Abstract
Rapid within-species evolution can alter community structure, yet the mechanisms underpinning this effect remain unknown. Populations that rapidly evolve large amounts of phenotypic diversity are likely to interact with more species and have the largest impact on community structure. However, the evolution of phenotypic diversity is, in turn, influenced by the presence of other species. Here, we investigate how microbial community structure changes as a consequence of rapidly evolved within-species diversity using Pseudomonas fluorescens as a focal species. Evolved P. fluorescens populations showed substantial phenotypic diversification in resource-use (and correlated genomic change) irrespective of whether they were pre-adapted in isolation or in a community context. Manipulating diversity revealed that more diverse P. fluorescens populations had the greatest impact on community structure, by suppressing some bacterial taxa, but facilitating others. These findings suggest that conditions that promote the evolution of high within-population diversity should result in a larger impact on community structure.
Subject terms: Microbial ecology, Community ecology
Introduction
It is well documented that evolution can occur over ecological timescales [1–3], and the impact of evolutionary change on ecological processes is likely to be especially important in microbial communities, where generation times are short and population sizes are large [4]. Contemporary, rapid evolution can alter species interactions [5] and the evolutionary trajectories of interacting species [6], which will contribute to community-level changes in assembly, composition and functioning [7–9]. Despite the importance of eco-evolutionary dynamics and their impact on community processes, the mechanisms through which contemporary evolution alters communities, and how abiotic and biotic conditions alter the magnitude of these eco-evolutionary impacts, remains unknown.
Rapid evolutionary diversification into resource specialists (which has been observed both in vitro [10] and in vivo [5]) could play a key role in driving changes in community structure and function [11]. Within a trophic level of a community, a more diversified population is likely to interact with more species, and hence would be expected to have a greater impact on community composition [12, 13]. Conditions that promote adaptive diversification could therefore result in greater consequences of rapid evolution on community composition.
Competition—both intra- and interspecific—is a key driver of adaptive diversification. Populations evolving in the absence of strong interspecific competition have been shown to diversify to fill available abiotic niches to reduce intraspecific competition [14, 15]. Consequently, after mixing with a novel community, populations that have diversified in the absence of interspecific competition are likely to experience greater niche-overlap with interspecific competitors. This would strongly impact community composition. Interspecific competition can also promote adaptive diversification [16], but likely in ways that reduce niche overlap with interspecific competitors (character displacement) [15, 17], although niche differentiation may be limited when competition is for essential resources [9]. Thus, populations that diversified in the presence of interspecific competitors are expected to have a lesser impact on the structure of a novel community.
Another factor that may alter the effect of a species on community composition is within-species coevolution. Populations of a species which coevolved together are more likely to have higher intraspecific diversity [18, 19]. As such, coevolved focal species populations are likely to show greater niche overlap and interact with more community members. As a consequence, the effects of diversity—both positive and negative—on community composition are likely to be stronger when genotypes are strongly coevolved [10, 18, 20].
Here, we investigate how the rapid evolution of diversity in a focal species impacts the structure of a natural microbial community. We build on previous work which demonstrated adapting Pseudomonas fluorescens to a novel potting soil environment prior to interacting with the natural community resulted in major changes in community composition[3]: an example of an “evolutionary priority effect” [8, 21, 22]. Here, we attempted to disentangle various mechanisms through which pre-adaptation of a focal species could alter subsequent community composition. We pre-adapted the soil bacterium Pseudomonas fluorescens to compost for 6 weeks and then tracked how these pre-adapted populations affected the natural microbial community over another 6 weeks. Pre-adaptation was done in both the presence and absence of the natural microbial community to examine the importance of interspecific competition in the evolution of within-species diversity. When measuring the focal species’ impact on the natural microbial community, we manipulated (1) pre-adaptation history (with or without the natural microbial community), (2) focal species diversity (1–24 clones), and (3) whether clones had pre-adapted in the same population (sympatric of allopatric pre-adaptation).
Methods
Pre-adaptation of Pseudomonas fluorescens
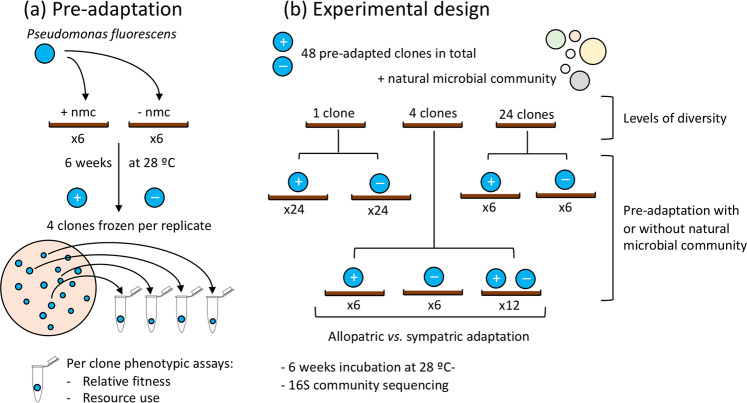
Isogenic Pseudomonas fluorescens SBW25 was pre-adapted to compost, following similar methods to those used previously [3]. The focal species contained a LacZ genetic marker and a gentamicin cassette that allowed us identify our focal species from the rest of the natural microbial community [23]. Specifically, a single clone of P. fluorescens was grown overnight at 28 °C in King’s Medium B (KB) and then ~ 5 × 107 cells were inoculated into 12 replicate sterile microcosms. These microcosms consisted of 90 mm round petri-dishes containing 30 g twice-autoclaved compost (Verve, John Innes No2, UK). Half of these microcosms (n = 6) were also inoculated with a 2 mL compost wash (20 g of compost per 100 mL M9 buffer) containing the natural microbial community (Fig. 1a), and the remainder with an equal volume of M9 salts solution. Microcosms were then incubated for 6 weeks at 28 °C. At the end of this pre-adaptation phase, 1 g of compost was taken per microcosm and added to 6 mL of M9 solution in 30 mL glass vials. Vials were shaken for 2 h at 28 °C at 180 r.p.m, after which supernatants were frozen at −80 °C in glycerol (final concentration 25%) for future assays. Each soil wash was plated onto LB agar with X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside; 90 µg/mL) and gentamicin (30 µg/mL) to allow the LacZ marked and gentamicin resistant P. fluorescens to be identified [24]. Four clones were picked from each microcosm and individually grown overnight in KB broth at 28 °C and then stored in glycerol (final concentration ~25%) at −80 °C. This resulted in 48 clones of LacZ marked P. fluorescens that had been pre-adapted to the compost environment, half of which in the presence of a community (4 clones from 6 replicates of each; Fig. 1a).
Fig. 1. Overview of experimental design.
a Populations of Pseudomonas fluorescens were pre-adapted to grow in compost in the presence (+ nmc) and absence (− nmc) of the natural microbial community. After 6 weeks, clones were isolated and frozen. b Isolated clones were assembled into different levels of diversity (1–24 clones) and different pre-adaptation histories and left for another 6 weeks in compost alongside the natural microbial community to investigate how pre-adaptation changes the effect of a focal species on the resident microbial community.
Experimental setup
To disentangle the mechanisms important in determining how pre-adaptation of a focal species impacts community composition, we manipulated focal species diversity, pre-adaptation history and whether focal species clones coevolved together to determine the relative importance of these factors in determining the impact of a focal species on soil microbial community structure. We imposed three levels of focal species diversity: a single clone, four clones and 24 clones (Fig. 1b; Table S1). Pooling clones across replicates for the 24-clone treatment was the best way to capture as much diversity as possible using the factorial design. Within each level of diversity, the focal species had either been pre-adapted with or without the microbial community. For the microcosms inoculated with a mixture of 4 clones, we also manipulated whether the clones had pre-adapted in the same or different microcosms. Twelve microcosms were inoculated with clones from the same pre-adapted population (i.e., sympatric adaptation; pre-adapted with [n = 6] and without [n = 6] the natural microbial community). In addition, twelve microcosms were inoculated with four randomly picked clones, ensuring that no two clones from the same replicate were inoculated into the same microcosm (i.e., allopatric adaptation). For the allopatric populations, clones were chosen irrespective of their pre-adaptation history.
Experiments were conducted in square petri dishes (10 × 10 cm) containing ~75 g twice-autoclaved compost. Pre-adapted clones and the LacZ ancestor were grown shaking overnight in KB broth at 28 °C. Each microcosm was then inoculated with a total of ~1 × 107 cells of P. fluorescens, resuspended in M9 buffer, with the abundance of each clone normalised based on the diversity treatment. This was added alongside 50 µL of the soil wash containing the resident community (prepared as above) and was further diluted in M9 buffer to a total volume of 2 mL. This inoculum was added to each microcosm alongside 8 mL of sterile deionised H2O to maintain soil moisture during the experiment. In addition to these treatments, 6 negative control microcosms were initiated that consisted of just the compost community. Experimental microcosms were incubated at 28 °C for 6 weeks at 80% relative humidity. After 6 weeks, soil samples were collected (2 g) and vortexed for 1 minute with sterile beads and 12 mL of M9 buffer. The resultant soil washes were used to determine P. fluorescens density by plating onto KB agar supplemented with gentamicin and X-gal (see above) and for DNA isolations to profile communities.
Catabolic profiling of the focal species
To investigate whether pre-adapted clones had diversified their resource use during pre-adaptation, the catabolic profiles of each of the 48 P. fluorescens clones was measured using Biolog EcoPlates (Biolog, USA). Every plate had 3 replicate sets of wells containing 31 different carbon sources (Fig. S1), allowing bacterial growth to be measured on multiple substrates. Each clone was grown shaking for 2 days from a freezer stock in 6 mL of KB broth at 28 °C. 1 mL of each culture was spun down in Eppendorf tubes at 15,000 g and resuspended in 1 mL of M9 salts solution to ensure minimal carryover of nutrients into the Biolog plates. Starting densities of P. fluorescens clones were normalised to an estimated OD600 of 0.0015 (~600,000 cells mL−1) and 150 µL (~90,000 cells) was inoculated into each well. Plates were incubated at 28 °C for 3 days. Each plate was placed in a plastic box with a moist sponge at the bottom to prevent evaporation of media from the wells which may confound measurements of optical density (OD). OD (590 & 600 nm wavelengths) was measured as a proxy for density of Pseudomonas fluorescens using a plate reader (Biotek Instruments Ltd). Readings of OD were taken with the lid off at an average of every 12 h. Five clones of the ancestral LacZ P. fluorescens strain were isolated from an overnight stock and used to assess the diversity of the ancestral population.
Sequencing of the focal species
To confirm that phenotypic variation in resource use reflected genomic evolution, we did whole genome sequencing of all clones used in the experiment. Each of the 48 clones and the ancestor were grown overnight in KB broth (28 °C at 180 r.p.m). The next day, the cultures were diluted in M9 salts buffer to ensure they had equal densities as measured by OD600. Total DNA extraction (1.2 mL per sample) was performed using the Qiagen Blood and Tissue kit following the manufacturer’s instructions. An Illumina HiSeq 2000 sequencer was used to generate 100 base pair (bp) paired end reads from a 500 bp insert library at the Centre for Genomic Research (Liverpool, UK). Reads were trimmed for the presence of Illumina adaptor sequences using Cutadapt (v1.2.1) [25]. The reads were further trimmed with a minimum quality score of 20. Reads shorter than 10 bp after trimming were removed. Trimmed reads were mapped to the P. fluorescens SBW25 reference genome with bwa-mem (v0.7.17-r1188) [26]. Of the 49 sequenced clones, 45 mapped well to the reference genome (mean read mapping percentage = 98.4%); the other 4 (mean read mapping percentage = 0.06%) were removed from further processing and analysis. Variants were called using freebayes (v1.3.2-dirty) [27] with ploidy set to 1 (-p 1) and vcffilter from vcflib [28] was used to only keep variants with a quality score >20. Further hard filtering steps were taken to try and retain only high confidence variants (depth within 2 standard deviations of the mean depth across all positions within a sample, depth >2, quality:depth >2, end placement probability >20, strand balance probability >20, number of alternate observations on the forward strand >20). We then removed the two variants that were present in the ancestor, which resulted in 10 unique genetic changes being identified across the remaining 44 clones.
Characterisation of the microbial community
After the 6-week experimental period, microbial communities were characterised using 16S rRNA gene amplicon sequencing. At the end of the experiment, DNA of each compost microcosm was extracted from the soil washes using the MO BIO PowerSoil DNA extraction kit (MO BIO Laboratories Inc. Carlsbad) following the manufacturer’s instructions. A 254 bp conserved fragment from the V4 hypervariable region was targeted using N501f and N701r primers with a pool of indexed primers suitable for multiplex sequencing with Illumina technology. Sequencing of amplicons of the V4 region of the 16 S rRNA gene using the Illumina MiSeq 16S Ribosomal RNA Gene Amplicons Workflow was undertaken by the Centre for Genomic Research (Liverpool, UK). We then processed and analysed the sequence data in R (v 3.5.1) using the packages “dada2”, “phyloseq” and “phangorn”. Following the standard full stack workflow [29], we estimated error rates, inferred and merged sequences, constructed a sequence table, removed chimeric sequences and assigned taxonomy. During processing, forward and reverse reads were truncated between 25 and 250 nucleotide positions due to poor quality scores. Assembled amplicon sequence variants (ASVs) were assigned taxonomy using the Ribosomal Database Project [30]. We estimated the phylogenetic tree using the R package “phangorn” [31] to allow for the calculation of weighted Unifrac distance between communities. To do this, we first constructed a neighbour-joining tree, and then fit a Generalized time-reversible with Gamma rate variation maximum likelihood tree using the neighbour-joining tree as a starting point. We then removed any reads that had not been assigned to at least the phylum level (583 of 5521 unique ASVs). To identify the focal species, we manually blasted the 7 different obtained ASVs for the genus Pseudomonas against the SBW25 16S sequence. This ASV was removed from every sample before most downstream analyses to ensure that any differences in community composition were not driven by changes in the read abundance of the focal species. Processing and filtering steps resulted in 1 sample being removed from downstream analysis (clonal diversity = 4, pre-adaptation history = with community), with the remaining data having a maximum number of reads in a sample of 204,796, minimum of 28,591 and mean of 94,494. As the negative control and the natural microbial community samples did not contain any of the focal species, P. fluorescens, they were removed after completion of data cleaning.
Statistical analyses
Analysing resource use of the focal species
The analysis of resource use splits phenotypic variation (VP) of the 4 clones isolated from each replicate population into clonal variation (VC), environmental variation (VE) and clone-by-environment variation (VCE). As growth in Biolog plates does not necessarily represent growth in the compost environment, we concentrate on relative measures of growth (VC and VCE) rather than absolute measures of growth (VE). Differences in VP, VC and VCE between treatments and the ancestor would indicate that the pre-adaptation phase altered resource-use and would strongly suggest our diversity treatments reflect real differences in resource-use diversity. Measurements of OD in blank wells (inoculated with M9) were subtracted from the OD readings and negative corrected values of OD were removed. We visually examined all time points and chose a set of measurements and wavelength where exponential growth had occurred but before stationary growth and the formation of biofilms inflated the values of OD on some substrates (Fig. S1). Consequently, we used the measurements of OD590 after ~45 h of growth. Two of the 48 pre-adapted clones failed on the biolog plates and were removed from the analysis. The number of substrates was filtered to remove substrates where no growth was observed (defined as OD590 < 0.05). This left 18 carbon substrates from which analysis of resource-use was applied, with metrics being calculated per population.
All metrics were calculated at a population level. Phenotypic variation, VP, was calculated as the average (by taking the mean) euclidean distance between catabolic profiles across all pairs of clones [32]. Our calculation of within-population clonal variation was the average variance of clone performance on each substrate [33], and environmental variance as the average clone performance across all substrates. We were particularly interested in clone-by-environment variation, as this captures the extent to which clones diversified into resource use specialists: a key assumption for the hypothesis that diversity is a key mechanism by which rapid evolution affects community composition. More specifically, clone by environment interactions can be decomposed into responsiveness and inconsistency (Fig. S2) [33–35]. Responsiveness, R, indicates differences in environmental variances between clones within a population.
| 1 |
where G is the number of clones tested within a population and σi and σj are the standard deviations of environmental responses of each clone tested. A relatively high responsiveness value would mean that some clones within that population are generalists and some clones are specialists (i.e. one clone performs equally well on substrate A and B, while another clone performs much better on substrate A relative to B). Resource specialisation is quantified by inconsistency, I,
| 2 |
where pij is the correlation of performance across substrates between each pair of clones. High inconsistency means negative correlations between clones across environments (i.e. one clone performs better on substrate A than B, and vice versa for another clone). In instances of high inconsistency and high responsiveness, clones take advantage of different resources, and some clones are specialists, and some are generalists (Fig. S2). Each variance component was calculated for the four clones isolated from each replicate; differences between pre-adaptation treatments (with or without the microbial community) were analysed using linear models. It was not possible to include the LacZ ancestor in this analysis as there was only a single replicate population. Instead, Mann-Whitney U tests of each variance component was compared to a null hypothesis where the mean is no different from the value of the variance component in the LacZ ancestral population. For the multiple testing of different variance components, the false discovery rate (fdr) method of adjusting p values was used [36].
Analysing genomic evolution of the focal species
We evaluated genomic diversity of the focal species by calculating alpha diversity using a modified version of the Hardy-Weinberg equilibrium, such that , where i is the position of each variant, p is the proportion of the variant and q is 1 − p. This was calculated for each population by pooling the variants across clones and calculating each variant’s frequency. We also looked at genomic diversity of the 24 clone mixes by pooling clones across pre-adaptation treatments. Differences between pre-adaptation treatments (with or without the microbial community) were analysed using linear models. It was not possible to include the 24 clone mixes into this analysis as they are only a single point. Instead, within each pre-adaptation treatment, Mann-Whitney U tests of each genomic diversity of 4 clones was compared to a null hypothesis where the mean is no different from the value of genomic diversity present in the 24 clone pool.
Analysing the impact of the focal species on the natural microbial community
The 16S sequencing data was split up to investigate the effect of focal species diversity, pre-adaptation history and coevolution of the focal species on community composition. To investigate community change, we looked at changes in community composition, alpha and beta diversity. Each analysis used dissimilarity indices to quantify the compositional dissimilarity between communities. We used the weighted Unifrac distance [37] as our measure of compositional dissimilarity between communities, which weights the branches of the phylogenetic tree based on the relative abundance of each ASV. Differences in composition between compost microbial communities were analysed using the R packages “phyloseq” [38], “vegan” [39], and “ape” [40]. Permutational ANOVA tests were run using the “adonis” function in the “vegan” package in R with 9999 permutations and differences in group dispersion (which test for differences in beta-diversity between treatments) were analysed using the “betadisper” function from the same package. The proportion of variance explained by each principal component was calculated using the function “pcoa” in the “ape’” package, where we applied the cailliez correction to account for negative eigenvalues [41].
First, we tested whether pre-adaptation history (evolved with or without the microbial community) altered the impact of the focal species on community composition. To do this we split the data at each level of diversity (i.e., single clone, four clones or 24 clones), and did a separate permutational ANOVA and homogeneity of variance test, with pre-adaptation history as the predictor variable. For the individual clones, we first checked whether there was any impact of population origin on community composition. For each clone, we calculated the mean weighted Unifrac distance to each sympatric and allopatric clone, and there was no effect of sympatry or allopatry on average distance between clones (Fig. S3).
Next, we looked for an overall impact of diversity on community composition. We pooled the different pre-adaptation history treatments at each level of diversity, so we had four levels of diversity: LacZ ancestor (n=6), single clone (n = 48), four clones (n = 24), and 24 clones (n = 12). A single permutational ANOVA tested whether there was an overall impact of diversity on community composition. Following this, we ran pairwise permutational ANOVAs between all pairs of diversity treatments to disentangle which treatments were causing any observed effect of diversity as a whole. This was done by subsetting the data into each pair of diversity treatments, running a permutational ANOVA on each subset, and extracting the R2 value and p value, which was adjusted using the fdr method. We also looked for an effect of diversity on community composition within pre-adaptation treatments. (Pairwise) permutational ANOVAs were run as above.
Within the four clone mixes, we investigated whether clones that coevolved together (sympatric adaptation) had a different effect on community composition than mixes consisting of clones from different communities (allopatric adaptation). We subsetted the data to just include the four clone mixes, and then ran a permutational ANOVA with allopatric vs. sympatric focal species population as a predictor variable.
To identify which ASVs differed in abundance between diversity levels, we fitted a negative binomial generalised linear model to the data using the “DESeq” function in the R package “DESeq2” [42, 43]. This method calculated significant differences in the abundance of ASVs using Wald tests and p value correction using the fdr method. For this analysis, we left the ASV assigned to focal species in each sample as it was important to know how common it was and whether it changed in abundance. Based on the paired permutational ANOVAs, we grouped the levels of diversity into high (24 clones) and low (the LacZ ancestor, individual clone and four clone mixes). This method used the raw counts and we selected only the 100 most common ASVs across all samples as the weighted Unifrac distance reduces the impact of low abundant ASVs. We tested whether changes in abundance were correlated with phylogenetic distance from the focal species using a Pearson’s correlation test. Phylogenetic distance of each ASV to the focal species was calculated using the “distTips” function in the R package “adephylo” [44].
To investigate differences in alpha diversity between communities, we used the total number of ASVs as our metric of alpha diversity. We looked at whether pre-adaptation history impacted alpha diversity by splitting the data into levels of focal species diversity (individual clones, 4 clones and 24 clones) where all clones were either pre-adapted with or without the natural community. Separate linear models were ran on each subset of the data, with observed ASVs as the response and pre-adaptation history as the predictor. We further investigated whether focal species diversity impacted alpha diversity by pooling the different pre-adaptation history treatments at each level of diversity: LacZ ancestor (n = 6), single clone (n = 48), four clones (n = 24), and 24 clones (n = 12). We tested for differences in abundance between treatments using a linear model. Model selection and comparisons were done as for the analysis on the density of P. fluorescens.
Analysing the density of the focal species
The final density of P. fluorescens across treatments was analysed using linear models. The data was first log10 transformed and one replicate was removed where no P. fluorescens was cultured (pre-adapted without the community, the individual clones that also failed on the biolog plate). We looked at whether pre-adaptation history impacted final density of the focal species by splitting the data into levels of diversity (individual clones, 4 clones and 24 clones) where all clones were either pre-adapted with or without the natural community. Separate linear models were ran on each subset of the data, with log10 abundance g−1 soil as the response and pre-adaptation history as the predictor. Model selection was done using likelihood ratio tests and the fdr method was used to adjust p values. We further investigated whether diversity affected P. fluorescens density by pooling the different pre-adaptation history treatments at each level of diversity: LacZ ancestor (n = 6), single clone (n = 48), four clones (n = 24), and 24 clones (n = 12). We tested for differences in abundance between treatments using a linear model. Model selection was done as above and comparisons were done between individual treatments using the R package “emmeans” [45]. Despite the unbalanced sample sizes between treatments, within group variance did not correlate with sample size (spearman’s rank correlation between within-group variance and number of samples within that group: p = 0.42), thus justifying our use of standard linear models. All analyses were undertaken in R (v3.6.1) [46] and all plots were made using the package “ggplot2” [47].
Results
Pre-adaptation to compost results in higher focal species functional diversity
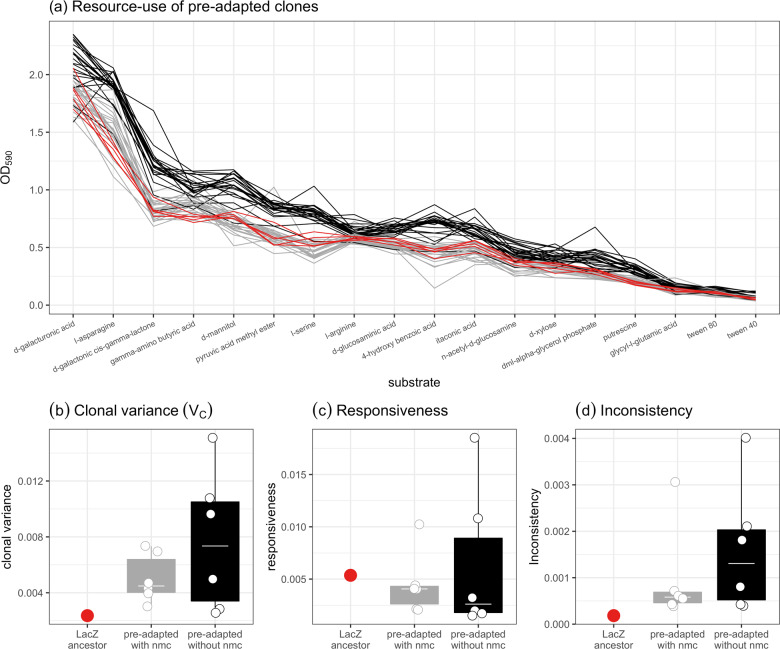
Isolated clones grew differentially on different carbon substrates and there was variation between individual clones and across treatments (Figs. 2a and S4). Pre-adaptation resulted in higher phenotypic variation (Wilcoxon rank sum: padj = 0.0006) and clonal variation (Wilcoxon rank sum: padj = 0.0006) compared to the ancestral population, with every pre-adapted population having higher values than the ancestral population (Fig. 2b). However, there was no difference in phenotypic or clonal variation (Figs. 2b and 3a) between populations pre-adapted with or without the microbial community.
Fig. 2. Differences in resource use between clones.
a Ranked substrate fitness curves for ancestral clones (red) and clones pre-adapted with (grey) and without (black) the natural microbial community. Substrates are ranked by the mean value across all clones. Increased separation between responses indicates greater clonal variation, while increased slope differences indicate greater environmental variation. Moreover, differences in rankings (some clones being better on the second substrate than the first), represent instances of resource specialisation. b Pre-adapted populations had higher clonal variance than the LacZ ancestor. c There was no difference in responsiveness between treatments or the LacZ ancestor, indicating no generalist vs. specialist trade-offs due to pre-adaptation (d) Inconsistency was higher in the pre-adapted treatments regardless of the presence of the microbial community, indicating that pre-adaptation resulted in increased resource specialisation. In (b–d), tops and bottoms of the bars represent the 75th and 25th percentiles of the data, the white lines are the medians, and the whiskers extend from their respective hinge to the smallest or largest value no further than 1.5*interquartile range. Points represent the mean value per population.
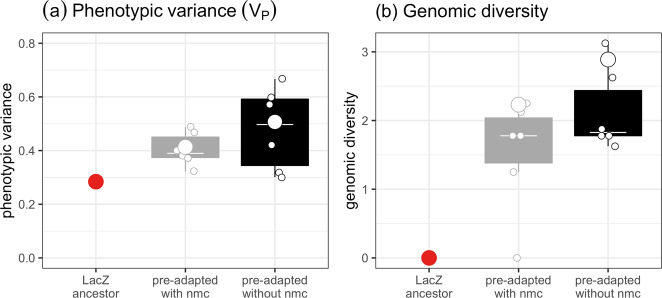
Fig. 3. Phenotypic variation in resource use after pre-adaptation reflects associated genomic changes.
a Differences in phenotypic variation of resource use between populations. b Differences in genomic diversity between populations. Tops and bottoms of the bars represent the 75th and 25th percentiles of the data, the white lines are the medians, and the whiskers extend from their respective hinge to the smallest or largest value no further than 1.5*interquartile range. Small points represent the diversity of a single population (four clones). The large points for pre-adapted populations represents the phenotypic and genomic diversity of the 24 clone pools.
To examine the evolution of diversity further, we looked for any clonal x environment interaction in the diversity of resource-use across populations. We split VCE into inconsistency and responsiveness. Pre-adapted populations showed higher inconsistency (i.e. the degree to which different clones specialised on different substrates) than the ancestral population (Wilcoxon rank sum: padj = 0.0006), with every value being greater than the ancestral population (Fig. 2c). There was no difference between evolving with or without the natural microbial community (ANOVA comparing models with and without the presence of the community: F1,10 = 0.80, p = 0.39). Moreover, responsiveness (i.e. variance in the degree of generalism) did not differ between populations pre-adapted with and without the natural microbial community, and responsiveness was not higher in pre-adapted populations compared to the ancestral population (Wilcoxon rank sum: padj = 0.677); (Fig. 2d). Overall, pre-adapted populations had higher functional diversity than the ancestral population and this was not impacted by pre-adapting with or without the natural microbial community.
Patterns of phenotypic variation broadly align with patterns of genomic evolution
To look at whether increased diversity of resource use was associated with genomic changes, we sequenced all 49 clones (48 pre-adapted and 1 ancestor) used in the experiment after the pre-adaptation phase. Across all populations, all but one population contained higher genomic diversity than the individual clones or the ancestral clone (Fig. 3b; genomic diversity > 0). In both pre-adaptation treatments, genomic diversity of the 24 clone mixes was higher than 5 out of 6 of the 4 clone mixes (Wilcoxon rank test: p = 0.0625). The patterns of genomic diversity also broadly align with the patterns of phenotypic variance (Fig. 3), demonstrating that the diversification of resource use of the focal species was likely driven by rapid evolution. In line with this, there was no difference in genomic diversity between populations of P. fluorescens pre-adapted in the presence or absence of the natural microbial community (Fig. 3b; ANOVA comparing models with and without pre-adaptation history: F1,10 = 2.10, p = 0.178).
Focal species diversity drives the impact of a focal species on community composition
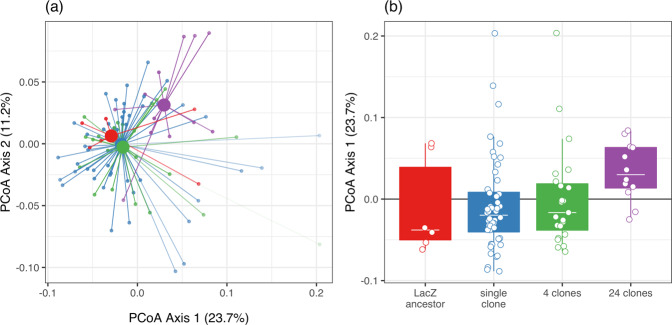
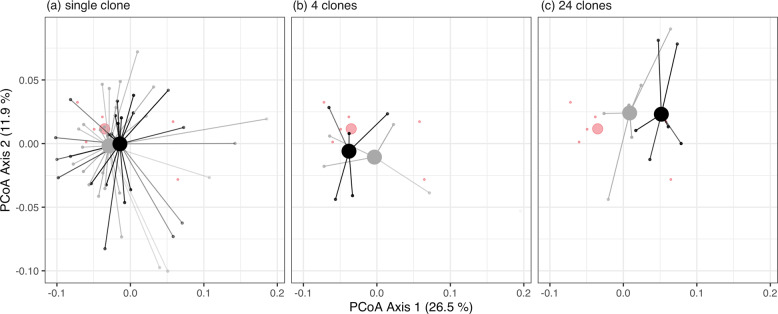
We looked for an overall effect of diversity on community composition by pooling across pre-adaptation treatments. Focal species diversity significantly altered community composition (PERMANOVA, F3,85 = 2.30, R2 = 0.08, p = 0.01; Fig. 4a). The first principal coordinate explained 23.7% of the total variation and partially separated the highest diversity treatment (24 clones) from the other levels of diversity (Fig. 4b): 83% of communities that contained 24 clones of the focal species mapped to a positive axis 1 score, compared to an average of 31% of single clone samples, 44% of four clone samples and 33% of samples inoculated with the LacZ ancestor. To determine whether this separation was significant, we ran the multiple pairwise permutational ANOVAs (see Methods, Table S2). Communities that were inoculated with the highest diversity treatment of the focal species, 24 clones of P. fluorescens, had a different composition than communities grown with a single pre-adapted clone (PERMANOVA: R2 = 0.08, padj = 0.01), four pre-adapted clones (PERMANOVA: R2 = 0.12, padj = 0.008), and the LacZ ancestral population (PERMANOVA: R2 = 0.17, padj = 0.036). None of the contrasts between any of the other diversity treatments were significant (PERMANOVAs: all padj > 0.05, Table S2). When looking at an effect of diversity within the same pre-adaptation treatment, similar results were found when populations were pre-adapted without the natural microbial community (high diversity changed community composition), but diversity did not impact community composition in populations pre-adapted with the natural microbial community (Table S3).
Fig. 4. Effect of diversity on community composition.
a Principal Coordinate (PCoA) plot of communities based on weighted-Unifrac distance. The percentage of variation explained is shown on each axis (calculated from the relevant eigenvalues). Increasing diversity (24 clones; purple) alters community composition, whereas individual (blue) and sets of 4 (green) pre-adapted clones had no consistent impact on community composition compared to the LacZ ancestor (red). b The separation of microcosms with diversity along PCoA axis 1 scores. In (a) each small point is a microcosm, large points are positions of centroids and lines connect individual microcosms to treatment centroids. In (b) points are individual microcosms, tops and bottoms of the bars represent the 75th and 25th percentiles of the data, the white lines are the medians, and the whiskers extend from their respective hinge to the smallest or largest value no further than 1.5*interquartile range.
As expected from the phenotypic assays, there was no effect of pre-adaptation history on community composition (based on weighted Unifrac distances) at any level of focal species diversity (Fig. 5; Table S4; multiple PERMANOVAs: all p values > 0.05). Moreover, there was no difference in the community composition between those that evolved sympatrically or allopatrically (PERMANOVA: F1,21 = 0.6, R2 = 0.03, p = 0.63). In summary, these results show that it is the evolution of diversity in the pre-adapted population that resulted in pre-adapted P. fluorescens impacting community composition differently compared to the ancestral P. fluorescens.
Fig. 5. Effect of pre-adaptation on community structure at different levels of diversity.
Across levels of diversity, from (a) a single clone, (b) 4 clones and (c) 24 clones, there was no effect of pre-adaptation history of the focal species with (grey) or without (black) the community on subsequent community structure. Large points represent centroids of treatments and small points individual microcosms. Lines connect individual microcosms to treatment centroids. The centroids and microcosms of the LacZ ancestor (red) are plotted across panels to help visualise the overall effect of diversity.
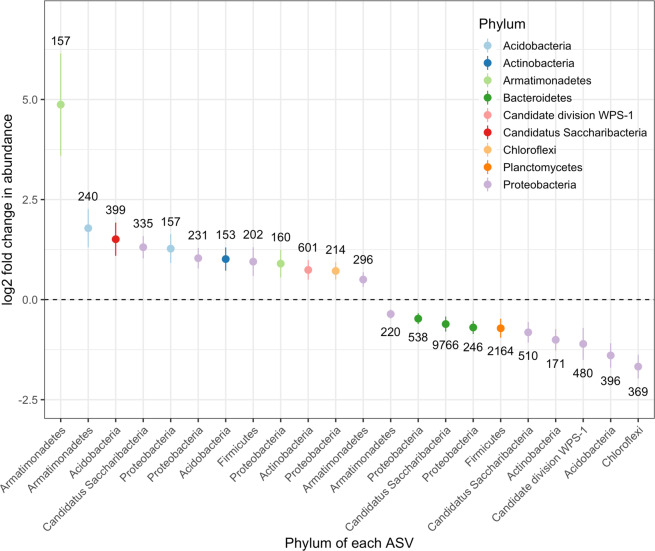
Of the 100 most common ASVs across all samples, 22 of these had significant changes in total abundance between low (LacZ ancestor, individual clone and 4 clone samples pooled) and high diversity (24 clone samples) treatments (Fig. 6; padj < 0.05), with 12 ASVs increasing and 10 decreasing in abundance. There was no correlation between phylogenetic distance from the focal species, P. fluorescens, and changes in taxon abundance (Pearson’s correlation between log2 fold abundance change and phylogenetic distance from P. fluorescens SBW25: t = 1.27, d.f. = 20, p = 0.22).
Fig. 6. Effect of diversity on the abundance of common bacterial ASVs.
The log2 fold change in raw abundance of the 100 most common ASVs from low diversity (pooled LacZ ancestor, individual and four clones) to high diversity (24 clones). Of the 100 most common ASVs tested, 22 significantly differed in abundance between low and high diversity, but no pattern was found between these ASVs and relatedness to the focal species, P. fluorescens. P. fluorescens was the 39th most abundant ASV. Points represent estimated log2 fold change in raw abundance of each ASV between low and high diversity samples, error bars are ± the standard error and the number by each point is the estimated mean number of that ASV in the low diversity samples.
Focal species are more abundant in the community after pre-adaptation
The ASV identified as the focal species was the 39th most abundant (of 4928 unique ASVs) across all samples. There was no effect of pre-adaptation history on final population density of the focal species across all levels of focal species diversity (all raw p values > 0.05). Consequently, we looked for an effect of diversity on population density (Fig. 7) and found that density varied across diversity treatments (ANOVA comparing models with and without diversity: F3,84 = 3.78, p = 0.013). This effect was driven by the final density of the LacZ ancestor (mean log10 density g−1 soil = 5.47, 95% CI = 5.06–5.87), which was significantly lower than that of the pre-adapted focal populations at each level of diversity (Fig. 7, Table S5). Diversity did not differentially affect the population density of pre-adapted Pseudomonas fluorescens (mean log10 abundance g−1 soil = 6.16, 95% CI = 6.05–6.27) (Fig. 7, Table S5).
Fig. 7. Density of the focal species at the end of the experiment.
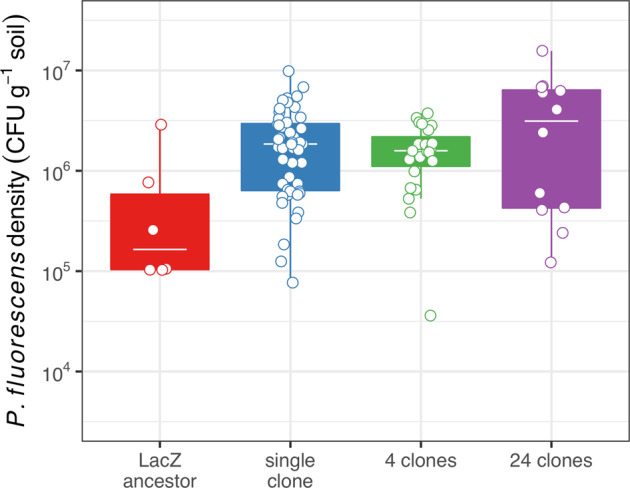
Tops and bottoms of the bars represent the 75th and 25th percentiles of the data, the white lines are the medians, and the whiskers extend from their respective hinge to the smallest or largest value no further than 1.5*interquartile range. Points represent the log10-transformed density of an individual replicate population.
There were few changes in beta-diversity and alpha diversity
There were no overall differences in between-community diversity (beta-diversity) across levels of diversity or pre-adaptation with or without the natural microbial community (all p values > 0.05). Moreover, there was little change in alpha diversity (Fig. S5) across levels of focal species diversity or pre-adaptation with or without the natural microbial community. Alpha diversity was lower in communities with 24 clones pre-adapted without the natural microbial community (ANOVA comparing models with and without the presence of the community: F1,10 = 11.38, padj = 0.02), but this effect of pre-adaptation was not observed at other levels of focal species diversity (Fig. S5). There was no overall effect of focal species diversity (ANOVA comparing models with and without the effect of focal species diversity: F3,85 = 1.34, p = 0.27).
Discussion
While pre-adaptation of a focal species has been shown to be as important as the purely ecological effects of the presence of the species [3, 8], the mechanisms underlying how and why pre-adaptation matters remain unknown. Here, we investigated these mechanisms by examining how pre-adaptation of a focal bacterial strain (P. fluorescens SBW25) in different biotic and abiotic environments altered the subsequent structure of a natural compost microbial community. We show that rapid evolution of diversity, rather than specific adaptation(s) per se, drove changes in community structure. Adaptation of the focal strain resulted in the evolution of increased diversity in resource use compared to the ancestor (Fig. 2). These changes in phenotypic diversity correlated with genetic changes (Fig. 3). In line with this, pre-adapted populations only changed community structure, relative to the effect of the ancestor, at high levels of evolved diversity (Fig. 4). This effect of diversity could not be explained by changes in focal population density (Fig. 7), strongly indicating that it was the evolved diversity, rather than concomitant changes in density, that underpinned this pattern. Pre-adaptation history or whether or not the focal species population coevolved together did not alter the impact of the focal species on subsequent community structure; nor did we find any impact of the focal species on community diversity metrics.
Our work recaptures key findings from previous studies demonstrating that pre-adaptation is important in shaping community composition [3]. Crucially, however, we show that increasing levels of diversity after pre-adaptation are the key feature that drives this effect. At low diversity (single or 4 clones), pre-adapted populations had no effect on the natural microbial community compared to the presence of P. fluorescens per se (Figs. 4 and 5). An effect of pre-adaptation on community composition was only seen at high levels of diversity (24 clones). At higher levels of genetic and phenotypic diversity, where clones of the focal species diversified to specialise on different resources, the focal species as a whole will likely interact with a larger number of other species [2, 12, 48]. However, quantifying which species of the natural microbial community the focal species directly interacted with was beyond the scope of this study. Of the 100 most common ASVs, 22 had significant changes in abundance at high levels of focal species diversity (Fig. 6), but whether or not an ASV significantly increased or decreased in abundance was not linked to how closely related the ASV was to the focal species. The lack of this link could be because relatedness is often a poor predictor of resource-use and competitive traits in microbes [49, 50]. Even though our study cannot tell us anything about the resource-use of these organisms, significant changes in their abundance still reflect that the focal species diversified to interact differently with a wide range of organisms.
Pre-adaptation increased niche differentiation (Fig. 2), which likely altered the impact of interspecific competition in multiple ways. First, by increasing niche overlap with a larger number of species, and second by reducing the impact of niche overlap on any single competing species because fewer individuals of the focal species compete for resources with it [12]. We see an overall effect of intraspecific diversity even in this highly diverse natural microbial community containing hundreds of unique species. Consequently, higher intraspecific diversity resulting in a larger impact on community structure is likely to be a general rule in eco-evolutionary systems.
Against expectations, pre-adaptation without the existing community (i.e. evolutionary priority effects) did not alter future community composition (Fig. 5). This is presumably because evolved diversity, based on catabolic profiles, did not differ when the community was present or absent (Fig. 2). Previous work using this experimental system reported greater diversification in the absence of the community [5], although this was based on the frequency of novel (wrinkly spreader) morphotypes, which rarely exceeded 10% of the population. Niche complementarity between morphotypes (i.e., smooth and wrinkly spreader) is much greater than within morphotypes [10, 51], so perhaps it is unsurprising that we do not see any impact of pre-adaptation on the catabolic profiles of the focal species.
We had expected coevolved genotypes of the focal species to have a greater impact on community composition than randomly assembled genotypes, given that we hypothesised greater diversity in the former. Our experimental design allowed us to investigate this as when four clones were inoculated, some were randomly assembled whereas others were from the same pre-adaptation replicate. We found no impact of focal species coevolution on community composition. The absence of an effect is however consistent with recent work showing that P. fluorescens evolves in a very parallel fashion (in vitro) with no evidence of genotypes being locally adapted to their populations [52].
Our experimental design specifically addressed how contemporary evolution affects community composition in the context of evolutionary priority effects: rapid local adaptation reinforcing the well-established ecological impact of colonisation order on community properties [8, 21, 22]. Of course, in most contexts, single species will not evolve in isolation but instead species would be simultaneously (co)evolving. It is unclear how this would change the impact of contemporaneous evolution, and would likely depend on whether such coevolution increased the scope for resource partitioning between species, or increased the chances for competitive exclusion due to intraspecific niche complementarity [48]. In summary, we show that the rapid evolution of diversity is important in determining the impact contemporary evolution of species on its competitors. Hence, abiotic and biotic conditions that promote adaptive diversification are likely to result in the largest impacts of pre-adaptation on community structure. Better understanding the conditions that promote adaptive diversification is therefore key in understanding instances where pre-adaptation is likely to strongly influence community dynamics.
Supplementary information
Acknowledgements
This work was funded by a NERC grant awarded to AB, SP, and RG.
Author contributions
AB, DP, AV, and EH conceived the study and designed the experiments. AV, DP, and EH conducted the experiments and DP analysed the data. SP and RG provided new perspectives. DP, EH, and AB wrote the first draft of the manuscript, with all authors contributing to the final version
Data availability
All data and R code used in the analysis is available on GitHub (https://git.io/JUOpE) and archived on Zenodo.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Daniel Padfield, Email: D.Padfield@exeter.ac.uk.
Elze Hesse, Email: E.Hesse@exeter.ac.uk.
Supplementary information
The online version of this article (10.1038/s41396-020-00755-3) contains supplementary material, which is available to authorized users.
References
- 1.Hairston NG, Jr, Ellner SP, Geber MA, Yoshida T, Fox JA. Rapid evolution and the convergence of ecological and evolutionary time. Ecol Lett. 2005;8:1114–27. [Google Scholar]
- 2.Ellner SP, Geber MA, Hairston NG., Jr Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol Lett. 2011;14:603–14. doi: 10.1111/j.1461-0248.2011.01616.x. [DOI] [PubMed] [Google Scholar]
- 3.Gómez P, Paterson S, De Meester L, Liu X, Lenzi L, Sharma MD, et al. Local adaptation of a bacterium is as important as its presence in structuring a natural microbial community. Nat Commun. 2016;7:12453. doi: 10.1038/ncomms12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckling A, Craig Maclean R, Brockhurst MA, Colegrave N. The beagle in a bottle. Nature. 2009;457:824–9. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- 5.Gómez P, Buckling A. Real-time microbial adaptive diversification in soil. Ecol Lett. 2013;16:650–5. doi: 10.1111/ele.12093. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lankau RA. Rapid evolutionary change and the coexistence of species. Annu Rev Ecol Evol Syst. 2011;42:335–54. [Google Scholar]
- 8.Pantel JH, Duvivier C, Meester LD. Rapid local adaptation mediates zooplankton community assembly in experimental mesocosms. Ecol Lett. 2015;18:992–1000. doi: 10.1111/ele.12480. [DOI] [PubMed] [Google Scholar]
- 9.Hart SP, Turcotte MM, Levine JM. Effects of rapid evolution on species coexistence. Proc Natl Acad Sci. 2019;116:2112–7. doi: 10.1073/pnas.1816298116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 11.Hughes AR, Inouye BD, Johnson MT, Underwood N, Vellend M. Ecological consequences of genetic diversity. Ecol Lett. 2008;11:609–23. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- 12.Bolnick DI, Amarasekare P, Araújo MS, Bürger R, Levine JM, Novak M, et al. Why intraspecific trait variation matters in community ecology. Trends Ecol evolution. 2011;26:183–92. doi: 10.1016/j.tree.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Violle C, Enquist BJ, McGill BJ, Jiang LIN, Albert CH, Hulshof C, et al. The return of the variance: intraspecific variability in community ecology. Trends Ecol Evol. 2012;27:244–52. doi: 10.1016/j.tree.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 14.Bolnick DI, Ingram T, Stutz WE, Snowberg LK, Lau OL, Paull JS. Ecological release from interspecific competition leads to decoupled changes in population and individual niche width. Proc R Soc B Biol Sci. 2010;277:1789–97. doi: 10.1098/rspb.2010.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey SF, Dettman JR, Rainey PB, Kassen R. Competition both drives and impedes diversification in a model adaptive radiation. Proc R Soc B Biol Sci. 2013;280:20131253. doi: 10.1098/rspb.2013.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jousset A, Eisenhauer N, Merker M, Mouquet N, Scheu S. High functional diversity stimulates diversification in experimental microbial communities. Sci Adv. 2016;2:e1600124. doi: 10.1126/sciadv.1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schluter D. Experimental evidence that competition promotes divergence in adaptive radiation. Science. 1994;266:798–801. doi: 10.1126/science.266.5186.798. [DOI] [PubMed] [Google Scholar]
- 18.Ellis CN, Traverse CC, Mayo-Smith L, Buskirk SW, Cooper VS. Character displacement and the evolution of niche complementarity in a model biofilm community. Evolution. 2015;69:283–93. doi: 10.1111/evo.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zee PC, Fukami T. Priority effects are weakened by a short, but not long, history of sympatric evolution. Proc R Soc B Biol Sci. 2018;285:20171722. doi: 10.1098/rspb.2017.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schluter D. Ecological character displacement in adaptive radiation. Am Nat. 2000;156:S4–S16. [Google Scholar]
- 21.Urban MC, De Meester L. Community monopolization: local adaptation enhances priority effects in an evolving metacommunity. Proc R Soc B Biol Sci. 2009;276:4129–38. doi: 10.1098/rspb.2009.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Meester L, Vanoverbeke J, Kilsdonk LJ, Urban MC. Evolving perspectives on monopolization and priority effects. Trends Ecol Evol. 2016;31:136–46. doi: 10.1016/j.tree.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Luján AM, Gómez P, Buckling A. Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol Lett. 2015;11:20140934. doi: 10.1098/rsbl.2014.0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Brien S, Hesse E, Luján A, Hodgson DJ, Gardner A, Buckling A. No effect of intraspecific relatedness on public goods cooperation in a complex community. Evolution. 2018;72:1165–73. doi: 10.1111/evo.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 26.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997 2013.
- 27.Garrison E, Marth G Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:12073907 2012.
- 28.Garrison E Vcflib: A C++ library for parsing and manipulating VCF files. GitHub https://www.githubcom/ekg/vcflib 2012.
- 29.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Research 2016;5:1492. [DOI] [PMC free article] [PubMed]
- 30.Maidak BL, Cole JR, Lilburn TG, Parker CT, Jr, Saxman PR, Stredwick JM, et al. The RDP (ribosomal database project) continues. Nucleic Acids Res. 2000;28:173–4. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2010;27:592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall AR, Colegrave N. How does resource supply affect evolutionary diversification? Proc R Soc B Biol Sci. 2006;274:73–78. doi: 10.1098/rspb.2006.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venail PA, MacLean RC, Bouvier T, Brockhurst MA, Hochberg ME, Mouquet N. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature. 2008;452:210. doi: 10.1038/nature06554. [DOI] [PubMed] [Google Scholar]
- 34.Robertson A. Experimental design on the measurement of heritabilities and genetic correlations: biometrical genetics. Biometrics. 1959;15:219–26. [Google Scholar]
- 35.Barrett RD, MacLean RC, Bell G. Experimental evolution of pseudomonas fluorescens in simple and complex environments. Am Naturalist. 2005;166:470–80. doi: 10.1086/444440. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 37.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–72. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:631–7. [Google Scholar]
- 40.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–8. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 41.Cailliez F. The analytical solution of the additive constant problem. Psychometrika. 1983;48:305–8. [Google Scholar]
- 42.Love M, Anders S, Huber W. Differential analysis of count data–the DESeq2 package. Genome Biol. 2014;15:10–1186. [Google Scholar]
- 43.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS computational Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jombart T, Balloux F, Dray S. Adephylo: new tools for investigating the phylogenetic signal in biological traits. Bioinformatics. 2010;26:1907–9. doi: 10.1093/bioinformatics/btq292. [DOI] [PubMed] [Google Scholar]
- 45.Lenth R Emmeans: Estimated marginal means, aka least-squares means. R Package Version 2018; 1.
- 46.R Core Team. R: A language and environment for statistical computing. 2013.
- 47.Wickham H ggplot2: elegant graphics for data analysis. 2016. Springer.
- 48.Vellend M. The consequences of genetic diversity in competitive communities. Ecology. 2006;87:304–11. doi: 10.1890/05-0173. [DOI] [PubMed] [Google Scholar]
- 49.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–5. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 50.Narwani A, Alexandrou MA, Herrin J, Vouaux A, Zhou C, Oakley TH, et al. Common ancestry is a poor predictor of competitive traits in freshwater green algae. PLoS ONE. 2015;10:e0137085. doi: 10.1371/journal.pone.0137085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckling A, Kassen R, Bell G, Rainey PB. Disturbance and diversity in experimental microcosms. Nature. 2000;408:961. doi: 10.1038/35050080. [DOI] [PubMed] [Google Scholar]
- 52.Castledine M, Buckling A, Padfield D. A shared coevolutionary history does not alter the outcome of coalescence in experimental populations of Pseudomonas fluorescens. J Evol Biol. 2019;32:58–65. doi: 10.1111/jeb.13394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and R code used in the analysis is available on GitHub (https://git.io/JUOpE) and archived on Zenodo.