol Biosci. 2019;6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kloepfer K, Hagn F. Beyond detergent micelles: The advantages and applications of non‐micellar and lipid‐based membrane mimetics for solution‐state NMR. Progr NMR Spectrosc. 2019;114‐115:271–283. [DOI] [PubMed] [Google Scholar]
- 121. Bibow S, Bohm R, Modaresi SM, Hiller S. Detergent titration as an efficient method for NMR resonance assignments of membrane proteins in lipid‐bilayer nanodiscs. Analyt Chem. 2020;92:7786–7793. [DOI] [PubMed] [Google Scholar]
- 122. Schuster M, Deluigi M, Pantic M, et al. Optimizing the α1B‐adrenergic receptor for solution NMR studies. Biochim Biophys Acta Biomembranes. 1862;2020:183354. [DOI] [PubMed] [Google Scholar]
- 123. Ahuja S, Jahr N, Im SC, et al. A model of the membrane‐bound cytochrome b5‐cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem. 2013;288:22080–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang M, Huang R, Im SC, Waskell L, Ramamoorthy A. Effects of membrane mimetics on cytochrome P450‐cytochrome b5 interactions characterized by NMR spectroscopy. J Biol Chem. 2015;290:12705–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhang M, Le Clair SV, Huang R, et al. Insights into the role of substrates on the interaction between cytochrome b5 and cytochrome P450 2B4 by NMR. Sci Rep. 2015;5:8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhang M, Huang R, Ackermann R, et al. Reconstitution of the Cytb5‐CytP450 complex in nanodiscs for structural studies using NMR spectroscopy. Ang Chemie Int Ed. 2016;55:4497–4499. [DOI] [PubMed] [Google Scholar]
- 127. Bai J, Wang J, Ravula T, et al. Expression, purification, and functional reconstitution of 19F‐labeled cytochrome b5 in peptide nanodiscs for NMR studies. Biochim Biophys Acta Biomembranes. 2020;1862:183194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Frueh V, Zhou Y, Chen D, et al. Application of fragment‐based drug discovery to membrane proteins: Identification of ligands of the integral membrane enzyme DsbB. Chem Biol. 2010;17:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sevrioukova IF, Poulos TL. Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand. J Biol Chem. 2012;287:3510–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Kinetics of dithionite‐dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ebel RE, O'Keeffe DH, Peterson JA. Substrate binding to hepatic microsomal cytochrome P‐450. Influence of the microsomal membrane. J Biol Chem. 1978;253:3888–3897. [PubMed] [Google Scholar]
- 132. Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35:556–558. 560, 562‐563. [DOI] [PubMed] [Google Scholar]
- 133. Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. [DOI] [PubMed] [Google Scholar]
- 134. Duan H, Civjan NR, Sligar SG, Schuler MA. Co‐incorporation of heterologously expressed Arabidopsis cytochrome P450 and P450 reductase into soluble nanoscale lipid bilayers. Arch Biochem Biophys. 2004;424:141–153. [DOI] [PubMed] [Google Scholar]
- 135. Denisov IG, Grinkova YV, Baas BJ, Sligar SG. The ferrous‐dioxygen intermediate in human cytochrome P450 3A4—Substrate dependence of formation and decay kinetics. J Biol Chem. 2006;281:23313–23318. [DOI] [PubMed] [Google Scholar]
- 136. Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4—Linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. [DOI] [PubMed] [Google Scholar]
- 137. Das A, Grinkova YV, Sligar SG. Redox potential control by drug binding to cytochrome P 450 3A4. J Am Chem Soc. 2007;129:13778–13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Grinkova YV, Denisov IG, Waterman MR, Arase M, Kagawa N, Sligar SG. The ferrous‐oxy complex of human aromatase. Biochem Biophys Res Commun. 2008;372:379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Denisov IG, Grinkova YV, McLean MA, Sligar SG. The one‐electron autoxidation of human cytochrome P450 3A4. J Biol Chem. 2007;282:26865–26873. [DOI] [PubMed] [Google Scholar]
- 140. McClary WD, Sumida JP, Scian M, Paco L, Atkins WM. Membrane fluidity modulates thermal stability and ligand binding of cytochrome P4503A4 in lipid nanodiscs. Biochemistry. 2016;55:6258–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zarate‐Perez F, Hackett JC. Conformational selection is present in ligand binding to cytochrome P450 19A1 lipoprotein nanodiscs. J Inorg Biochem. 2020;209:111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhao J, Das A, Schatz GC, Sligar SG, Van Duyne RP. Resonance localized surface plasmon spectroscopy: Sensing substrate and inhibitor binding to cytochrome P450. J Phys Chem C. 2008;112:13084–13088. [Google Scholar]
- 143. Das A, Zhao J, Schatz GC, Sligar SG, Van Duyne RP. Screening of type I and II drug binding to human cytochrome P450‐3A4 in nanodiscs by localized surface plasmon resonance spectroscopy. Anal Chem. 2009;81:3754–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Plucinski L, Ranjan Gartia M, Arnold WR, et al. Substrate binding to cytochrome P450‐2J2 in nanodiscs detected by nanoplasmonic Lycurgus cup arrays. Biosens Bioelectron. 2016;75:337–346. [DOI] [PubMed] [Google Scholar]
- 145. Sloan CD, Marty MT, Sligar SG, Bailey RC. Interfacing lipid bilayer nanodiscs and silicon photonic sensor arrays for multiplexed protein‐lipid and protein‐membrane protein interaction screening. Anal Chem. 2013;85:2970–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Frank DJ, Denisov IG, Sligar SG. Analysis of heterotropic cooperativity in cytochrome P450 3A4 using α‐naphthoflavone and testosterone. J Biol Chem. 2011;286:5540–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Denisov IG, Grinkova YV, Baylon JL, Tajkhorshid E, Sligar SG. Mechanism of drug‐drug interactions mediated by human cytochrome P450 CYP3A4 monomer. Biochemistry. 2015;54:2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Denisov IG, Mak PJ, Grinkova YV, et al. The use of isomeric testosterone dimers to explore allosteric effects in substrate binding to cytochrome P450 CYP3A4. J Inorg Biochem. 2016;158:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Denisov IG, Baylon JL, Grinkova YV, Tajkhorshid E, Sligar SG. Drug‐drug interactions between atorvastatin and dronedarone mediated by monomeric CYP3A4. Biochemistry. 2018;57:805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Denisov IG, Grinkova YV, Nandigrami P, Shekhar M, Tajkhorshid E, Sligar SG. Allosteric interactions in human cytochrome P450 CYP3A4: The role of phenylalanine 213. Biochemistry. 2019;58:1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Mak PJ, Denisov IG, Grinkova YV, Sligar SG, Kincaid JR. Defining CYP3A4 structural responses to substrate binding. Raman spectroscopic studies of a nanodisc‐incorporated mammalian cytochrome P450. J Am Chem Soc. 2011;133:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Das A, Weigle AT, Arnold WR, Kim JS, Carnevale LN, Huff HC. CYP2J2 molecular recognition: A new axis for therapeutic design. Pharmacol Ther. 2020;215:107601/1–107601/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Arnold WR, Baylon JL, Tajkhorshid E, Das A. Asymmetric binding and metabolism of polyunsaturated fatty acids (PUFAs) by CYP2J2 epoxygenase. Biochemistry. 2016;55:6969–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sligar SG. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. [DOI] [PubMed] [Google Scholar]
- 155. Fisher MT, Scarlata SF, Sligar SG. High‐pressure investigations of cytochrome P‐450 spin and substrate binding equilibria. Arch Biochem Biophys. 1985;240:456–463. [DOI] [PubMed] [Google Scholar]
- 156. Ost TWB, Clark J, Mowat CG, et al. Oxygen activation and electron transfer in flavocytochrome P 450 BM3. J Am Chem Soc. 2003;125:15010–15020. [DOI] [PubMed] [Google Scholar]
- 157. Das A, Sligar SG. Modulation of the cytochrome P450 reductase redox potential by the phospholipid bilayer. Biochemistry. 2009;48:12104–12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Liu K‐C, Hughes JMX, Hay S, Scrutton NS. Liver microsomal lipid enhances the activity and redox coupling of colocalized cytochrome P 450 reductase‐cytochrome P 450 3A4 in nanodiscs. FEBS J. 2017;284:2302–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Das A, Varma SS, Mularczyk C, Meling DD. Functional investigations of thromboxane synthase (CYP5A1) in lipid bilayers of nanodiscs. Chembiochem. 2014;15:892–899. [DOI] [PubMed] [Google Scholar]
- 160. Luthra A, Denisov IG, Sligar SG. Temperature derivative spectroscopy to monitor the autoxidation decay of cytochromes P450. Anal Chem. 2011;83:5394–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gantt SL, Denisov IG, Grinkova YV, Sligar SG. The critical iron‐oxygen intermediate in human aromatase. Biochem Biophys Res Commun. 2009;387:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Davydov R, Hoffman BM. Active intermediates in heme monooxygenase reactions as revealed by cryoreduction/annealing, EPR/ENDOR studies. Arch Biochem Biophys. 2011;507:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Mak PJ, Duggal R, Denisov IG, Gregory MC, Sligar SG, Kincaid JR. Human cytochrome CYP17A1: The structural basis for compromised Lyase activity with 17‐hydroxyprogesterone. J Am Chem Soc. 2018;140:7324–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Gregory MC, Denisov IG, Grinkova YV, Khatri Y, Sligar SG. Kinetic solvent isotope effect in human P450 CYP17A1‐mediated androgen formation: Evidence for a reactive peroxoanion intermediate. J Am Chem Soc. 2013;135:16245–16247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Khatri Y, Gregory MC, Grinkova YV, Denisov IG, Sligar SG. Active site proton delivery and the lyase activity of human CYP17A1. Biochem Biophys Res Commun. 2014;443:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Mak PJ, Gregory MC, Denisov IG, Sligar SG, Kincaid JR. Unveiling the crucial intermediates in androgen production. Proc Natl Acad Sci U S A. 2015;112:15856–15861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Liu Y, Denisov I, Grinkova Y, Sligar S, Kincaid JR. P450 CYP17A1 variant with a disordered proton shuttle assembly retains peroxo‐mediated lyase efficiency. Chemistry. 2020. 10.1002/chem.202003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Kijac AZ, Li Y, Sligar SG, Rienstra CM. Magic‐angle spinning solid‐state NMR spectroscopy of nanodisc‐embedded human CYP3A4. Biochemistry. 2007;46:13696–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pandey MK, Vivekanandan S, Ahuja S, et al. Cytochrome‐P450‐cytochrome‐b5 interaction in a membrane environment changes 15N chemical shift anisotropy tensors. J Phys Chem B. 2013;117:13851–13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Huang R, Yamamoto K, Zhang M, et al. Probing the transmembrane structure and dynamics of microsomal NADPH‐cytochrome P450 oxidoreductase by solid‐state NMR. Biophys J. 2014;106:2126–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Mahajan M, Ravula T, Prade E, Anantharamaiah GM, Ramamoorthy A. Probing membrane enhanced protein‐protein interactions in a minimal redox complex of cytochrome‐P450 and P450‐reductase. Chem Comm. 2019;55:5777–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Gentry KA, Anantharamaiah GM, Ramamoorthy A. Probing protein‐protein and protein‐substrate interactions in the dynamic membrane‐associated ternary complex of cytochromes P450, b(5), and reductase. Chem Commun. 2019;55:13422–13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Goluch ED, Shaw AW, Sligar SG, Liu C. Microfluidic patterning of nanodisc lipid bilayers and multiplexed analysis of protein interaction. Lab Chip. 2008;8:1723–1728. [DOI] [PubMed] [Google Scholar]
- 174. Benjamin CJ, Wright KJ, Hyun S‐H, et al. Nonfouling NTA‐PEG‐based TEM grid coatings for selective capture of histidine‐tagged protein targets from cell lysates. Langmuir. 2016;32:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Trahey M, Li MJ, Kwon H, Woodahl EL, McClary WD, Atkins WM. Applications of lipid nanodiscs for the study of membrane proteins by surface plasmon resonance. Curr Protoc Protein Sci. 2015;81:29.13.21–29.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. [DOI] [PubMed] [Google Scholar]
- 177. Nath A, Koo PK, Rhoades E, Atkins WM. Allosteric effects on substrate dissociation from cytochrome P450 3A4 in nanodiscs observed by ensemble and single‐molecule fluorescence spectroscopy. J Am Chem Soc. 2008;130:15746–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Nath A, Trexler AJ, Koo P, Miranker AD, Atkins WM, Rhoades E. Single‐molecule fluorescence spectroscopy using phospholipid bilayer nanodiscs. Methods Enzymol. 2010;472:89–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Laursen T, Singha A, Rantzau N, et al. Single molecule activity measurements of cytochrome P450 oxidoreductase reveal the existence of two discrete functional states. ACS Chem Biol. 2014;9:630–634. [DOI] [PubMed] [Google Scholar]
- 180. Bavishi K, Li D, Eiersholt S, et al. Direct observation of multiple conformational states in cytochrome P450 oxidoreductase and their modulation by membrane environment and ionic strength. Sci Rep. 2018;8:6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Redhair M, Clouser AF, Atkins WM. Hydrogen‐deuterium exchange mass spectrometry of membrane proteins in lipid nanodiscs. Chem Phys Lipids. 2019;220:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Treuheit NA, Redhair M, Kwon H, et al. Membrane interactions, ligand‐dependent dynamics, and stability of cytochrome P4503A4 in lipid nanodiscs. Biochemistry. 2016;55:1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Roy J, Adili R, Kulmacz R, Holinstat M, Das A. Development of poly unsaturated fatty acid derivatives of aspirin for inhibition of platelet function. J Pharmacol Exp Ther. 2016;359:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Orlando BJ, McDougle DR, Lucido MJ, et al. Cyclooxygenase‐2 catalysis and inhibition in lipid bilayer nanodiscs. Arch BiochemBiophys. 2014;546:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Cruz F, Edmondson DE. Kinetic properties of recombinant MAO‐A on incorporation into phospholipid nanodisks. J Neural Transm. 2007;114:699–702. [DOI] [PubMed] [Google Scholar]
- 186. Ro SY, Schachner LF, Koo CW, et al. Native top‐down mass spectrometry provides insights into the copper centers of membrane‐bound methane monooxygenase. Nat Commun. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Stix R, Song J, Banerjee A, Faraldo‐Gómez JD. DHHC20 Palmitoyl‐transferase reshapes the membrane to Foster catalysis. Biophys J. 2020;118:980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. McLean MA, Gregory MC, Sligar SG. Nanodiscs: A controlled bilayer surface for the study of membrane proteins. Annu Rev Biophys. 2018;47:107–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Haverstick DM, Glaser M. Influence of proteins on the reorganization of phospholipid bilayers into large domains. Biophys J. 1989;55:677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Morrissey JH, Pureza V, Davis‐Harrison RL, Sligar SG, Ohkubo YZ, Tajkhorshid E. Blood clotting reactions on nanoscale phospholipid bilayers. Thromb Res. 2008;122:S23–S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Medfisch SM, Muehl EM, Morrissey JH, Bailey RC. Phosphatidylethanolamine‐phosphatidylserine binding synergy of seven coagulation factors revealed using nanodisc arrays on silicon photonic sensors. Sci Rep. 2020;10:17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Morrissey JH, Pureza V, Davis‐Harrison RL, et al. Protein‐membrane interactions: Blood clotting on nanoscale bilayers. J Thromb Haemostasis. 2009;7:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Morrissey JH, Tajkhorshid E, Sligar SG, Rienstra CM. Tissue factor/factor VIIa complex: Role of the membrane surface. Thromb Res. 2012;129(Suppl 2):S8–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. [DOI] [PubMed] [Google Scholar]
- 195. Ye F, Hu GQ, Taylor D, et al. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Anderson LR, Owens TW, Naylor MJ. Structural and mechanical functions of integrins. Biophys Rev. 2014;6:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Ye X, McLean MA, Sligar SG. Conformational equilibrium of talin is regulated by anionic lipids. Biochim Biophys Acta. 2016;1858:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ye X, McLean MA, Sligar SG. PIP2 modulates the affinity of talin‐1 for phospholipid bilayers and activates its auto‐inhibited form. Biochemistry. 2016;55:5038–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. [DOI] [PubMed] [Google Scholar]
- 200. McLean MA, Stephen AG, Sligar SG. PIP2 influences the conformational dynamics of membrane‐bound KRAS4b. Biochemistry. 2019;58:3537–3545. [DOI] [PubMed] [Google Scholar]
- 201. Lee K‐Y, Fang Z, Enomoto M, et al. Two distinct structures of membrane‐associated homodimers of GTP‐ and GDP‐bound KRAS4B revealed by paramagnetic relaxation enhancement. Ang Chemie Int Ed. 2020;59:11037–11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Yahalom A, Davidov G, Kolusheva S, et al. Structure and membrane‐targeting of a Bordetella pertussis effector N‐terminal domain. Biochim Biophys Acta Biomembranes. 2019;1861:183054. [DOI] [PubMed] [Google Scholar]
- 203. Walker LR, Marty MT. Revealing the specificity of a range of antimicrobial peptides in lipid nanodiscs by native mass spectrometry. Biochemistry. 2020;59:2135–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ghazarian H, Hu W, Mao A, et al. NMR analysis of free and lipid nanodisc anchored CEACAM1 membrane proximal peptides with Ca2+/CaM. Biochim Biophys Acta, Biomembranes. 2019;1861:787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Velasco PT, Heffern MC, Sebollela A, et al. Synapse‐binding subpopulations of Aβ oligomers sensitive to peptide assembly blockers and scFv antibodies. ACS Chem Nerosci. 2012;3:972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Klein WL. Synaptotoxic amyloid‐β oligomers: A molecular basis for the cause, diagnosis, and treatment of Alzheimer's disease? J Alzheimers Dis. 2013;33(Suppl 1):S49–S65. [DOI] [PubMed] [Google Scholar]
- 207. Marty MT, Wilcox KC, Klein WL, Sligar SG. Nanodisc‐solubilized membrane protein library reflects the membrane proteome. Anal Bioanal Chem. 2013;405:4009–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Wilcox KC, Marunde MR, Das A, et al. Nanoscale synaptic membrane mimetic allows unbiased high throughput screen that targets binding sites for Alzheimer's‐associated abeta oligomers. PLoS One. 2015;10:e0125263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. DiChiara T, DiNunno N, Clark J, et al. Alzheimer's toxic amyloid beta oligomers: Unwelcome visitors to the Na/K ATPase alpha3 docking station. Yale J Biol Med. 2017;90:45–61. [PMC free article] [PubMed] [Google Scholar]
- 210. Lo Bu R, Clark J, DiChiara T, Sligar SG, Klein WL. SMPL synaptic membranes: Nanodisc‐mediated synaptic membrane mimetics expand the toolkit for drug discovery and the molecular cell biology of synapses. Neuromethods. 2018;141:227–250. [Google Scholar]


