Abstract
Gut microbiota are essential for host health and survival, but we are still far from understanding the processes involved in shaping their composition and evolution. Controlled experimental work under lab conditions as well as human studies pointed at environmental factors (i.e., diet) as the main determinant of the microbiota with little evidence of genetic effects, while comparative interspecific studies detected significant phylogenetic effects. Different species, however, also differ in diet, feeding behavior, and environmental characteristics of habitats, all of which also vary interspecifically, and, therefore, can potentially explain most of the detected phylogenetic patterns. Here, we take advantage of the reproductive strategy of avian brood parasites and investigate gut microbiotas (esophageal (food and saliva) and intestinal) of great spotted cuckoo (Clamator glandarius) and magpie (Pica pica) nestlings that grow in the same nests. We also estimated diet received by each nestling and explored its association with gut microbiota characteristics. Although esophageal microbiota of magpies and great spotted cuckoos raised within the same environment (nest) did not vary, the microbiota of cloacal samples showed clear interspecific differences. Moreover, diet of great spotted cuckoo and magpie nestlings explained the microbiota composition of esophageal samples, but not of cloaca samples. These results strongly suggest a genetic component determining the intestinal microbiota of host and parasitic bird species, indicating that interspecific differences in gut morphology and physiology are responsible for such interspecific differences.
Subject terms: Microbial ecology, Community ecology
Introduction
The study of microbiotas associated with different environments, including animal hosts, has become one of the most fruitful areas of biological research during the last decade [1–3]. Reasons include that microorganisms should have profound impact on animal life and evolution, particularly those in close contact with animals, as is the case of gut microbiota [4, 5]. Gut microbiota shares intricate relationships with their hosts and are essential for host health and survival [6, 7]. For instance, it plays a key role in the development of a healthy immune system [8, 9], brain physiology and behavior [10], host protection from pathogenic infections [11], nutrient absorption [12], and provisioning of hosts with essential or otherwise inaccessible nutrients from food [13]. Despite the importance of gut microbiotas for animal life, we are still far from a general understanding of the processes involved in shaping their composition and evolution [14].
Controlled experimental work under lab conditions with animal models, as well as human studies, clearly detected strong environmental components determining gut microbiota with little evidence of genetic effects [15, 16]. Diet is, perhaps, the main environmental factor since experimental modifications of diets resulted in drastic changes in gut microbiota [17–19]. Social and physical environments are also important as both experimentally cohoused mice [20, 21] and cohabiting unrelated humans [22, 23] showed convergent gut microbiota composition. Studies of wild animals have also reported important variation in gut microbiotas in relation to, for instance, seasonal and habitat differences [24, 25]. Effects of genetic factors determining gut microbiota at the intraspecific level are considered rather weak [18, 26, 27]. Conversely, most evidence of genetic factors influencing gut microbiota comes from comparative interspecific studies showing consistent species-specific differences [15, 28, 29] or even phylogenetic covariation between animals and their gut microbial communities [30–32]. Different species, however, also differ in diet, feeding behavior and environmental characteristics of inhabited areas, and, therefore, most of the phylogenetic patterns may also be driven by environmental factors that vary interspecifically, making environmental factors co-vary with phylogenetic differences [32, 33]. This makes it difficult to disentangle the effects of environmental and genetic (i.e., phylogenetic) factors on gut microbiota.
To cope with this problem, Knowles et al. [15] explored the effect of habitat (i.e., location) and common evolutionary history (i.e., species identity) in species of three widespread mammal genera, some of them living in sympatry with each other. This approach allowed them to statistically separate the effects of habitat and species, concluding that the latter was dominant over the former factor [15]. Interestingly, interspecific differences in gut microbiota positively covaried with dietary differences, and, thus, the detected genetic component may also reflect the species-specific dietary niche of hosts. However, gut morphology and physiological characteristics of hosts are usually closely related to dietary niche [33–35]. Consequently, effects of gut microbiota that are a priori attributable to dietary niche might be overinterpreted, and should be controlled not only for host phylogeny but also for gut morphology [28]. Thus, the genetic effect of gut microbiota of animals due to anatomical or physiological characteristics should be determined after controlling for the effect of diet, sociality, and physical environment. Avian brood parasitism model systems bring us a golden opportunity to dissect this relationship [36].
Brood parasitism is a form of reproductive strategy by which parasites lay their eggs in the nests of other species so that when their eggs hatch in the host’s nest, the host adults subsequently rear their offspring [37]. Brood parasites and their hosts are usually phylogenetically distant species and, during growth, nestlings of both species receive a similar diet in identical social and physical environments [38]. In addition, brood parasite and host adults may differ in their dietary niche and consequently in gut morphology and physiology. In this case, interspecific differences in gut microbiota of cuckoo and host nestlings would be hardly explained by environmental factors [36]. The brood parasite—host system formed by the great spotted cuckoo (Clamator glandarius) (hereafter cuckoo) and its magpie (Pica pica) host fits all these characteristics. They are phylogenetically distant species [39], brood parasite and host nestlings may develop together within the same nest and are fed with a similar diet by adult magpies [40, 41]. Moreover, while adult magpies have an omnivorous diet [42], adult great spotted cuckoos mainly feed on hairy caterpillars [38], and, consequently, morphology of their guts differs even at the fledgling stage [43]. We already know that the gut microbiota of cuckoo nestlings differs from that of host nestlings [36, 43] and is intermediate between those of cuckoo adults and magpie nestlings [36]. However, the cuckoo and magpie nestlings used for comparison in these previous studies did not share their nests with nestlings of the other species. Thus, it is possible that differences in diet and/or in transmitted microbiota from adult magpies to nestlings were partially responsible for the detected interspecific differences, and, as a result, the genetic component of gut microbiota may have been overinterpreted.
To demonstrate further differences in the genetic component of the gut microbiota of cuckoo and magpie nestlings, we here experimentally moved cuckoo eggs among magpie nests so that the cuckoo eggs hatched a few days later than magpie eggs in these nests. This procedure dramatically increased the likelihood of nestlings of both species surviving until the fledging stage while sharing identical social and physical environments [41]. Moreover, we collected bacterial samples from the cloaca and from the nestlings’ beak (esophagus), which included those of saliva and food received from magpie adults. These samples allowed us to explore for the first time the possibility that interspecific differences detected in these esophageal microbiotas explained previously detected differences in cloacal microbial communities. Within this framework, a genetic component determining interspecific differences in the gut microbiota will be identified if esophageal microbiotas of cuckoo and host siblings do not vary, but clear differences in cloacal microbiota are detected. This prediction assumes that cuckoos and magpies that share the same nests are fed a similar diet that bestows similar food associated microbiotas. Here, we tested these predictions and assumptions of genetic component explaining interspecific differences in gut microbiotas of magpies and great spotted cuckoos. Bacterial groups responsible for interspecific differences in gut microbiotas of magpies and great spotted cuckoos have been described elsewhere [36], and, thus, here we concentrate on exploring the associations between esophageal and intestinal bacterial communities, as well as the association between microbiotas and diet.
Materials and methods
Study area and species
The study was performed in the Hoya de Guadix (37°18′ N, 3°11′ W), southern Spain; a high-altitude plateau where magpie nests are frequently parasitized by great spotted cuckoos [44]. The vegetation is sparse with oak (Quercus ilex), almond (Prunus dulcis) and pine (Pinus halepensis and Pinus pinaster) trees where a relatively high density of magpie nests is present.
Magpies lay one egg per day until they complete a typical clutch size of six or more eggs. Normally, magpies start to incubate when the 4th egg is laid, and their eggs hatch after 21 days of incubation. The great spotted cuckoo is a non-evictor cuckoo, and it can share a nest with foster siblings. Nonetheless, the incubation period of cuckoo eggs is 5–6 days shorter than that of magpie eggs [41]. Thus, in the case that brood parasitism occurs before magpie incubation starts, cuckoo eggs hatch some days before those of magpies, which confers a great advantage to cuckoo nestlings when competing for food with foster siblings during the nestling period [40, 45]. In most cases, magpie nestlings are outcompeted by cuckoo nestlings unless brood parasitism occurs after magpies finish laying their eggs [46]. The nestling periods also differ according to species, with cuckoos spending 16–18 days and magpies 21–23 days in the nest [47].
Fieldwork
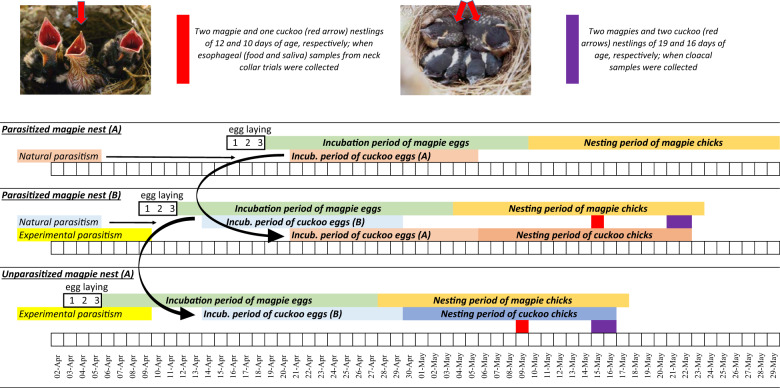
At the beginning of April 2018, we searched intensively on a weekly basis for new magpie nests in the study area, which allowed us to detect new magpie nests during the egg laying stage. In the case of brood parasitism, surveillance of magpie nests allowed us to envisage hatching time of magpie and cuckoo eggs. To maximize the number of nests where great spotted cuckoo and magpie nestlings developed together, we translocated cuckoo and magpie eggs among nests to adjust expected hatching day of magpie chicks to be two days in advance of cuckoo eggs (Fig. 1). We did so in eight magpie nests from which both great spotted cuckoos and magpies successfully fledged.
Fig. 1. Time schedule scheme and fieldwork protocol followed to synchronizing great spotted cuckoo and magpie eggs hatching within the same magpie nest.
Incubation period of great spotted cuckoo eggs is on average four days shorter than that of magpie eggs. Magpie females lay one egg per day and start to incubate with the fourth egg. Thus, cuckoo eggs detected in magpie nests before incubation started were moved (black arrows connected two nests) to magpie nests where the expected hatching date of host eggs will occur in advance of or the same day as the experimental cuckoo egg. Date of collecting esophageal (saliva and food) and cloacal samples are shown in red and purple colors, respectively (color figure online).
When magpie and cuckoo nestlings in experimental nests were about 12 and 10 days old, respectively, we collected esophageal samples, i.e., the food and the saliva they received from adult parents, by means of neck-collar trials [48] (Fig. 1). Briefly, we used plastic zip ties of three millimeters width and of different color for nestling recognition. These neck collars were carefully placed around nestlings’ necks in such a way that it allowed normal respiration, but ingestion of food provided by adult magpies was prevented. Once neck collars were adjusted, we cut the remaining length of zip ties and returned the chicks to the nest. After ~2 h, we returned to the experimental nest, collected food samples from nestlings’ esophagus, and carefully cut the zip ties with scissors. Since adult magpies transported food to the nests in their gular cavity and regurgitated food to begging nestlings, nestlings received not only food, but also saliva from their parents. Therefore, the liquid part of the collected samples consisted of a mixture of adult magpie and nestling saliva. Samples were immediately transferred into 500 µL of lysis buffer (50 mM Tris-HCl, 0.5% SDS, 2 mM EDTA, 100 mM NaCl) and kept at 4 °C in a portable fridge until arrival at the laboratory a few hours later, where the samples were frozen at −20 °C for further analysis. Collar trials were only performed in experimental nests (number of nests = 8, number magpie nestlings sampled = 17, number of cuckoo nestling sampled = 8).
Cloacal samples were collected from magpie and cuckoo nestlings close to the time of abandoning their nests (15–16 and 18–19 days old cuckoos and magpies, respectively). Only the two heavier host nestlings and all cuckoo nestlings found in magpie nests were sampled. These samples were collected by injecting and repipetting 500 µL of sterile phosphate buffer (0.1 M Na2HPO4 and 0.1 M NaH2PO4, pH 7.4) in nestlings’ cloaca using sterile tips and an automatic micropipette. Collected samples were stored in 500 µL of lysis buffer (50 mM Tris-HCl, 0.5% SDS, 2 mM EDTA, 100 mM NaCl), and kept at 4 °C in a portable fridge until the portable fridge was transported back to the laboratory a few hours later. Samples were then stored at −20 °C until further analysis.
Sample preparation
For food item identifications, samples were laid out on a petri dish where food items were separated from each other. Since we were interested in characterizing bacterial communities of food, to avoid contamination, manipulation of prey items was conducted under aseptic conditions, with previously sterilized tweezers while wearing gloves previously washed with ethanol. Once food items were separated from each other, we took photos that were later used for identification. Whenever possible, invertebrate animal food was classified to the family level via visual inspection (ESM—Fig. 1). Detected vegetable food items were in all cases green barley grain. Identification of some small pieces of food items was not possible and they were labelled as ‘unidentified’.
Esophageal samples contained solid tissues in suspension with buffer lysis and saliva from adult and nestling birds. The liquid content (hereafter saliva) were separated from the solid one and kept in different 2 mL microfuge tubes. Collar samples containing solid tissues (hereafter, esophageal samples) and cloaca samples were first mechanically homogenized using zirconia ceramic beads (for a detailed description of the followed protocol see Electronic Supplementary Material (ESM1—Material and method extension).
DNA amplification for high throughput sequencing
We amplified the region spanning from V6 to V8 of the bacterial gene 16S RNA following procedure described in Ruiz-Rodríguez et al. [36]. First PCR was carried out using two sets of primers, B969F_bar1 and BA1406R and B969F_bar2 and BA1406R (for a detailed description of the protocol of PCR amplifications see Electronic Supplementary Material (ESM1—Material and method extension).
Sequence analyses
Sequence data were analyzed with QIIME2 v2018.8 (Quantitative Insights Into Microbial Ecology) [49, 50]. Primer trimming and sequence quality filtering were performed using default parameters. Paired sequences were joined following default parameters in QIIME2. We then used Deblur, a sub-Operational Taxonomic Unit (sOTU) to further remove sequencing errors and to create the sub-OTU table [51] with a fragment length set at 380 bp. Afterwards, sequences were aligned and followed by building a de novo phylogenetic tree following Janssen et al. [52]. Taxonomy assignment was performed based on Greengenes 13_08 with a similarity of 99% [53]. Lastly, nonbacterial, chloroplastidial, and mitochondrial sequences were removed from the sub-OTU table as well as non-taxonomically assigned sequences at the phylum level.
Sequences from next-generation sequencing techniques may include reagent contaminants due to DNA extraction, amplification, and library preparation kits. This may influence the characterization of bacterial communities, mainly of those with very small microbial biomass [54, 55]. However, we analyzed cloacal and esophageal samples, which contained relatively high bacterial biomass. More importantly, we were not interested on characterizing microbiotas, but exploring expected differences between microbiotas that had been characterized with identical protocols, which would have included, if any, identical reagent contaminants. Finally, we used negative control during DNA extraction and PCR amplifications. Those negative controls were also pooled with our samples. Those controls produced few sequences after high throughput sequencing, and all of them were excluded for subsequent analyses after sequence quality filtering. Thus, reagent contaminants would negligibly influence our result.
Statistical analyses and sample sizes
We examined the effect of genetic and nongenetic components in the determination of gut microbiota by exploring the influence of species identity and gut site in alpha and beta microbial diversity [56]. Alpha diversity is the diversity within a sample, while beta diversity measures the between samples differences.
Alpha diversity of gut microbiotas was explored by means of Shannon’s diversity index, which considers the number of detected OTUs and abundance of each of them [57]. Interspecific differences were explored in General Linear Mixed Models (GLMM) by full crossing the two independent variables, i.e., species identity (magpie and cuckoo) and gut region (saliva, food, and cloaca) as the independent factors, and nest identity as random factors. The model also included interactions between fixed and random factors to account for the repeated measurement nature (i.e., within nest comparisons) of our data sets. Residuals were visually inspected for normality and homoscedasticity, justifying the use of parametric tests. Alpha diversity statistical tests were performed in Statistica 10.0.
Beta diversity matrices were calculated using weighted and unweighted UniFrac distance [58, 59] based on a rarefied OTU table at 4000 sequence per sample. Principal Coordinate Analysis (PCoA) plot was generated using these UniFrac distance matrices. While weighted Unifrac weighs sequence according to the number of sequence found per OTU in a given sample, unweighted UniFrac simply counts the absence or presence of an OTUs in a given sample. In other words, for any given sample, weighted UniFrac gives more importance to the most abundant bacteria while unweighted UniFrac gives more importance to bacteria found in minority. Both weighted and unweighted UniFrac distance matrices were then used in nonparametric multivariate statistical test, PERMANOVA, implemented in PRIMER-7 (Anderson, 2001) with 9999 permutations. Briefly, the models testing for interspecific and gut location differences included these two factors as fixed effects, and nest identity nested within the interaction of fixed factors as random effects. To explore interspecific differences of microbiota associated to the cloaca, saliva and food of magpies and great spotted cuckoos, we ran three separate models that included species and nest identity as the fixed and the random factors, respectively. Similarly, to explore within species differences in microbiota due to gut locations, we ran two sets of models, one for great spotted cuckoos and another for magpies that included gut location and nest identity as fixed and random factors, respectively. Post-hoc comparisons between pairs of gut locations were explored in separate models with two-level fixed factor (i.e., pairwise). By including nest identities as random factor, all these PERMANOVAs assumed within-nests comparisons and, thus, only considered nests with the two species and the three gut locations, which drastically reduces degrees of freedom. However, because the effects of fixed factors rarely depended on nest identity (see ‘Results’), to increase statistical power, we also ran models that did not include the random factor and showed results in Electronic Supplementary Material (ESM—Table 1).
The associations between diet and microbiotas were explored by means of Mantel tests in the R environment (Version 3.6.1). Matrices of dissimilarities of diet were calculated based on Bray Curtis dissimilarity index. Matrices of dissimilarities of species identity were built with 0’s (when the pairwise of individual was of the same species) and 1’s (when the pairwise of individual was of different species). As matrices of similarities of bacterial communities, we used both weighted and unweighted UniFrac distance matrices. Afterward, by using the R package ‘ecodist’ version 1.2.3 [60], we performed autocorrelation analyses (Mantel tests with 9999 permutations), with matrices of bacterial communities at different locations as dependent variables, and matrices of species identity and diet as explanatory variables. We estimated the strength of the correlation (Beta values), and whether these values differed significantly from zero (P values). Unfortunately, nestlings were not individually marked at the time of sampling their diet and, thus, we were not able to match diet with cloaca samples at the level of the individual. To solve this problem, we randomly matched cloaca and diet samples within the same nests and species. We did so four times and, because results did not vary qualitatively, we used average estimates and associated P values.
We sampled microbiota of 33 magpie and 22 great spotted cuckoo nestlings in 18 magpie nests. In ten of these magpie nests, great spotted cuckoo and magpie nestlings grew together, in six only magpies, and in two nests only cuckoos developed. As a rule, only two magpies and all cuckoo nestlings were sampled per nest, the cloaca was always sampled, but saliva and food were only sampled in eight nests containing cuckoo and magpie nestlings. Bacterial DNA amplification and sequencing failed for some samples and, thus, samples sizes shown in the ESM (ESM—Table 2) differed among nests, species, and sample location.
Results
Gut microbiota of great spotted cuckoo nestlings were not significantly more diverse than those of the magpie host nest mates, independently of gut location (GLMM, F = 0.18, df = 1, 11.8, P = 0.68). Moreover, independently of species (GLMM, interaction between species and gut location, F = 0.03, df = 2, 10.6, P = 0.97), cloacal microbiota tends to be more diverse than the microbiota of saliva or food (GLMM, F = 3.28, df = 2, 10.6, P = 0.064, Fig. 2).
Fig. 2. Shannon index values of alpha diversity of microbiota of cloaca, food and saliva samples of great spotted cuckoo and magpie nestlings.
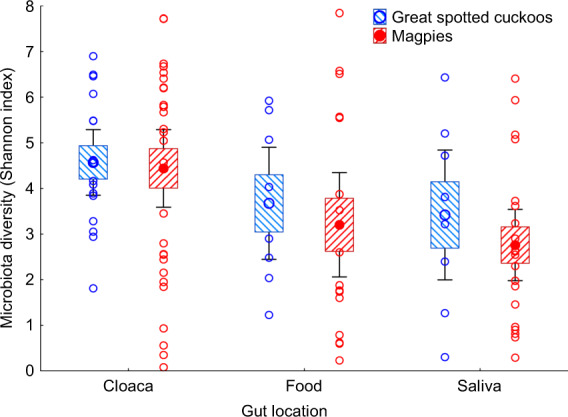
Values are weighted means ± Standard Errors (Boxes), 95% CI (Whiskers). We also show raw data collected from great spotted cuckoo and magpie nestlings.
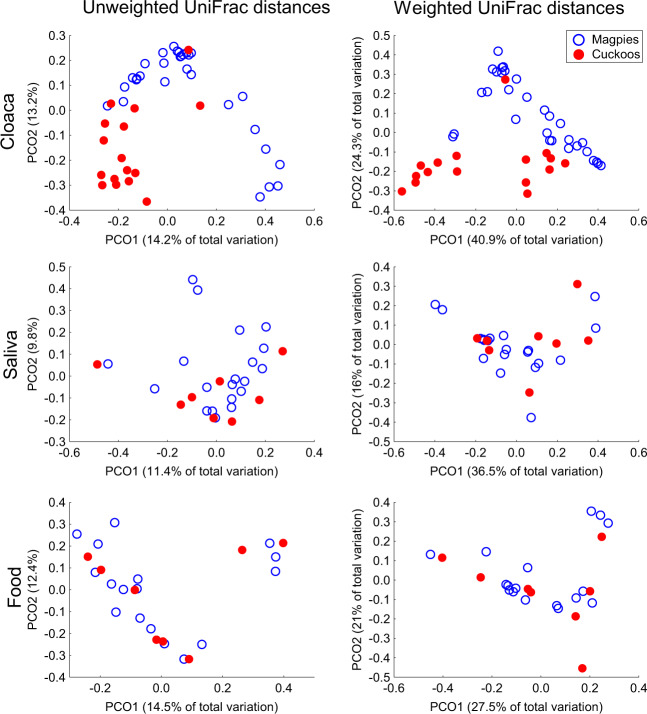
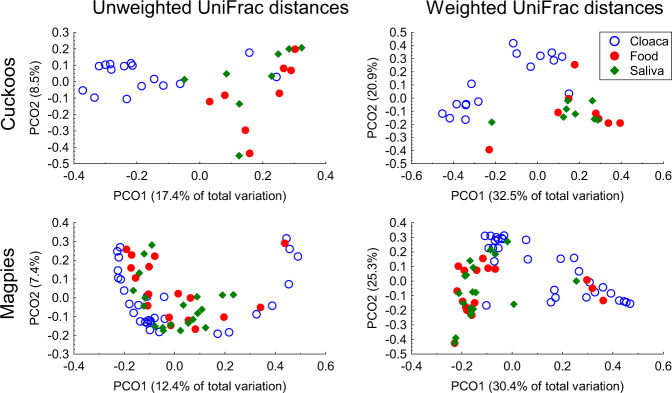
In beta diversity analyses, we found that microbiota of nestlings reared in magpie nests depended on species identity (magpie vs. great spotted cuckoo) and gut location (cloaca, saliva, and food) (Species × Gut location comparisons in Table 1, P < 0.001, and ESM—Table 1). The effect of species identity was, therefore, due to the microbiota detected when comparing cloacal samples, since microbiota of esophageal samples (saliva and food) did not differ interspecifically (see species comparisons in Table 1). Accordingly, principal coordinate analyses on both unweighted and weighted UniFrac distances showed clear clustering of cloacal samples (P < 0.012), but those of saliva (P > 0.5) or food samples (P > 0.5) of both species were distributed randomly between species (Table 1, ESM—Table 1, and Fig. 3). Intraspecific differences due to gut microbiota locations reached statistical significance in both great spotted cuckoo (P < 0.003) and magpie (only for weighted UniFrac distances, P = 0.004) nestlings (Table 1, ESM—Table 1, and Fig. 4). These differences were mainly due to those between cloacal microbiota and microbiota of esophageal (both saliva and food) samples (Table 1 and ESM—Table 1), which were more apparent in cuckoo than in magpie samples (Fig. 4).
Table 1.
PERMANOVAs exploring the effects of species identity (Sp) on the gut microbiota of different locations (G loc., saliva, food and cloaca; both as fixed (Fixed) factors) based on weighted and unweighted Unifrac distance matrixes of cloacal (CL) and esophageal (saliva—Sa; and food—Pr) samples of great spotted cuckoos and magpies that shared identical environmental conditions (i.e. nest identity, as random (Rnd) factor) during development.
| Unweighted UniFrac | Weighted UniFrac | ||||||
|---|---|---|---|---|---|---|---|
| Pseudo F | df | P | Pseudo F | df | P | ||
| Species × gut location comparisons | |||||||
| Species | Fixed | 1.66 | 1, 50 | 0.0076 | 3.49 | 1, 50 | 0.0045 |
| G loc. | Fixed | 2.36 | 2, 50 | 0.0001 | 7.79 | 2, 50 | 0.0001 |
| Sp × G loc. | Fixed | 1.45 | 2, 50 | 0.0074 | 2.31 | 2, 50 | 0.0098 |
| Nest (Sp × G loc.) | Rnd | 1.17 | 50, 43 | 0.0001 | 1.21 | 50, 43 | 0.0521 |
| Species comparisons | |||||||
| Cloacal samples | |||||||
| Species | Fixed | 2.85 | 1, 8 | 0.0109 | 4.96 | 1, 8 | 0.0111 |
| Nest | Rnd | 1.12 | 17, 8 | 0.0826 | 1.3 | 17, 8 | 0.1242 |
| Nest × species | Rnd | 1.17 | 8, 19 | 0.1052 | 1.88 | 8, 19 | 0.0093 |
| Esophageal samples | |||||||
| Saliva | |||||||
| Species | Fixed | 0.95 | 1, 6 | 0.5315 | 0.56 | 1, 6 | 0.6126 |
| Nest | Rnd | 1.12 | 7, 6 | 0.1669 | 0.51 | 7, 6 | 0.9837 |
| Nest × Species | Rnd | 0.83 | 6, 13 | 0.9308 | 0.87 | 6, 13 | 0.6346 |
| Food | |||||||
| Species | Fixed | 0.88 | 1, 5 | 0.5857 | 0.21 | 1, 5 | 0.9397 |
| Nest | Rnd | 1.61 | 7, 5 | 0.0009 | 1.56 | 7, 5 | 0.0617 |
| Nest × species | Rnd | 1.01 | 5, 11 | 0.4836 | 0.97 | 5, 11 | 0.4859 |
| Gut location comparisons | |||||||
| Great spotted cuckoos | |||||||
| G loc. | Fixed | 2.03 | 2, 11 | 0.0026 | 3.84 | 2, 11 | 0.0012 |
| Nest | Rnd | 1.46 | 11, 11 | 0.0157 | 1.25 | 11, 11 | 0.2788 |
| Nest × G loc. | Rnd | 1.12 | 11, 7 | 0.2847 | 0.84 | 11, 7 | 0.7161 |
| G loc. (Cl–Sa) | Fixed | 1.97 | 1, 11 | 0.0102 | 4.90 | 1, 11 | 0.0037 |
| Nest | Rnd | 1.30 | 11, 6 | 0.0132 | 1.42 | 11, 6 | 0.0899 |
| Nest × G loc | Rnd | 1.21 | 5, 6 | 0.0772 | 1.10 | 5, 6 | 0.3688 |
| G loc. (Cl–Pr) | Fixed | 2.51 | 1, 11 | 0.0017 | 4.02 | 1, 11 | 0.0086 |
| Nest | Rnd | 1.35 | 11, 6 | 0.0055 | 1.36 | 11, 6 | 0.1376 |
| Nest × G loc | Rnd | 1.36 | 5, 6 | 0.0126 | 1.40 | 5, 6 | 0.1531 |
| G loc. (Sa–Pr) | Fixed | 1.28 | 1, 6 | 0.1251 | 0.96 | 1, 6 | 0.4424 |
| Nest | Rnd | 1.47 | 6, 6 | 0.0273 | 0.58 | 6, 6 | 0.8534 |
| Nest × G loc. | Rnd | 0.77 | 6, 2 | 0.9464 | 0.23 | 6, 2 | 0.9969 |
| Magpies | |||||||
| G loc. | Fixed | 1.29 | 2, 13 | 0.1469 | 3.03 | 2, 13 | 0.0040 |
| Nest | Rnd | 1.23 | 15, 13 | 0.0028 | 1.13 | 15, 13 | 0.2504 |
| Nest × G loc. | Rnd | 1.14 | 13, 36 | 0.0575 | 1.23 | 13, 36 | 0.1118 |
| G loc. (Cl–Sa) | Fixed | 1.55 | 1, 7 | 0.0228 | 5.03 | 1, 7 | 0.0054 |
| Nest | Rnd | 1.04 | 15, 7 | 0.2667 | 0.98 | 15, 7 | 0.5259 |
| Nest × G loc. | Rnd | 1.12 | 7, 26 | 0.0911 | 1.15 | 7, 26 | 0.2613 |
| G loc. (Cl–Pr) | Fixed | 1.34 | 1, 6 | 0.1092 | 2.63 | 1, 6 | 0.0463 |
| Nest | Rnd | 1.22 | 15, 6 | 0.0097 | 1.21 | 15, 6 | 0.1552 |
| Nest × G loc. | Rnd | 1.31 | 6, 24 | 0.0100 | 1.71 | 6, 24 | 0.0164 |
| G loc. (Sa–Pr) | Fixed | 0.87 | 1, 6 | 0.7303 | 0.66 | 1, 6 | 0.7049 |
| Nest | Rnd | 1.60 | 7, 6 | 0.0001 | 1.42 | 7, 6 | 0.0653 |
| Nest × G loc. | Rnd | 0.93 | 6, 22 | 0.7914 | 0.90 | 6, 22 | 0.6292 |
Factors associated with P values lower than 0.1 are in bold font. Variables in bold font are statistically significant at the 10% level.
Fig. 3. Between species comparisons.
Principal coordinate analyses on both unweighted and weighted UniFrac distances of microbiotas of great spotted cuckoo (Cuckoos) and Magpie nestlings from cloaca and esophageal (saliva and food) samples. We show percentage of variance explained by the two first axes.
Fig. 4. Among locations comparisons.
Principal coordinate analyses on both unweighted and weighted UniFrac distances of microbiotas of great spotted cuckoo (Cuckoos) and magpie nestlings from cloaca and esophageal (saliva and food) samples. We show percentage of variance explained by the two first axes.
Finally, diet collected from great spotted cuckoos and their magpie foster siblings explained differences in gut microbiota at different locations after controlling for the effect of species identity (Table 2). The strength of the positive associations, however, varied depending on the location and the diversity index used to estimate distance matrices of microbiota. In terms of unweighted UniFrac differences, diet of great spotted cuckoo and magpie nestlings explained microbiota composition of esophageal samples (Saliva: P = 0.049; Food: P = 0.001), but not of cloaca samples (P = 0.32).
Table 2.
Mantel tests exploring the association between Bray Curtis distance matrices of food (i.e., diet) collected from great spotted cuckoo and magpie nestlings and their gut sampled microbiotas (saliva, food, and cloaca) estimated as weighted and unweighted UniFrac distance matrices.
| Unweighted UniFrac | Weighted UniFrac | |||
|---|---|---|---|---|
| Beta | P | Beta | P | |
| Cloacal samples | ||||
| Species | 0.039 | 0.035 | 0.141 | 0.002 |
| Diet | 0.045 | 0.322 | 0.087 | 0.337 |
| Esophageal samples | ||||
| Saliva | ||||
| Species | 0.002 | 0.881 | 0.004 | 0.927 |
| Diet | 0.060 | 0.049 | 0.131 | 0.225 |
| Food | ||||
| Species | −0.024 | 0.046 | −0.006 | 0.869 |
| Diet | 0.131 | 0.001 | 0.040 | 0.665 |
Matrices of species identity (same or different species) were also included in the models to control the expected association for the effect of species identity. Factors associated with P values lower than 0.1 are in bold font.
Discussion
Although esophageal microbiota of magpies and great spotted cuckoos raised within the same environment (nest) did not vary significantly, the microbiota of cloacal samples showed clear interspecific differences. Analyses were performed by comparing microbiotas of great spotted cuckoos with those of their magpie foster siblings, with which they shared environmental conditions and parental care from the same adults. Thus, interspecific differences can only be interpreted as being the result of genetic factors determining gut microbiota. Moreover, diet of great spotted cuckoo and magpie nestlings explained microbiota composition of esophageal samples, but not of cloaca samples, pointing out the well-known role of diet determining the gut microbiota. Below, we discuss the importance of these results, possible caveats of the statistical inferences, and the advantages of using brood parasitism as a model system to disentangle genetic and environmental factors determining gut microbiota, and to explore mechanisms underlining interspecific differences as well as functional consequences.
Given the importance of gut microbiota for animal life, including brain physiology and behavior of hosts [10], determining the importance of environmental and genetic factors explaining the gut microbiota of animals is a major topic not only in microbiology, but also in ecology and evolution [3]. Most support for a genetic component of gut microbiota in animals came from comparative studies where species-specific differences and phylogenetic relatedness influences were considered as evidence of genetic factors determining gastro-intestinal microbiota [30, 32, 35]. However, different species also varied in environmental conditions that they experience and their lifestyles, including those related to habitat characteristics, foraging behavior and diet; all of which being well-known environmental factors determining gut microbiota of animals.
To overcome such problems when exploring genetic influences, several recently published papers carried out comparative analyses while controlling for interspecific differences in diet composition [28, 32], habitat exploitation [15, 36], and/or host physiology [32]. Briefly, Amato et al. [28] compared the gut microbiota of 18 species of wild nonhuman primates that represented a range of gut morphological specialization. They concluded that the influence of host phylogeny was much stronger than that of host dietary niche, mainly because of the importance of host physiology as a determinant of gut microbiota. The rationale of the study is that the represented species do not share identical environment and, thus, authors statistically controlled for host geographic locations and actual dietary intake. In contrast, Knowles et al. [15] explored the microbiota and diet of sympatric small mammal species across multiple habitats, which allowed for statistically disentangling environmental (habitat and diet) and genetic (species identity) components of gut microbiota. They also found that diet shapes microbiota but due to species-specific diet, they concluded in favor of a large influence of genetic factors explaining gut microbiota of small mammals. However, another study of artiodactyls that also explored the gut microbiota of pairs of sympatric species across the Americas failed to detect phylogenetic effects explaining gut microbiota, although each geographic area displayed a unique gut microbiota composition [61]. Finally, Ruiz-Rodríguez et al. [36] took advantage of the brood parasitism model system and detected that gut microbiota of magpie nestlings differed from that of great spotted cuckoo nestlings although both species were reared in different nests of the same species. Here, we went one step further and experimentally forced cuckoos and magpies to develop within the same nests, so that they share the same parents under identical environmental conditions. Our results confirm interspecific differences in cloacal microbiota, therefore, suggesting that previous findings were not due to magpies and great spotted cuckoos developing in different magpie nests.
Several nonexclusive possibilities might explain interspecific differences after controlling for environmental conditions. The first one is related to the existence of processes of host-symbiont co-diversification due to vertical inheritance of microbiota; a hypothesis predicting that gut microbiota diversification should increase with phylogenetic distances for which some studies found support [15, 62, 63]. However, vertical transmission of gut microbiota cannot explain the detected interspecific differences between magpies and cuckoos because brood parasitic nestlings do not have contact with cuckoo adults until nest independence [64].
Another possibility is that closely related species, or individuals of the same species, are more likely to share genetic or behavioral mechanisms that allow horizontal acquisition of similar bacteria from the environment [5]. This mechanism includes dietary preferences, which cannot explain the detected interspecific differences in cloacal microbiotas of cuckoos and magpies because of our results and previous research [40] showed that magpies and great spotted cuckoos received a similar diet from magpie adults. Moreover, the microbiota of food collected from esophageal samples did not differ between species and, thus, external environmental conditions related to diet cannot explain interspecific differences in the intestinal microbiota of these two species. Interestingly, and in agreement with this inference, among-individual differences in diet were positively related to differences in esophageal microbiota, but among-individual differences in diet did not associate with differences in intestinal microbiotas of these species. These results, on the one hand, point to the importance of diet determining esophageal microbiotas, and, on the other, suggest that similar diets might result in significantly different intestinal microbiota when comparing different species. Therefore, these results strongly suggest a genetic component of intestinal microbiota that is independent of external environmental factors including diet.
The interspecific differences should therefore be explained by anatomical or physiological characteristics that influence gut microbiota, and that differ between magpies and great spotted cuckoos. Innate and adaptive immune system, gut morphology and mucus characteristics can differentially select components of the intestinal microbiota [28, 30, 65–67]. We know that great spotted cuckoos and magpies differ in their immune system [68]; that cellular immunity of magpies and body condition of great spotted cuckoos predicted cloacal microbiota [69]; and that, different from magpies, the intestine morphology of cuckoos is shorter and contains a relative large caecum [43]. These interspecific morphological and physiological differences are therefore likely explanations for the detected interspecific differences in the intestinal microbiota that deserve further investigation. Future studies could, for instance, explore the effect of the immune system by experimentally enhancing cellular immune response [70], and exploring differential interspecific effects in intestinal microbiotas of magpies and of great spotted cuckoos developing under identical environmental conditions. The study system would also allow exploring the consequences of having different intestinal microbiota but feeding similar diets in terms of, for instance, nutrient absorption capacity. Intestinal digestibility of great spotted cuckoo nestlings is less efficient than that of magpie host nestlings [71], so this might be related to the detected differences in their intestinal microbiota. The association between intestinal microbiotas and nutrients in the feces of cuckoo and host nestlings growing in the same nest would allow detecting bacteria responsible for the interspecific differences in nutrient absorption. Shotgun metagenomics and multi-omic methodologies applied to the cuckoo-host system will allow us to understand the causes and consequences of the detected interspecific differences, and, thus, exploring genetic mechanisms behind the genetic effect of microbiota while controlling for environmental conditions.
To summarize, our results strongly suggest a genetic component determining the intestinal microbiota of the study species while controlling for the effect of several environmental components including both abiotic and biotic conditions related to nest of rearing as parental identity, behavior and diet. The fact that interspecific differences in esophageal microbiota were not strong strengthens the possibility that interspecific differences in gut morphology and physiology are responsible for the observed differences in gut microbiota. Similar to laboratory experiments, the reproductive strategy of brood parasitism allows researchers to locate genetically unrelated individuals under identical environmental conditions and diet, where exploration of the influence of the genetic component on gut microbiota under natural conditions is made possible. This approach escapes from the undesired effects of captivity in gut microbiota [28, 72, 73], and, thus, brood parasitism should be considered a model system for exploring genetic and environmental factors affecting gut microbiota of animals as well as mechanisms and related consequences.
Supplementary information
Acknowledgements
We thank Manuel Martin-Vivaldi Martínez and Jarkko Rutila for discussion on the interest of exploring genetic and environmental factors of gut microbiotas of brood parasitic and host nestlings. The research group benefits from facilities, including an apartment, provided by the city hall of Guadix where a small lab to quickly process the samples was installed. Financial support was provided by the Spanish Ministerio de Ciencia, Innovación y Universidades and European (FEDER) funds (CGL2017-83103-P). All procedures were conducted under licence from the Environmental Department of the Regional Government of Andalucía, Spain (reference SGYB/FOA/AFR). All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chop Yan Lee, Juan Manuel Peralta-Sánchez
Supplementary information
The online version of this article (10.1038/s41396-020-0719-y) contains supplementary material, which is available to authorized users.
References
- 1.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–63. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parfrey LW, Moreau CS, Russell JA. Introduction: the host-associated microbiome: pattern, process and function. Mol Ecol. 2018;27:1749–65. doi: 10.1111/mec.14706. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA, Ochman H, Hammer TJ. Evolutionary and ecological consequences of gut microbial communities. Annu Rev Ecol Evol Syst. 2019;50:451–75. doi: 10.1146/annurev-ecolsys-110617-062453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colston TJ, Jackson CR. Microbiome evolution along divergent branches of the vertebrate tree of life: what is known and unknown. Mol Ecol. 2016;25:3776–800. doi: 10.1111/mec.13730. [DOI] [PubMed] [Google Scholar]
- 6.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne HP, Neville BA, Forster SC, Lawley TD. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15:531–43. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018;554:255–9. doi: 10.1038/nature25437. [DOI] [PubMed] [Google Scholar]
- 9.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherwin E, Bordenstein SR, Quinn JL, Dinan TG, Cryan JF. Microbiota and the social brain. Science. 2019;366:eaar2016. doi: 10.1126/science.aar2016. [DOI] [PubMed] [Google Scholar]
- 11.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015;26:493–501. doi: 10.1016/j.tem.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowles SCL, Eccles RM, Baltrūnaitė L. Species identity dominates over environment in shaping the microbiota of small mammals. Ecol Lett. 2019;22:826–37. doi: 10.1111/ele.13240. [DOI] [PubMed] [Google Scholar]
- 16.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–5. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmody Rachel N, Gerber Georg K, Luevano Jesus M, Jr., Gatti Daniel M, Somes L, Svenson Karen L, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seedorf H, Griffin Nicholas W, Ridaura Vanessa K, Reyes A, Cheng J, Rey Federico E, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–66. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand F, Nguyen TLA, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2014;2:25. doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurice CF, Cl Knowles S, Ladau J, Pollard KS, Fenton A, Pedersen AB, et al. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015;9:2423–34. doi: 10.1038/ismej.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren T, Boutin S, Humphries MM, Dantzer B, Gorrell JC, Coltman DW, et al. Seasonal, spatial, and maternal effects on gut microbiome in wild red squirrels. Microbiome. 2017;5:163. doi: 10.1186/s40168-017-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Chen L, Zhao N, Xu X, Xu Y, Zhu B. Of genes and microbes: solving the intricacies in host genomes. Protein Cell. 2018;9:446–61. doi: 10.1007/s13238-018-0532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 28.Amato KR, G. Sanders J, Song SJ, Nute M, Metcalf JL, Thompson LR, et al. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13:576–87. doi: 10.1038/s41396-018-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida AH, Ochman H. Rates of gut microbiome divergence in mammals. Mol Ecol. 2018;27:1884–97. doi: 10.1111/mec.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:e2000225. doi: 10.1371/journal.pbio.2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kartzinel TR, Hsing JC, Musili PM, Brown BRP, Pringle RM. Covariation of diet and gut microbiome in African megafauna. Proc Natl Acad Sci USA. 2019;116:23588–93. doi: 10.1073/pnas.1905666116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, González A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol. 2014;23:1301–17. doi: 10.1111/mec.12501. [DOI] [PubMed] [Google Scholar]
- 35.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz-Rodríguez M, Martín-Vivaldi M, Martínez-Bueno M, Soler JJ. Gut microbiota of great spotted cuckoo nestlings is a mixture of those of their foster magpie siblings and of cuckoo adults. Genes. 2018;9:381. doi: 10.3390/genes9080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies NB. Cuckoo adaptations: trickery and tuning. J Zool. 2011;284:1–14. [Google Scholar]
- 38.Payne RB. The cuckoos. New York: Oxford University Press; 2005. [Google Scholar]
- 39.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–73. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 40.Soler M, Martínez JG, Soler JJ, Møller AP. Preferential allocation of food by magpie Pica pica to great spotted cuckoo Clamator glandarius chicks. Behav Ecol Sociobiol. 1995;37:7–13. [Google Scholar]
- 41.Soler JJ, Martínez JG, Soler M, Møller AP. Coevolutionary interactions in a host-parasite system. Ecol Lett. 2001;4:470–6. [Google Scholar]
- 42.Birkhead TR. The Magpies. The ecology and behaviour of black-billed and yellow-billed magpies. London: T & A D Poyser; 1991. [Google Scholar]
- 43.Ruiz-Rodríguez M, Lucas FS, Heeb P, Soler JJ. Differences in intestinal microbiota between avian brood parasites and their hosts. Biol J Linn Soc. 2009;96:406–14. [Google Scholar]
- 44.Soler JJ, Martin-Galvez D, De Neve L, Soler M. Brood parasitism correlates with the strength of spatial autocorrelation of life history and defensive traits in Magpies. Ecology. 2013;94:1338–46. doi: 10.1890/12-1350.1. [DOI] [PubMed] [Google Scholar]
- 45.Moreno-Rueda G, Soler M, Soler JJ, Martínez JG, Pérez-Contreras T. Rules of food allocation between nestlings of the black-billed magpie Pica pica, a species showing brood reduction. Ardeola. 2007;54:15–25. [Google Scholar]
- 46.Soler M, Soler JJ, Martínez JG. Duration of sympatry and coevolution between the great spotted cuckoo (Clamator glandarius) and its primary host, the magpie (Pica pica). In: Rothstein SI, SK Robinson SK, editors. Parasitic Birds and their hosts, studies in coevolution. Oxford: Oxford University Press; 1998. p. 113–28.
- 47.Soler M, Soler JJ. Growth and development of great spotted cuckoos and their magpie host. Condor. 1991;93:49–54. [Google Scholar]
- 48.Martín-Gálvez D, Pérez-Contreras T, Soler M, Soler JJ. Benefits associated with escalated begging behaviour of black-billed magpie nestlings overcompensate the associated energetic costs. J Exp Biol. 2011;214:1463–72. doi: 10.1242/jeb.050153. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–00116. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen S, McDonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3:e00021–00018. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, et al. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–3. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 56.Whittaker RH. Evolution and measurement of species diversity. Taxon. 1972;21:213–51. [Google Scholar]
- 57.Shannon CE. A mathematical theory of communication. Bell Labs Tech J. 1948;27:379–423. [Google Scholar]
- 58.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–85. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22:i07. [Google Scholar]
- 61.Moeller A, Suzuki T, Lin D, Lacey E, Wasser S, Nachman M. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc Natl Acad Sci USA. 2017;114:13768–73. doi: 10.1073/pnas.1700122114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, et al. Cospeciation of gut microbiota with hominids. Science. 2016;353:380–2. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun. 2017;8:14319. doi: 10.1038/ncomms14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soler M, Soler JJ. Innate versus learned recognition of conspecifics in great spotted cuckoos Clamator glandarius. Anim Cogn. 1999;2:97–102. [Google Scholar]
- 65.Donaldson GP, Ladinsky MS, Yu KB, Sanders JB, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360:795–800. doi: 10.1126/science.aaq0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 67.Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. 2017;7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soler JJ, Møller AP, Soler M, Martínez JG. Interactions between a brood parasite and its host in relation to parasitism and immune defence. Evol Ecol Res. 1999;1:189–210. [Google Scholar]
- 69.Ruiz-Rodríguez M, Soler JJ, Lucas FS, Heeb P, Palacios M, Martín-Gálvez D, et al. Bacterial diversity at the cloaca relates to an immune response in magpie Pica pica and to body condition of great spotted cuckoo Clamator glandarius nestlings. J Avian Biol. 2009;40:42–8. [Google Scholar]
- 70.Soler JJ, De Neve L, Pérez-Contreras T, Soler M, Sorci G. Trade-off between immunocompetence and growth in magpies: an experimental study. Proc R Soc Lond B Biol Sci. 2003;270:241–8. doi: 10.1098/rspb.2002.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soler M, Rubio LA, Perez-Contreras T, Ontanilla J, De Neve L. Intestinal digestibility of great spotted cuckoo nestlings is less efficient than that of magpie host nestlings. Biol J Linn Soc. 2017;122:675–80. [Google Scholar]
- 72.Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, et al. Captivity humanizes the primate microbiome. Proc Natl Acad Sci USA. 2016;113:10376–81. doi: 10.1073/pnas.1521835113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kohl K, Skopec M, Dearing MD. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Cons Physiol. 2014;2:cou009. doi: 10.1093/conphys/cou009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.