Abstract
We have previously reported that recombinant adeno-associated virus serotype 3 (AAV3) vectors transduce human liver tumors more efficiently in a mouse xenograft model following systemic administration. Others have utilized AAV8 vectors expressing miR-26a and miR-122 to achieve near total inhibition of growth of mouse liver tumors. Since AAV3 vectors transduce human hepatic cells more efficiently than AAV8 vectors, in the present studies, we wished to evaluate the efficacy of AAV3-miR-26a/122 vectors in suppressing the growth of human hepatocellular carcinoma (HCC) cells in vitro, and human liver tumors in a mouse model in vivo. To this end, a human HCC cell line, Huh7, was transduced with various multiplicities of infection (MOIs) of AAV3-miR-26a or scAAV3-miR-122 vectors, or both, which also co-expressed a Gaussia luciferase (GLuc) reporter gene. Only a modest level of dose-dependent growth inhibition of Huh7 cells (~12-13%) was observed at the highest MOI (1x105 vgs/cell) with each vector. When Huh7 cells were co-transduced with both vectors, the extent of growth inhibition was additive (~26%). However, AAV3-miR-26a and scAAV3-miR-122 vectors led to ~70% inhibition of growth of Huh-derived human liver tumors in a mouse xenograft model in vivo. Thus, the combined use of miR-26a and scAAV3-miR-122 delivered by AAV3 vectors offers a potentially useful approach to target human liver tumors.
Keywords: AAV vectors, miRNA, gene therapy, liver cancer
Introduction
Liver cancer is predicted to be the sixth most commonly diagnosed cancer, and the fourth leading cause of cancer death worldwide reported in 2018, with about 841,000 new cases and 782,000 deaths annually [1]. During the past two decades, the incidence of liver cancer in the US has tripled while the 5-year survival rate has remained below 12% [2]. Hepatocellular carcinoma (HCC) is an aggressive tumor with poor prognosis, which accounts for 85%-90% of primary liver cancers [3, 4]. HCC is a highly vascularized tumor with high incidence, largely attributable to resistance to treatment, tumor recurrence and metastasis [5]. The early diagnosis of HCC is difficult, which results in high 5-year postoperative recurrence rate and poor 5-year survival rate [6, 7]. Consequently, the identification of specific and sensitive biomarkers for the diagnosis and prognosis of HCC is particularly crucial, and novel therapeutic strategies are needed to target this disease.
MicroRNAs (miRNAs) are a class of small, endogenous, single-stranded, noncoding RNAs, approximately 22 nucleotides in length, which were first discovered in 1993 [8]. miRNAs regulate gene expression by directly binding with the 3’-untranslated region (3’UTR) of target mRNAs, causing translational inhibition, mRNA cleavage and degradation [9]. miRNAs are involved in critical biological, physiological and pathological processes, including angiogenesis, metastasis, inflammation, autophagy, proliferation, migration, differentiation, apoptosis and oncogenesis [10-12]. Therefore, miRNAs are considered a powerful tool for identification and characterization of tumor genesis.
miR-26a, which is located on human chromosome 3, is frequently downregulated in HCC tissues [13]. Downregulated miR-26a have reduced ability to regulate cell cycle proteins like cyclins D/E and cyclin dependent kinases 2/4/6 [14]. miR-26a can regulate apoptosis and suppress metastasis of HCC by targeting interleukin-6-Stat3 pathway [15]. miR-26a can inhibit angiogenesis of HCC through targeting PIK3C2α/Akt/HIF-1α/VEGFA and hepatocyte growth factor-cMet pathway [16, 17]. The overall survival rate of HCC patients with poor miR-26a expression was shorter than those with high miR-26a expression [18].
miR-122, which is a conserved and abundant liver-specific miRNA, accounts for 70% of the hepatic miRNA in the adult mouse [19]. miR-122 can modulate anti-apoptotic molecules such as Bcl-2, Bcl-w, Bcl-xl and Mcl-1 [14]. miR-122 can induce apoptosis and suppress proliferation in HCC by targeting the Wnt/β-catenin pathway [20]. miR-122 can inhibit tumor growth and proliferation by targeting cyclin G1/E2F1 and TCF-4 respectively [21-23]. miR-122 can also inhibit angiogenesis and metastasis through suppressing expression of tumor necrosis factor-α-converting enzyme and ADAM17, a key protein involved in metastasis [24, 25].
Adeno-associated virus (AAV) is a small, non-enveloped, nonpathogenic human parvovirus, which can enhance gene expression and facilitate efficient transduction [26]. AAV8 was used to deliver miR-26a to suppress tumorigenesis in inducible-cMYC mice [27]. AAV-mediated delivery of miR-122 decreased tumorigenicity in a tet-o-MYC; LAP-tTAT mouse model [28]. Recombinant AAV (rAAV) vectors are currently being used in many gene therapy clinical trials, such as such as in Leber’s congenital amaurosis [29-32], lipoprotein lipase deficiency [33], Hemophilia B [34-39], aromatic L-amino acid decarboxylase deficiency [40], choroideremia [41], Leber hereditary optic neuropathy [42], hemophilia A [37], and spinal muscular atrophy [43]. Based on our previous studies, rAAV3 could transduce human HCC cells highly efficiently both in vitro and in vivo with high efficiency [44, 45]. Consequently, the combination of rAAV3 with miRNA expression cassettes or inhibitors, are valuable tools of miRNA-based gene therapy for human HCC.
In the present study, we determined the effects of rAAV3-miR-26a and rAAV3-miRNA-122 vectors, either alone, or in combination, on the growth of a human HCC cell line (Huh7) in vitro. We also evaluated the efficacy of the combined use of rAAV3-miR-26a and rAAV3-miRNA-122 vectors on Huh7 cells-induced liver tumors in a murine xenograft model in vivo. The results suggest that rAAV3-miR-26a/122 vectors may prove to be useful in the potential gene therapy of human liver cancer.
Materials and Methods
Cell lines and cultures
Human embryonic kidney, HEK293, and human hepatocellular carcinoma, Huh7, cells were purchased from American Type Culture Collection (Manassas, VA), and HepaRG cells, terminally differentiated hepatic cells derived from a human hepatic progenitor cell line that retain many of the characteristics of primary human hepatocytes, were obtained from ThermoFisher Scientific, Waltham, MA. Huh7 cells, stably transfected with a firefly luciferase expression plasmid (Huh-FLuc) were generated as described previously [46]. Cells were maintained in complete Dulbecco Modified Eagle Medium (DMEM, Mediatech, Manassas,VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO) and 1% penicillin and streptomycin (P/S, Lonza, Walkersville, MD). Cells were grown as adherent cultures in a humidified atmosphere at 37°C in 5% CO2, subcultured after treatment with trypsin-Versene mixture (Lonza, Walkersville, MD) for 2 to 5 min at room temperature, washed, and resuspended in complete DMEM.
AAV vectors and vector production
Self-complementary AAV3 vectors expressing the Gaussia luciferase (GLuc) reporter gene under the control of the chicken β-actin promoter (scAAV3-CBAAp-GLuc) and those containing miRNA-26a (scAAV3-CBAAp-miRNA-26a-GLuc) or miRNA-122 (scAAV3-CBAAp-miRNA-122-GLuc) were generated using standard cloning methods. All viral vectors were packaged using the polyethyleneimine-mediated triple-plasmid transfection protocol [47]. Briefly, HEK293 cells were co-transfected with three plasmids using polyethyleneimine (PEI, linear, MW 25000, Polysciences, Inc., Warrington, PA), and medium was replaced after 6 hours transfection. Cells were harvested after 72 hours transfection, lysed by 3 rounds of freeze-thaw and digested with Benzonase (EMD Millipore, Darmstadt, Germany) at 37 °C for 1 hour. AAV vectors were purified by iodixanol (Sigma, St. Louis, MO) gradient ultracentrifugation followed by ion exchange column chromatography (HiTrap Q HP, 5ml, GE Healthcare, Piscataway, NJ), washed with phosphate-buffered saline (PBS) and concentrated by centrifugation using centrifugal spin concentrators (Apollo; 150 kDa cutoff, 20 ml capacity, Orbital Biosciences, Topsfield, MA).
Quantitative DNA slot blot analyses
The physical genomic titers of highly purified scAAV vector stocks were determined by quantitative DNA slot blot analysis as described previously [48], with modification. Briefly, 10 μl of vector stock was digested with Benzonase (EMD Millipore, Darmstadt, Germany) at 37°C for 1 hour. An equal volume of 100 mM NaOH was added, followed by incubation at 65°C for 30 min. A known quantity of plasmid DNA was denatured in the same manner for use as a reference standard for quantitation. Denatured DNA samples were loaded in two-fold serial dilutions onto Immobilon-NY+ membranes (Millipore, Bedford, MA). After UV cross-linking, the membranes were prehybridized for 1 hour at 42°C in a hybridization solution containing 6×SSC, 100 μg/ml denatured herring sperm DNA, 0.5% sodium dodecyl sulfate (SDS), and 5×Denhardt’s reagent. Subsequently, the membranes were hybridized with Cy3-labled DNA probe in a hybridization solution at 42°C for 18 to 20 hours. Membranes were washed twice with wash solution I (2×SSC, 0.1% SDS) at room temperature for 15 min, twice with wash solution II (0.5×SSC, 0.1% SDS) at 42°C for 15 min, and then exposed to Amersham Typhoon RGB Biomolecular Imager (GE Healthcare, Chicago, IL) at room temperature.
Western blot assays
Western blot assays were performed as previously described [49]. The following antibodies were used: anti-AAV capsid antibody B1 (1:1,000 dilution; ARP, Waltham, MA); horseradish peroxidase-conjugated secondary antibodies (1:2,000 dilution; Cell Signaling Technology, Inc., Danvers, MA).
AAV vector transduction in vitro
Cells were seeded in 12-well plates at a concentration of 1x104 (Huh7), 1x104 or 4x104 (Huh7-FLuc), and 1x105 (HepaRG) cells per well in complete DMEM medium. After incubation of 24 hours, cells were mock-transduced or transduced with AAV3 vectors for 2 hours. AAV infections were performed in serum- and antibiotic-free DMEM medium at a multiplicities of infection (MOIs) of 1x102, 1x103, 1x104 , and 1x105 vgs/cell in various experiments.
Quantitation of cell numbers, and FLuc and GLuc gene expression in vitro
At 3-, 6-, 9-, and 12-days post-infection, culture media supernatants were harvested for Gaussia luciferase (GLuc) activity using BioLux® Gaussia Luciferase Flex Assay Kits (New England Biolabs, Inc., Ipswich, MA). Mock-transduced and vector-transduced cells were washed with PBS, and cell lysates were prepared using 1×passive lysis buffer. Firefly luciferase (FLuc) activity was determined using an injector-equipped luminometer (BMG Labtech, FLUOstar Optima, Cary, NC), as recommended by the manufacturer. Cell numbers were determined using a Cell Counting Kit-8 (CCK8, Dojindo Laboratories, Tokyo, Japan).
Analysis of vector genome copy numbers
The vector genomes were extracted by Proteinase K digestion followed by phenol/chloroform purification and DNA precipitation by ethyl alcohol [50]. Vector genome copy numbers were determined by qPCR analysis with the primers listed below. GLuc-F: GGAGGTGCTCAAAGAGATGG; GLuc-R: TTGAACCCAGGAATCTCAGG. Data from three independent experiments were used to quantitate the vector genome copy numbers.
RNA extraction and quantitative real-time PCR assays
Total RNA was isolated from Huh7-FLuc cells, using Aurum™ Total RNA Mini Kit (BIO-RAD, Hercules, CA), and cDNA was synthesized using iScript Reverse Transcription Kit (BIO-RAD, Hercules, CA) according to the manufacturer’s specifications. The expression levels of miRNAs were calculated using relative quantification 2-ΔΔCT method after normalization with endogenous control U6 snRNA. All PCR reactions were performed in triplicates. The primers used in the qRT-PCR assays are described in Table 1.
Table 1:
qRT-PCR primers used in this study.
| Name | Sequence (5’-3’) |
|---|---|
| miR-26a-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCTA |
| miR-26a-F | TTCAAGTAATCCAGGATAGGCT |
| miR-26a-R | AAGTTCATTAGGTCCTATCCGA |
| miR-122-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAACA |
| miR-122-F | GGCTGGAGTGTGACAATGGTG |
| miR-122-R | AGTGCAGGGTCCGAGGTATT |
| U6 snRNA-RT | ACCATCACATCTCATTTTGC |
| U6 snRNA-F | CTCGCTTCGGCAGCACATATACT |
| U6 snRNA-R | ACGCTTCACGAATTTGCGTGTC |
Animal handling
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) and performed according to the guidelines for animal care specified by the Animal Care Services (ACS) at the University of Florida (Gainesville, FL). Six to 10-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained by ACS at the University of Florida College of Medicine (Gainesville, FL). Huh7-FLuc cells were mock-transduced or transduced with AAV vectors at an of MOI of 100,000 vgs/cell for 2 hours. Five million Huh7-FLuc cells mixed with matrigel (Corning, Bedford, MA) at a 1:1 ratio, and were injected subcutaneously in a total volume of 200 μl in mice on the ventral side of the neck between shoulder blades. Animals were kept in sterile cages until the end of the experiments.
In vivo bioluminescence imaging
Firefly luciferase activity was analyzed at day 7, 10, 13, 16, 23, and 30 after injections using a Xenogen IVIS Lumina System equipped with a cooled couple-charged device (CCD) camera (Caliper Life Sciences, Hopkinton, MA). Mice were anesthetized with oxygen containing 4% isoflurane (Phoenix Pharmaceuticals, St. Joseph, MO) for induction, and 2% for maintenance. Mice were injected intraperitoneally with luciferin substrate (Caliper Life Sciences, Hopkinton, MA) at a dose of 0.15 mg/g of body weight. Mice were placed in a light-tight chamber and in vivo bioluminescence imaging was acquired at 5 minutes after the substrate injection. Images were analyzed by the Living Image 3.2 software (Caliper Life Sciences, Hopkinton, MA). Signal intensity was quantified as photons/second/cm2/steridian (p/sec/cm2/sr). All procedures were performed according to the principles of the National Research Council’s Guide for the Care and Use of Laboratory Animals. All efforts were made to minimize suffering of the animals challenged with Huh7-FLuc cells. Animals were monitored daily and humanely euthanized when tumor size reached 1.5 cm in diameter.
Statistical analysis
All results are presented as mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism 7 software (GraphPad Software, Inc., San Diego, CA). Differences between groups were identified by grouped-unpaired two-tailed distribution of Student's T-test. A value of P < 0.05 were considered statistically significant.
Results
Transduction with scAAV3-miRNA vectors leads to vector dose-dependent growth inhibition of human liver cancer cells in vitro
We have previously reported that human hepatocellular carcinoma cell lines, including Huh7 [51], can be efficiently transduced with AAV3 vectors [44, 45]. Using AAV8 vectors, others have observed remarkable efficacy of miR-26a and miR-122 in inhibiting the growth of mouse liver tumors [27, 28]. We evaluated the efficacy of scAAV3-miR-26a and scAAV3-miRNA-122 vectors in suppressing the growth of Huh7 cells. scAAV3 vectors expressing only the GLuc reporter gene were used as an appropriate control. These vectors are depicted schematically in Fig. 1A. Titers of the recombinant scAAV3 vectors were determined by quantitative DNA slot blot assays by Cy3-labeled DNA probe specific to the full-length viral genome (Fig. 1B). Analysis of the viral capsid on SDS-polyacrylamide (SDS-PAGE) gels, followed by Western blotting, also revealed the conventional VP1: VP2: VP3 protein ratio of 1:1:10 (Fig. 1C) indicating that each of the three vectors was of similar quality.
Figure 1:

Construction and characterization of scAAV3 vectors. A. Schematic structures of the scAAV3-CBA-GLuc, scAAV3-CBA-miRNA-26a-GLuc, and scAAV3-miRNA-122-CBA-GLuc vector genomes. B. DNA slot blot analysis for vector titer determination. C. Western blot analysis of viral proteins for vector quality.
Huh7 cells were either mock-transduced or transduced with each of the three scAAV3 vectors at multiplicities of infection (MOIs) ranging from 1x102 to 1x105 vgs/ cell under identical conditions. GLuc activity was determined 72 hrs post-transduction. These results, shown in Fig. 2A, indicate that the extent of transgene expression was similar from each of the vectors, and correlated with the MOIs. However, at high MOIs, scAAV3-miRNA-26a and scAAV3-miRNA-122 vectors, but not the control scAAV3-GLuc vector, led to a modest inhibition of cell growth. Significant levels of growth inhibition were observed at the highest vector doses used. Nearly identical results were obtained with Huh-FLuc cells (Figure 3). Thus, all subsequent studies were performed using Huh7-FLuc cells and an MOI of 1x105 vgs/cell.
Figure 2:
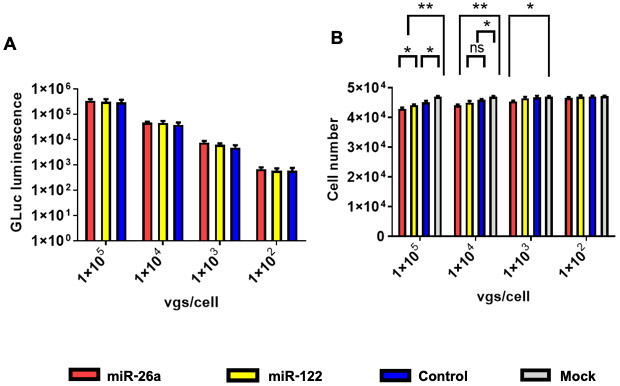
scAAV3-miRNA vectors-mediated growth inhibition of Huh7 human hepatocellular carcinoma cells. A. Equivalence of dose-dependent transduction efficiency of scAAV3 vectors. B. Growth inhibition of Huh7 cells with scAAV3-miRNA vectors. Data are presented as mean ± SD. *P < 0.05, **P < 0.01; ns, statistically not significant.
Figure 3:

scAAV3-miRNA vectors-mediated growth inhibition of Huh7 cells stably expressing the firefly luciferase (FLuc) reporter gene. A. Equivalence of dose-dependent transduction efficiency of scAAV3 vectors. B. Growth inhibition of Huh7-FLuc cells with scAAV3-miRNA vectors. C. Inhibition of dose-dependent FLuc gene expression by scAAV3-miRNA vectors. Data are presented as mean ± SD. *P < 0.05, **P < 0.01; ns, statistically not significant.
scAAV3-miRNA vector-mediated growth inhibition of HCC cells is transient
Since rAAV vector genomes in general have previously been shown to remain episomal [52], the miRNA-26a- and miRNA-122-mediated observed growth inhibition of Huh7 cells would not be expected to be long-lasting due to the rapidly proliferating nature of these cells, and as a consequence, the diminution of the vector would occur with time. We experimentally tested this possibility by transducing Huh7-FLuc cells with each of the three vectors as described above, and determined GLuc expression levels at day 3, day 6, day 9, and day 12 post-transduction. As can be seen in Figure 4A, there was a slight overall decrease, but no significant differences in GLuc expression levels were observed among the three groups. However, both FLuc expression levels (Figure 4B) and total cell numbers (Figure 4C) increased over time, and beyond day 6, there were no significant differences between the experimental and the control groups. Concurrently, the peak levels of the vector genome copy numbers at day 3 declined rapidly beyond day 6 (Figure 4D), indicating that the loss of the inhibitory activity of scAAV3-miRNA vectors was indeed due to vector dilution. This conclusion was further corroborated by evaluating expression levels of each of the two miRNAs in Huh7-FLuc cells. The results indicated that peak expression levels miR-26a (Figure 5A) and miR-122 (Figure 5B) also declined over time.
Figure 4:
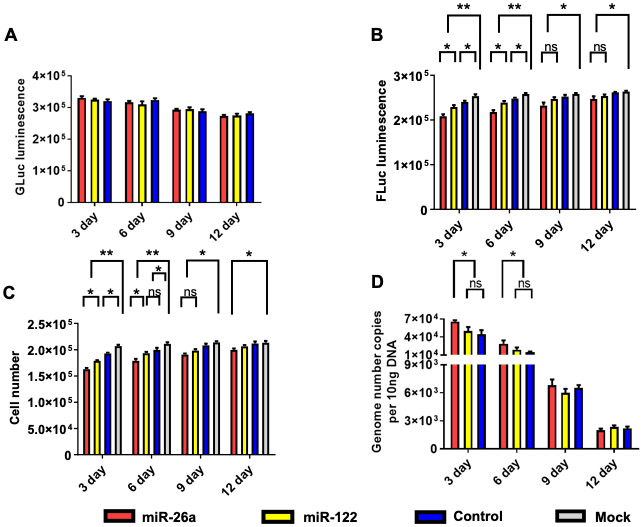
scAAV3-miRNA vectors-mediated growth inhibition of Huh7-FLuc cells correlates with diminution of transgene expression and loss of viral vector genomes as a function of time. A. GLuc expression, B. FLuc expression, C. Total cell numbers, and D. Vector genome copy numbers. Cells were transduced with 1x105 vgs/cell. Data are presented as mean ± SD. *P < 0.05, **P < 0.01; ns, statistically not significant.
Figure 5:
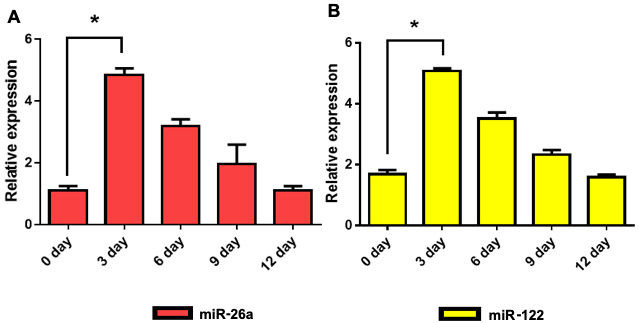
Loss of vector genome copy numbers correlates with expression levels of miRNAs in Huh7-FLuc cells. A. miR-26a, and B. miR-122. Cells were transduced with 1x105 vgs/cell. All data were normalized to expression levels of U6-small nuclear RNA. *P < 0.05.
Additive effect of scAAV3-miRNA-26a and scAAV3-miRNA-122 vectors on the growth inhibition of human HCC cells
Since only modest level of growth inhibition (~12–13%) of Huh7 cells was achieved with either scAAV3-miRNA-26a or scAAV3-miRNA-122 (Figures 1-3), it was of interest to examine whether co-transduction with both vectors could lead to a more significant degree of inhibition. To this end, Huh7-FLuc cells were either mock-transduced or transduced with scAAV3-miR-26a-GLuc or with scAAV3-miR-122-GLuc vectors alone, or co-transduced with scAAV3-miR-26a-GLuc+scAAV3-miR-122-GLuc vectors. Transductions with scAAV3-CBA-GLuc, or with scAAV3-miR-26a-GLuc+scAAV3-CBA-GLuc, or with scAAV3-miR-122-GLuc+scAAV3-CBA-GLuc vectors were used as appropriate controls. GLuc, FLuc, total cell numbers, and vector genome copy numbers were determined 72 hrs post-transductions as described above. These results are shown in Figure 6, and can be summarized as follows: (i) No significant differences was observed in the extent of GLuc expression among each group (Figure 6A); (ii) FLuc expression level in cells transduced with scAAV3-miR-26a-GLuc+scAAV3-miRNA-GLuc vectors was significantly lower than those transduced with each of the vectors alone (Figure 6B); (iii) Transduction with scAAV3-miR-26a-GLuc+scAAV3-miR-122-GLuc vectors had a significantly higher inhibitory effect (~26%), as determined by total cell numbers, compared with that with each of the scAAV3-miRNA vectors alone (~12-13%) (Figure 6C); which also correlated with the vector genome copy numbers (Figure 6D). Thus, the combined use of the two scAAV3-miRNA vectors would appear to have a higher therapeutic potential.
Figure 6:
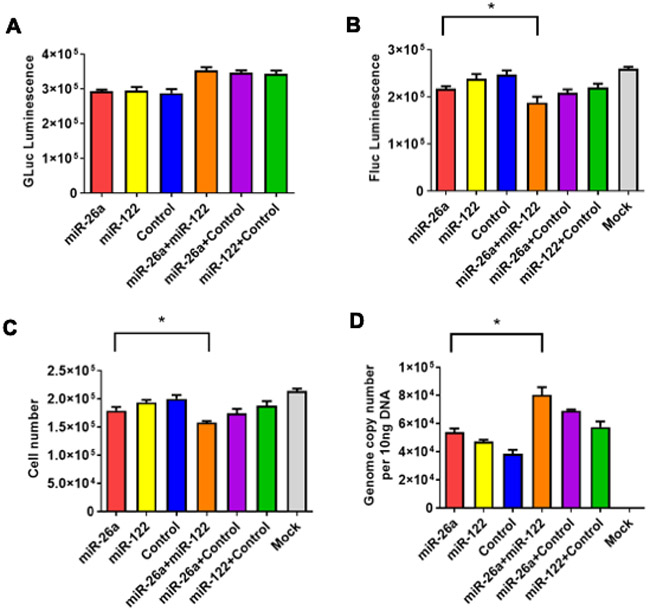
Growth inhibition of Huh7-FLuc cells with scAAV3-miRNA vectors is additive, and correlates with diminution of transgene expression and loss of viral vector genomes. A. GLuc expression, B. FLuc expression, C. Total cell numbers, and D. Vector genome copy numbers. Cells were transduced with s total of 1x105 vgs/cell. Data are presented as mean ± SD. *P < 0.05.
scAAV3-miRNA vectors fail to inhibit the growth of HepaRG cells
To ascertain that the observed growth inhibition of HCC cells mediated by scAAV3-miRNA vectors was specific, it was important to examine the effect of these vectors on non-transformed hepatic cells. HepaRG cells are terminally differentiated hepatic cells, derived from a human hepatic progenitor cell line and retain many of the characteristics of primary human hepatocytes. These cells have been used to study various aspects of liver functions [53]. HepaRG cells were either mock-transduced or transduced with scAAV3-CBA-GLuc vectors or co-transduced with scAAV3-miR-26a-GLuc+scAAV3-miR-122-GLuc vectors as described for Huh7-FLuc cells. GLuc expression levels and total cell numbers were evaluated 72 hrs post-transduction. It is evident that no significant differences in GLuc expression (Figure 7A) or total cell numbers (Figure 7B) were observed.
Figure 7:
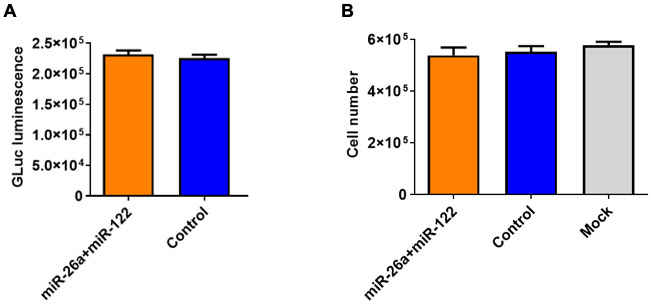
Lack of growth inhibition of HepaRG cells by scAAV3-miRNA vectors. A. GLuc expression and B. Total cell numbers. Cells were transduced with a total of 1x105 vgs/cell.
scAAV3-miRNA vectors suppress human liver tumor growth in a mouse xenograft model in vivo
Since cell culture studies in vitro alone are inadequate to evaluate the therapeutic potential of AAV-miRNA vectors, it was important to assess what effect, if any, transduction of Huh7-FLuc cells with AAV3-miRNA vectors had on the growth of human liver tumors induced by Huh7-FLuc in a mouse xenograft model. To this end, the following three groups of mice were set up for in vivo experiments: Group 1, injected with Huh7-FLuc cells only; Group 2, injected with Huh7-FLuc cells transduced with scAAV3-miR-26a-GLuc+scAAV3-miRNA-122-GLuc vectors; and Group 3, Huh7-FLuc cells transduced with scAAV3-CBA-GLuc vectors. Following subcutaneous injections of these cells in NSG mice (n=6 for each group), whole-body bioluminescence imaging was performed on day 7, 10, 13, 16, 23, and 30 post-injections. The day 7 results, shown in Figure 8, document that tumor growth of Huh7-FLuc cells transduced with scAAV3-miRNA vectors was significantly inhibited (P=0.009), compared with those induced by Huh7-FLuc cells, or Huh7-FLuc cells transduced with the control vector (P=0.23).
Figure 8:
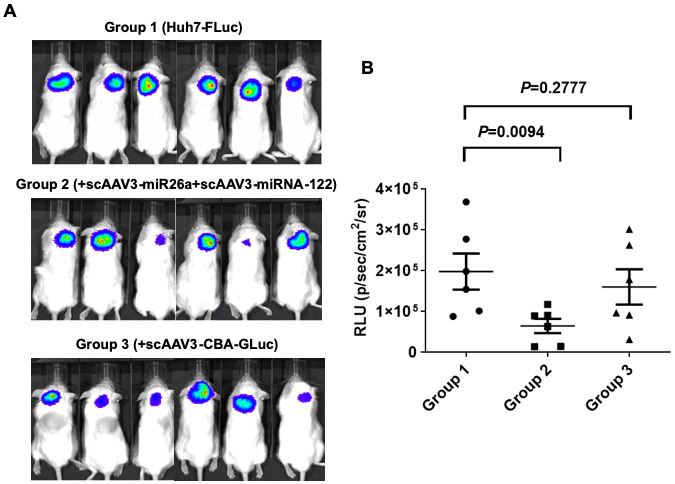
Suppression of growth of tumors induced by Huh7-FLuc cells transduced with scAAV3-miRNA vectors in vivo. A. Whole-body bioluminescence images of NSG mice 7-days post-injections of Huh7-FLuc cells (Group 1), and those transduced with scAAV3-miRNA-26a+scAAV3-miRNA-122 (Group 2), or scAAV3-CBA-GLuc (Group 3) vectors (n=6 for each Group). B. Quantitation of the luminescence signal intensity 7-days post-injections.
However, when tumor growth was monitored by whole-body bioluminescence in vivo imaging for up to 30 days post-injections, there were no significant differences among the 3 groups beyond day 10 (Figure 9), as expected, presumably due to the rapid rate of tumor growth, and consequently, the vector dilution. There were no significant differences in the weights of the mice among the three groups until 30 days post-injections (Figure 10) when the experiment was terminated due to the growth of liver tumors approaching the size of 1.5 cm. These results suggest that the combined use of scAAV3-miRNA-26a and scAAV3-miRNA-122 may prove to be an effective strategy to limit the early stages of human liver tumor growth.
Figure 9:
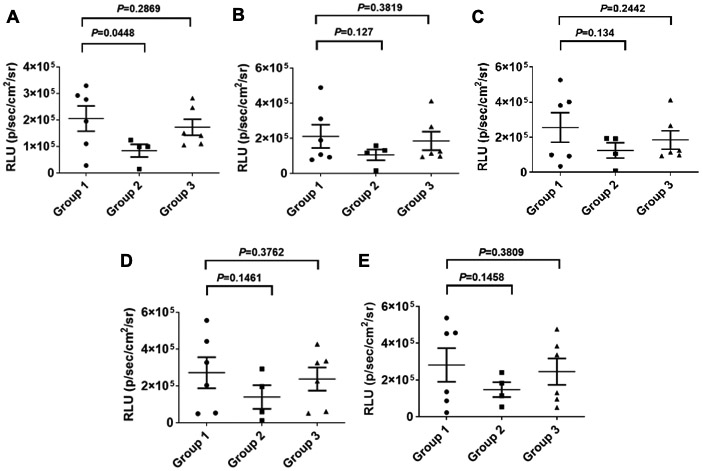
Suppression of growth of tumors induced by Huh7-FLuc cells transduced with scAAV3-miRNA vectors in vivo as a function of time. Quantitation of luminescence signal intensities at various time-points. A. Day 10, B. Day 13, C. Day 16, D. Day 23, and E. Day 30. Group 1: Huh7-FLuc cells; Group 2; scAAV3-miRNA-26a+scAAV3-miRNA-122 vectors; Group 3: scAAV3-CBA-GLuc vectors.
Figure 10:

Average weights of NSG mice injected with Huh7-FLuc cells (Group 1), and those transduced with scAAV3-miRNA-26a+scAAV3-miRNA-122 (Group 2), or scAAV3-CBA-GLuc (Group 3) vectors as a function of time. n=6 for each Group.
Discussion
We first reported that among the 10 most commonly used AAV serotype vectors, AAV3 vectors were the most efficient in transducing human hepatoblastoma (Huh6) and hepatocellular carcinoma (Huh7, HepG2) cell lines in vitro [54]. Subsequently, we also documented the remarkable tropism of AAV3 vectors for human liver tumors in a mouse xenograft model in vivo [46, 55]. Although we demonstrated some efficacy of AAV3 vectors expressing a plant gene, tricosanthin (TCS), in inhibiting human liver tumor growth in a mouse xenograft model in vivo [46], the extent of growth inhibition was neither absolute nor long-lasting [56].
In our quest to further develop more efficient AAV3 vectors for targeting human liver tumors, in the present studies, we explored the possibility of utilizing specific miRNAs for this purpose. Various miRNA-based-therapeutic approaches such as miRNA inhibition, miRNA replacement and oncolytic therapy have previously been employed [57]. More specifically, miRNA-26a and miRNA-122 have been utilized in preclinical mouse models (tet-o-MYC; LAP-tTAT and xenograft) by miRNA replacement and oncolytic therapy approaches [27, 28, 58]. In addition, miRNA-122 has also been used in a phase 2 clinical trial for the treatment of Hepatitis C virus (HCV) [59]. The endogenous expression of liver specific microRNA, miR-122, is more abundant than expression of miR-26a in Huh7-FLuc cells, which is consistent with previously published studies [60, 61].
In our studies, scAAV3-miR-26a and scAAV3-miR-122 vectors alone mediated a modest yet significant level of growth inhibition in Huh7. The observed growth inhibition observed was vector dose-dependent, but transient, due to rapidly proliferating nature of these cells and dilution of the vector genomes over time. Interestingly, however, no growth inhibition of HepaRG cells was observed under identical conditions (Figure 7), and the extent of growth inhibition of Huh7 cells-derived liver tumors was significantly more than that observed with a control vector (Figure 8), albeit short-lived, which is consistent with our previously published study [46]. Although additional studies are warranted to gain an insight into miRNA-26a- and miRNA-122-mediated growth inhibition, it is well known that miRNA-26a can silence p21, p27, and p57 tumor suppressor proteins and downregulate oncogenic proteins in the G1 cycle phase, cyclins and cyclin-dependent kinases (CDKs) [14]. miRNA-122 is also known to promote apoptosis by downregulating the expression of anti-apoptotic B-cell leukemia/lymphoma 2 (Bcl-2) family member proteins [62]. In addition, miRNA-122, which is a potential target for human liver cancer therapy/chemotherapy, can bind to the 3’-UTR of Bcl-2 family mRNAs to negatively regulate transcription according to the results from TARGETSCAN (http://www.Targetscan.org) and PICTAR (http://pictar.bio.nyu.edu) [63].
Our current studies are distinct from previously published studies [27, 28] in two following aspects: (i) In previous studies, AAV8 vectors, known to transduce mouse hepatocytes extremely efficiently [64, 65], were used to deliver miR-26a and miRNA-122, respectively. Since we and others have documented that AAV3 vectors are more efficient than AAV8 vectors in transducing human hepatocytes [66-68], we used AAV3-miRNA vectors; (ii) In previous studies, AAV8 vectors were used to target mouse liver tumors [27, 28], whereas in our studies, we targeted human liver tumors, albeit in a mouse xenograft model, since the ultimate objective is to pursue the potential gene therapy of human liver cancer.
However, one of the limitations of our current studies is that we did not evaluate the therapeutic potential of scAAV3-miRNA vectors post-tumor development as we did with scAAV3-TCS vectors [46]. Future possibilities include the following (i) the use of capsid-modified next generation of AAV3 vectors, which are significantly more efficient in targeting and transducing human liver tumors in vivo; the use of a single AAV3 vector expressing both miRNA-26a and miRNA-122; (iii) the use of additional therapeutic gene(s) , such as TCS, also co-expressing miRNA-26a and miRNA-122; and the combined use of the most efficient therapeutic scAAV3 vector in combination with the FDA-approved chemotherapeutic agents, such as Sorafenib [69], for the optimal gene+drug therapy of human liver cancer. Moreover, given the episomal nature of the recombinant AAV genome, it is prudent to consider the development of recombinant AAV vectors capable of undergoing stable integration for the potential long-term gene therapy of human cancers in general, and liver cancer in particular. These limitations notwithstanding, it is noteworthy that in contrast to the use of the less efficient AAV8 vectors expressing miRNA-26a and miRNA-122 led to near total inhibition of growth of mouse liver tumors [27, 28], but not with the use of the more efficient AAV3 vectors (our current studies), which further underscores not only the potentially inherent differences between liver tumors of mouse and human origin, but also the limitations of mouse models of human liver cancer. We, nonetheless, propose to perform future studies with capsid-optimized AAV3 vectors, which we have previously reported to be significantly more efficient in targeting human liver tumors in xenograft mice [45, 46] than the wild-type AAV3 vectors that were used in our current studies.
Acknowledgments
This research was supported in part by Public Health Service grants R01 HL-097088, R41 AI-122735, and R21 EB-015684 from the National Institutes of Health; a grant from the Children’s Miracle Network; and support from the Kitzman Foundation (to A.S.). L.Y. was supported by the International Postdoctoral Exchange Fellowship Program 2018 by the Office of China Postdoctoral Council.
Footnotes
Conflict of Interest
AS is a cofounder of, and holds equity in, Lacerta Therapeutics, aaVective, KASHX Bio, and Nirvana Therapeutics, and is an inventor on several issued patents on recombinant AAV vectors that have been licensed to various gene therapy companies. All other authors declare no conflict of interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Crissien AM, Frenette C. Current management of hepatocellular carcinoma. Gastroenterol Hepatol (N Y). 2014;10:153–161. [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Wang N, Cheung F, Lao L, Li C, Feng Y. Chinese medicines for prevention and treatment of human hepatocellular carcinoma: current progress on pharmacological actions and mechanisms. J Integr Med. 2015;13:142–164. [DOI] [PubMed] [Google Scholar]
- 4.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. [DOI] [PubMed] [Google Scholar]
- 5.Greene CM, Varley RB, Lawless MW. MicroRNAs and liver cancer associated with iron overload: Therapeutic targets unravelled. World J Gastroentero. 2013;19:5212–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erridge S, Pucher PH, Markar SR, Malietzis G, Athanasiou T, Darzi A, et al. Meta-analysis of determinants of survival following treatment of recurrent hepatocellular carcinoma. Br J Surg. 2017;104:1433–1442. [DOI] [PubMed] [Google Scholar]
- 7.Ling CQ, Fan J, Lin HS, Shen F, Xu ZY, Lin LZ, et al. Clinical practice guidelines for the treatment of primary liver cancer with integrative traditional Chinese and Western medicine. J Integr Med. 2018;16:236–248. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambrost V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-4. Cell. 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Port JD, Sucharov C. Role of microRNAs in cardiovascular disease: Therapeutic challenges and potentials. J Cardiovasc Pharmacol. 2010;56:444–453. [DOI] [PubMed] [Google Scholar]
- 11.Pager CT, Wehner KA, Fuchs G, Sarnow P. MicroRNA-mediated gene silencing. Prog Mol Biol Transl Sci. 2009;90:187–210. [DOI] [PubMed] [Google Scholar]
- 12.Hammond SM. MicroRNA therapeutics: A new niche for antisense nucleic acids. Trends Mol Med. 2006;12:99–101. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Zhang K, Xu YJ, Gao YP, Li C, Wang R, et al. The role of microRNA-26a in human cancer progression and clinical application. Tumor Biol. 2016;37:7095–7108. [DOI] [PubMed] [Google Scholar]
- 14.Sartorius K, Sartorius B, Cheryl W, Chuturgoon A, Makarova J. The biological and diagnostic role of mirna’s in hepatocellular carcinoma. Front Biosci. 2018;23:1701–1720. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, et al. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. [DOI] [PubMed] [Google Scholar]
- 16.Chai Z, Kong J, Zhu X, Zhang Y, Lu L, Zhou J, et al. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2α/Akt/HIF-1α pathway in hepatocellular carcinoma. PLoS One. 2013;8:e77957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Zhang XF, Lu X, Jia HL, Liang L, Dong QZ, et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology. 2014;59:1874–1885. [DOI] [PubMed] [Google Scholar]
- 18.Ji JF, Shi J, Budhu A, Yu ZP, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Zhu XM, Wu LJ, Yang R, Yang ZR, Wang QF, et al. microRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int. 2012;32:752–760. [DOI] [PubMed] [Google Scholar]
- 21.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Hsu SH, Wang XM, Kutay H, Bid HK, Yu JH, et al. Reciprocal regulation of microRNA-122 and c-Myc in hepatocellular cancer: Role of E2F1 and TFDP2. Hepatology. 2014;59:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan CG, Wang CM, Tian C, Wang Y, Li L, Sun WS, et al. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26:1281–1286. [DOI] [PubMed] [Google Scholar]
- 24.Sun J, Lu HQ, Wang X, Jin HC. MicroRNAs in hepatocellular carcinoma: Regulation, function, and clinical implications. ScientificWorldJournal. 2013;2013:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. [DOI] [PubMed] [Google Scholar]
- 26.Li MX, Jayandharan GR, Li BZ, Ling C, Ma WQ, Srivastava A, et al. High-Efficiency transduction of fibroblasts and mesenchymal stem cells by Tyrosine-Mutant AAV2 vectors for their potential use in cellular therapy. Hum Gene Ther. 2010;21:1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu SH, Wang B, Kota J, Yu JH, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. [DOI] [PubMed] [Google Scholar]
- 30.Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr, Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, et al. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno–associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cideciyan AV, Aleman TS, Boye SL, Schwartz SB, Kaushal S, Roman AJ, et al. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc Natl Acad Sci U S A. 2008;105:15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaudet D, Méthot J, Déry S, Brisson D, Essiembre C, Tremblay G, et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther. 2013;20:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, et al. Adenovirus-associated virus vector-mediated gene transfer in Hemophilia B. N Engl J Med. 2011;365:2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of Factor IX gene therapy in Hemophilia B. N Engl J Med. 2014;371:1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miesbach W, Meijer K, Coppens M, Kampmann P, Klamroth R, Schutgens R, et al. Gene therapy with adeno-associated virus vector 5-human Factor IX in adults with Hemophilia B. Blood. 2018;131:1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George LA, Sullivan SK, Giermasz A, Rasko JEJ, Samelson-Jones BJ, Ducore J, et al. Hemophilia B gene therapy with a high-specific-activity Factor IX variant. N Engl J Med. 2017;377:2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rangarajan S, Walsh L, Lester W, Perry D, Madan B, Laffan M, et al. AAV5-Factor VIII gene transfer in severe Hemophilia A. N Engl J Med. 2017;377:2519–2530. [DOI] [PubMed] [Google Scholar]
- 39.Pasi KJ, Rangarajan S, Mitchell N, Lester W, Symington E, Madan B, et al. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for Hemophilia A. N Engl J Med. 2020;382:29–40. [DOI] [PubMed] [Google Scholar]
- 40.Hwu WL, Muramatsu S, Tseng SH, Tzen KY, Lee NC, Chien YH, et al. Gene therapy for aromatic L-amino acid decarboxylase deficiency. Sci Transl Med. 2012;4:134ra61. [DOI] [PubMed] [Google Scholar]
- 41.MacLaren RE, Groppe M, Barnard AR, Cottriall CL, Tolmachova T, Seymour L, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guy J, Feuer WJ, Davis JL, Porciatti V, Gonzalez PJ, Koilkonda RD, et al. Gene therapy for Leber hereditary optic neuropathy: Low- and medium-dose visual results. Ophthalmology. 2017;124:1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–1722. [DOI] [PubMed] [Google Scholar]
- 44.Ling C, Lu Y, Cheng B, McGoogan KE, Gee SWY, Ma WQ, et al. High-Efficiency transduction of liver cancer cells by recombinant adeno-associated virus serotype 3 vectors. J Vis Exp. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng BB, Ling C, Dai Y, Lu Y, Glushakova LG, Gee SWY, et al. Development of optimized AAV3 serotype vectors: Mechanism of high-efficiency transduction of human liver cancer cells. Gene Ther. 2012;19:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling C, Wang Y, Zhang YH, Ejjigani A, Yin ZF, Lu Y, et al. Selective in vivo targeting of human liver tumors by optimized AAV3 vectors in a murine xenograft model. Hum Gene Ther. 2014;25:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Joo KI, Wang P. Endocytic processing of adeno-associated virus type 8 vectors for transduction of target cells. Gene Ther. 2013;20:308–317. [DOI] [PubMed] [Google Scholar]
- 48.Kube DM, Srivastava A. Quantitative DNA slot blot analysis: Inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997;25:3375–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling C, Wang Y, Lu Y, Wang LN, Jayandharan GR, Aslanidi GV, et al. Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J Virol. 2015;89:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aslanidi GV, Rivers AE, Ortiz L, Song L, Ling C, Govindasamy L, et al. Optimization of the capsid of recombinant adeno-associated virus 2 (AAV2) vectors: The final threshold? PLoS One. 2013;8:e59142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 52.Nowrouzi A, Penaud-Budloo M, Kaeppel C, Appelt U, Le Guiner C, Moullier P, et al. Integration frequency and intermolecular recombination of rAAV vectors in non-human primate skeletal muscle and liver. Mol Ther. 2012;20:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem-Biol Interact. 2007;168:66–73. [DOI] [PubMed] [Google Scholar]
- 54.Glushakova LG, Lisankie MJ, Eruslanov EB, Ojano-Dirain C, Zolotukhin I, Liu C, et al. AAV3-mediated transfer and expression of the pyruvate dehydrogenase E1 alpha subunit gene causes metabolic remodeling and apoptosis of human liver cancer cells. Mol Genet Metab. 2009;98:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ling C, Li BZ, Ma WQ, Srivastava A. Development of optimized AAV serotype vectors for high-efficiency transduction at further reduced doses. Hum Gene Ther Method. 2016;27:143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang YH, Wang Y, Yusufali AH, Ashby F, Zhang D, Yin ZF, et al. Cytotoxic genes from traditional Chinese medicine inhibit tumor growth both in vitro and in vivo. J Integr Med. 2014;12:483–494. [DOI] [PubMed] [Google Scholar]
- 57.Qadir MI, Rizvi SZ. MiRNA in hepatocellular carcinoma: Pathogenesis and therapeutic approaches. Crit Rev Eukaryot Gene Expr. 2017;27:355–361. [DOI] [PubMed] [Google Scholar]
- 58.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122—a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. [DOI] [PubMed] [Google Scholar]
- 59.Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. [DOI] [PubMed] [Google Scholar]
- 60.Chang L, Li K, Guo T. miR-26a-5p suppresses tumor metastasis by regulating EMT and is associated with prognosis in HCC. Clin Transl Oncol. 2017;19:695–703. [DOI] [PubMed] [Google Scholar]
- 61.Fukuhara T, Kambara H, Shiokawa M, Ono C, Katoh H, Morita E, et al. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J Virol. 2012; 86:7918–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol Res. 2016;46:391–406. [DOI] [PubMed] [Google Scholar]
- 63.Yin J, Tang HF, Xiang Q, Yu J, Yang XY, Hu N, et al. MiR-122 increases sensitivity of drug-resistant BEL-7402/5-FU cells to 5-fluorouracil via down-regulation of bcl-2 family proteins. Pharmazie. 2011;66:975–981. [PubMed] [Google Scholar]
- 64.Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci U S A. 2003;100:6081–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarkar R, Tetreault R, Gao G, Wang L, Bell P, Chandler R, et al. Total correction of Hemophilia A mice with canine FVIII using an AAV 8 serotype. Blood. 2004;103:1253–1260. [DOI] [PubMed] [Google Scholar]
- 66.Li S, Ling C, Zhong L, Li M, Su Q, He R, et al. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol Ther. 2015;23:1867–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Bell P, Somanathan S, Wang Q, He Z, Yu H, et al. Comparative study of liver gene transfer with AAV vectors based on natural and engineered AAV capsids. Mol Ther. 2015;23:1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vercauteren K, Hoffman BE, Zolotukhin I, Keeler GD, Xiao JW, Basner-Tschakarjan E, et al. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol Ther. 2016;24:1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.El-Serag HB. Current status of Sorafenib use for treatment of hepatocellular carcinoma. Gastroenterol Hepatol (N Y). 2017;13:623–625. [PMC free article] [PubMed] [Google Scholar]


