Dear Editor,
The glucagon-like peptide-1 receptor (GLP-1R) belongs to class B G protein-coupled receptors (GPCRs). It is a clinically proven target for type 2 diabetes and obesity.1 Several peptidic drugs have been approved including one in pill form, the oral semaglutide. Despite these advances, oral non-peptidic medicines have been pursued across the pharmaceutical industry for improved patient compliance with reduced side effects such as nausea and vomiting.1 The discovery of non-peptidic drugs has been hampered for many years, primarily owing to the difficulty of mimicking the peptide–receptor interactions with small molecule non-peptidic agonists.
GLP-1R contains an extracellular N-terminal domain (NTD) and a seven-transmembrane helix bundle, with the endogenous peptide agonist GLP-1 (7–37) binding to both domains. A generally accepted two-step model of activation postulates that interactions between C-terminus of GLP-1 and the receptor NTD facilitate the peptide N-terminus binding deeply into the receptor transmembrane domain (TMD).2 It has been observed that partial agonist activity retained by short peptide fragments, GLP (9–36), GLP (15–36) and Ex4 (3–39), suggesting that the N-terminal regions are required for full activation of the receptor to trigger G protein signaling.2 However, it was also reported that the five N-terminal residues of peptide agonists are dispensable for both G protein and arrestin signaling when they are fused to the N-terminus of GLP-1R.3
To date, two inactive structures of GLP-1R and three complexes with peptide agonists have been reported for structure-based drug discovery.4–7 In addition, the structure of GLP-1R with a partial agonist of the non-peptidic small molecule has also been reported,8 showing that it binds at distinct location from the peptide ligand, which leads to conformational differences at the orthosteric binding pocket. The basis for GLP-1R activation by the full agonists of small molecules remained unknown. Here we determined the cryo-EM structure of GLP-1R–Gs complex bound with a non-peptidic full agonist, revealing a new binding mode of GLP-1R. Structural comparison with reported GLP-1R structures provides an insight into the mechanism of GLP-1R activation by a small molecule full agonist and a framework for small molecule drug discovery.
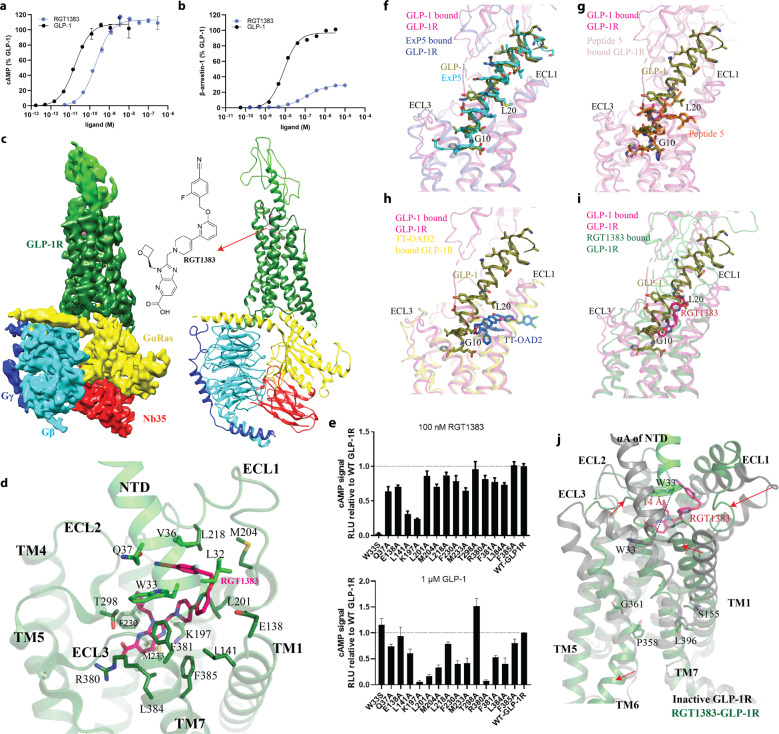
RGT1383 is a full agonist in G protein-mediated cAMP signaling with an EC50 value of 0.2 ± 0.04 nM and a partial agonist in β-arrestin-mediated signaling with maximal arrestin recruitment at ~30% (cf. 100% for GLP-1 &7–37) (Fig. 1a, b). We prepared RGT1383-bound GLP-1R–Gs complex for cryo-EM studies as described in Supplementary information, Data S1. 3D classification led to the cryo-EM reconstruction of the complex at a nominal resolution of 4.2 Å (Supplementary information, Fig. S1 and Table S1). The cryo-EM map allows the model building for most of the regions of the GLP-1R–Gs complex except the flexible α-helical domain (AHD) of Gα (Fig. 1c; Supplementary information, Fig. S2). The initial model of the complex was built by using the cryo-EM structure of a peptide-bound GLP-1R–Gs (PDB: 6B3J) and refined against the cryo-EM map.
Fig. 1. Structure of RGT1383–GLP-1R–Gs complex.
a, b Comparison of the effects between RGT1383 and GLP-1 on cAMP accumulation and β-arrestin recruitment. c Cryo-EM map and structure model of the complex. d The binding mode of RGT1383 in binding pocket of GLP-1R. e The mutagenesis analysis of residues in RGT1383-binding pocket of GLP-1R. Upper, 100 nM RGT1383; lower, 1 μM GLP-1. f–i Agonists occupy different positions in the orthosteric binding pocket. GLP-1 (deep olive), ExP5 (cyan), Peptide 5 (orange), TT-OAD2 (deep blue) and RGT1383 (hot pink) bound active GLP-1R structures are shown in light magenta, light blue, pink, yellow and deep green, respectively. j Structural comparison of RGT1383-bound GLP-1R with inactive GLP-1R (PDB code: 7C2E and 6LN2). Arrows show the extent of motion.
The small molecule agonist RGT1383 can be fitted into the cryo-EM map within the orthosteric pocket in the TMD (Supplementary information, Fig. S2). The structure model was built by computational docking and iterative manual building and refinement. RGT1383 is anchored in a tightly packed orthosteric binding pocket through interactions with E138 and L141 in TM1, K197 and L201 in TM2, M204 and L218 in ECL1, F230 and M233 in TM3, T298 in ECL2, R380, F381, L384 and F385 in TM7, as well as the L32, W33, V36 and Q37 in the NTD (Fig. 1d). These interactions between RGT1383 and the receptor were confirmed through mutagenesis studies using cAMP luciferase reporter assays (Fig. 1e; Supplementary information, Fig. S3). Most of these mutations maintain an expression level comparable to the wild-type GLP-1R9 but decrease the activation potency of RGT1383 on GLP-1R (Supplementary information, Fig. S3), in agreement with their direct interaction with the bound ligand. Notably, the cryo-EM map and mutagenesis studies suggest that W33 in NTD plays a crucial role in binding to RGT1383 (Fig. 1e), which was also observed in binding to non-peptide antagonist T-0632, and agonist peptide 5.5,10 The difference of W33 in humans and cognate residue S33 in rat GLP-1R determines the species specificity of small molecule ligands as RGT1383 only activates the human GLP-1R whereas GLP-1 can activate GLP-1R from both human and rat (Fig. 1e). The large interface between NTD and extracellular loops is also consistent with a previous mutational study that NTD plays an important role in the activation of GLP-1R.11 Compared to TT-OAD2-bound GLP-1R, a notable feature is the more extensive interactions between RGT1383 and residues in TM7, which induce inward displacements of the ECL3 and the extracellular ends of TM6–7 (Supplementary information, Fig. S4). This indicates that the previous observed outward movement of ECL3 in all other activated GLP-1R structures is probably not essential for the GLP-1R activation.
Compared to inactive GLP-1R, a remarkable difference among structures of the GLP-1-, ExP5-, Peptide 5-, TT-OAD2- and RGT1383-bound GLP-1R is the orientation of αA in NTD relative to receptor core (Supplementary information, Figs. S4, S5). Except the invisible NTD in the structure of the TT-OAD2–GLP-1R–Gs complex because of its high mobility,8 the αA of NTD in the GLP-1-, ExP5-, Peptide 5-, and RGT1383-bound receptors moves toward the ECL2 for 26, 26, 15 and 14 Å (measured from the Cα atoms of W33 or S33), respectively (Supplementary information, Figs. S4 and S5). In addition, these agonists occupy different positions in the orthosteric binding pocket and thereby differ in the types of interactions formed with the GLP-1R (Fig. 1f–i). The C-termini of peptide full-agonists, GLP-1 or ExP5, were clasped by the NTD with their N-termini inserted deeply into the transmembrane core of the receptor (Fig. 1f), while the most part of Peptide 5 overlapped with residues 7–20 of GLP-1(Fig. 1g). The non-peptide agonist TT-OAD2, a partial agonist for GLP-1R, with subtle overlap with other GLP-1R agonists, bound at a high-up position and embedded partly into the detergent micelle between extracellular TM2 and TM3 (Fig. 1h). Of interest, the full-agonist RGT1383 is almost completely overlapped with the position where residues 10–20 of GLP-1 occupied (Fig. 1i). The position of RGT1383 is much closer toward TM6 than the partial agonist TT-OAD2 (Fig. 1d, j; Supplementary information, Fig. S4), and this mode of RGT1383 binding induces the unwinding of this segment of TM6, a hallmark of class B GPCR activation. Therefore, the multiple active conformations of GLP-1R, induced by different agonist-binding modes and showed as diverse displacements of NTD, extracellular loops and TM segments, can transduce a common change at the extracellular portion of TM6, which subsequently induces the conformational changes in the intracellular face for Gs binding.
Structural comparison of the inactive full-length and NTD-truncated GLP-1R structures (PDB: 6LN2 and 5VEW) and molecular dynamics simulations7 reveal that NTD is dynamic but prefer an inactive conformation stabilized by the weak interactions between NTD and ECL1/3, and that the αA of NTD keeps extracellular TM1 and TM7 apart from each other. Compared to the two inactive structures, inward movement of TM1 was observed in all agonist-bound GLP-1R structures (Supplementary information, Fig. S4). In addition, it was reported that the simultaneous movement of TM1 and TM7 is mediated by a conserved polar interaction between the highly conserved S155 and the backbone of L396 in GLP-1R. Taken together, we propose a model shown as cartoon (Supplementary information, Fig. S6) that agonist-bound GLP-1R facilitates the movement of NTD toward ECL2, followed by inward movement of extracellular TM1 and rearrangement of TM7 which consequently lead to rearrangement of ECL3, downward rotation of extracellular half of TM6 (measured from P358) and subsequent outward movement of intracellular part of TM6 for the accommodation of Gs protein (Fig. 1j; Supplementary information, Fig. S5 and Videos. S1, S2).
In summary, we have determined the structure of the RGT1383, a small molecule full-agonist, bound GLP-1R in complex with Gs protein. Compared to previous reported agonist-bound GLP-1R structures, the most striking feature is that non-peptidic RGT1383 binds to a tightly packed orthosteric binding pocket owning to inward movement of ECL3 and the extracellular end of TM7. In addition, the W33 in NTD plays a critical role in the binding of RGT1383 to the receptor, providing insights into the activation mechanism of GLP-1R modulated by NTD and the species specificity of this series of small molecule agonists. We believe this structure provides a structural framework for designing orally bioavailable small molecule drugs to treat type 2 diabetes and obesity.
Accession codes
The cryo-EM density map and corresponding coordinate have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 7C2E, and EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-30274.
Supplementary information
Acknowledgements
The cryo-EM data were collected at Cryo-Electron Microscopy Research Center, Shanghai Institute of Material Medica. This work was partially supported by the Ministry of Science and Technology (China) grants (2018YFA0507002 to H.E.X.); Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to H.E.X.); CAS Strategic Priority Research Program (XDB08020303 to H.E.X.); Shanghai Science and Technology Committee (19ZR1467500 to H.M.); the Young Innovator Association of CAS (H.M.); SA-SIBS Scholarship Program (L.Z.), the Young Innovator Association of CAS (2018325 to L.Z.); the National Natural Science Foundation of China (31770796 to Y.J.); the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (2018ZX09711002 to Y.J.); the K.C. Wong Education Foundation (Y.J.).
Author contributions
H.M. carried out construct preparation, protein expression and purification and prepared the cryo-EM sample. H.M. and D.Y. collected the data. D.Y. processed cryo-EM data, modeled, refined and analyzed the structure, wrote manuscript with input from all authors. W.H. performed molecular docking. L.Z. designed the expression constructs for the compound–GLP-1R–Gs complex. X.W. purified the Nb35 nanobody and performed specimen screening by negative-stain EM. Y.J. assisted in construct preparation, protein expression, and purification. F.L. oversaw cell experiments. W.G. and X.S. assisted with model building. H.E.X. and W.Z. conceived of the project, W.Z. designed RGT1383, and H.E.X. supervised the overall structural studies and participated in manuscript writing.
Competing interests
The authors declare no competing interests.
Contributor Information
Daopeng Yuan, Email: Daopeng.yuan@qlregor.com.
H. Eric Xu, Email: Eric.xu@simm.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0384-8.
References
- 1.Htike ZZ, et al. Diabetes Obes. Metab. 2017;19:524–536. doi: 10.1111/dom.12849. [DOI] [PubMed] [Google Scholar]
- 2.Donnelly D. Br. J. Pharmacol. 2012;166:27–41. doi: 10.1111/j.1476-5381.2011.01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin Y, et al. Cell Discov. 2016;2:16042. doi: 10.1038/celldisc.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jazayeri A, et al. Nature. 2017;546:254–258. doi: 10.1038/nature22800. [DOI] [PubMed] [Google Scholar]
- 6.Liang YL, et al. Nature. 2018;555:121–125. doi: 10.1038/nature25773. [DOI] [PubMed] [Google Scholar]
- 7.Wu F, et al. Nat. Commun. 2020;11:1272. doi: 10.1038/s41467-020-14934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao P, et al. Nature. 2020;577:432–436. doi: 10.1038/s41586-019-1902-z. [DOI] [PubMed] [Google Scholar]
- 9.Graaf C, et al. Pharmacol. Rev. 2016;68:954–1013. doi: 10.1124/pr.115.011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibaduiza EC, Chen C, Beinborn M. J. Biol. Chem. 2001;276:37787–37793. doi: 10.1074/jbc.M106692200. [DOI] [PubMed] [Google Scholar]
- 11.Zhao LH, et al. J. Biol. Chem. 2016;291:15119–15130. doi: 10.1074/jbc.M116.726620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.