Noncoding “junk DNA”, which constitutes 98% of our genome, is now generally considered to play a fundamental role in the precise regulation of coding genes to establish cell identity. One feature of functional “junk DNA” is its accessibility. In cancer cells, aberrant chromatin accessibility is recognized as one of the major hallmarks. Understanding such events requires high-throughput screening, such as Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), which generates large-scale data that are puzzling for biologists. However, existing web tools only support cell line data and lack high-quality clinical phenotypes or matched transcriptomes. Here, we developed Shiny Pan-cancer Accessible Chromatin Explorer (SPACE) as an all-in-one web server encompassing 562,709 regulatory elements in 404 patients across 23 cancer types. To the best of our knowledge, SPACE is the first web application supporting ATAC-seq analysis with matched clinical phenotype and transcriptome data. The current version of SPACE supports the following: (i) searching of ranked regulatory elements (peaks); (ii) exploration of potential regulators of peaks; (iii) examination of the clinical relevance of selected peaks; (iv) prediction of peak-related pathways; (v) correlations of peaks with RNAs; and (vi) exploration of the role of peaks in the immune-related tumor microenvironment. Thus, SPACE is an operable and time-saving epigenetic resource for cancer biologists to examine the transcriptional and phenotypic consequences of DNA regulatory elements. SPACE is available at http://fun-science.club/SPACE.
In cancer cells, dynamic DNA accessibility orchestrates a precise regulatory network to foster oncogenes in cancer. State-of-the-art high-throughput technologies, such as ATAC-seq, can provide a precise map for understanding how chromatin states manipulate oncogenesis, which places ATAC-seq right in the limelight of the epigenetics community.1 However, existing online tools, such as Cistrome2 or ENCODE,3 are useful for cell-line-based analysis but do little in clinical sample-based analysis. Thus, there is a pressing need for developing a user-friendly resource of regulatory elements with high-quality phenotypes or matched transcriptomes. Most recently, newly published ATAC-seq datasets of TCGA samples make it possible to explore the clinical value of chromatin accessibility.1 However, large-scale data generated by ATAC-seq are hard for biologists to directly analyze. Linking the epigenome to matched transcriptional and clinical information requires programming skills and IT infrastructure.
Here, we developed SPACE (http://fun-science.club/SPACE) as an all-in-one web server to enable cancer biologists to analyze and visualize 562,709 accessible DNA elements from 404 patients across 23 cancer types. By integrating available datasets from public domains such as TCGA and ENCODE, SPACE allows users to search peaks (by gene or genomic regions) and further annotate peaks (super-enhancer, transcription factor (TF) binding, cis-regulatory, Expression quantitative trait loci (eQTL), and literature). With the matched transcriptome data, SPACE documents 17,638,113,605 peak-mRNA/lncRNA/miRNA pairs and their phenotypic association, such as prognosis or cancer subtype. Users can also predict peak-associated pathways and their role in the immune microenvironment. SPACE can serve as a useful resource to explore the roles of DNA regulatory elements and their associated transcriptional changes as well as the clinical relevance in cancer patients.
The user interface of SPACE was written in R Shiny. The detailed procedures of data processing are described on the document page of SPACE. In brief, SPACE supports an integrative analysis of ATAC-seq, RNA-seq, Chromatin Immunoprecipitation using sequencing (ChIP-seq), enhancers, 3D chromatin loops, literature mining, and eQTL analysis. Analysis results are returned to the web page and can be downloaded in PDF, XLS, CSV, and TXT formats. The workflow and typical output schema are shown in Fig. S1.
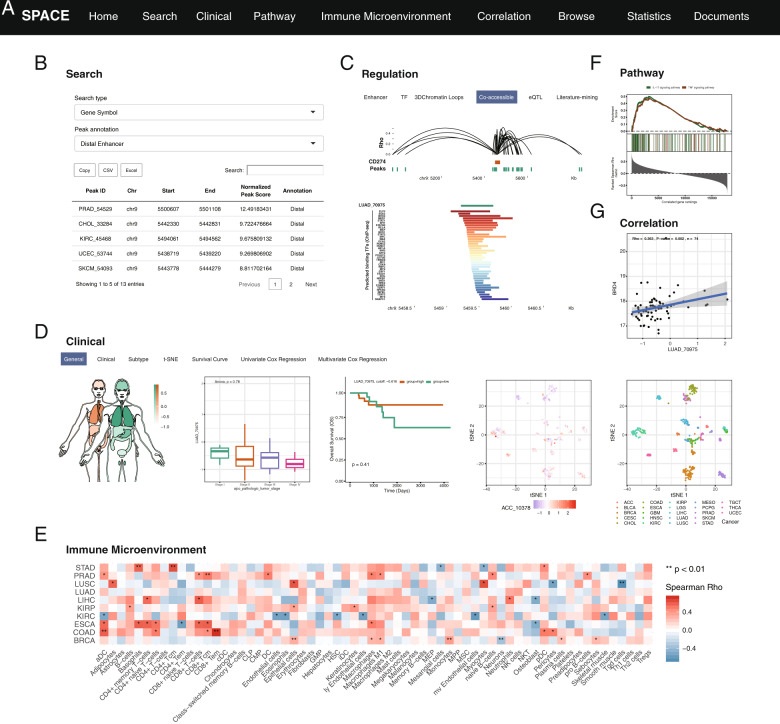
SPACE provides the following six analysis modules (Fig. 1a):
Search module: this module allows users to search ATAC-seq peaks (n = 562,709) according to gene symbols or genomic regions. In the search results, users can rank the queried peaks according to the normalized peak score, annotation (i.e., distal enhancer, promoter, and intron), and cancer type (Figs. 1b, S2a).
Regulation module: in this module, users can easily explore the regulatory role of peaks of their interest. Users can determine whether that peak overlaps with a super-enhancer or cancer-expressed enhancer (Figs. 1c, S2g). The TF panel integrates ChIP-seq data of TFs from the ENCODE project and other public ChIP-seq data from GEO (Fig. S2b). In the cis-regulatory panel, the server automatically screens the coaccessible peaks with the input gene and predicts the potential cis-regulatory peaks. In the eQTL panel, users can search the cis or trans eQTLs of peaks of their interest. The literature-mining panel integrates published experimentally supported enhancers and variants extracted from full-text publications.
Clinical module: this module integrates ATAC-seq profiles of 404 samples across 23 cancer types with matched transcriptome data and clinical phenotypes. Users can view the expression profiles of peaks and test whether those peaks are associated with cancer subtypes, patient prognosis, and clinical stages (Fig. 1d). The T-SNE panel allows users to determine the score distribution of peaks among 404 samples. Users can also conduct survival Cox regression analysis, which is widely used by clinicians.
Pathway module: this module computes the association between 20,932 precalculated pathway signatures and peaks. Users can also perform the gene set enrichment analysis of peak-related genes. SPACE provides a heat map, an interactive volcano plot, and scatter plots to visualize the results (Fig. 1e).
Immune microenvironment module: in this module, users can query peaks to compute their association with the immune microenvironment (64 immune cells or stromal cells, 24 immunoinhibitors, and 45 immunostimutators) (Fig. 1f).
Correlation module: users are able to explore potential cooperators (mRNA, lncRNA, and miRNA) of the inputted peak (Fig. 1g). We also documented 95 histones (16 subgroups such as chromatin remodeling) and 1664 TFs (72 subfamilies such as T-box and MYB).
Fig. 1.
Overview of SPACE. a Top navigation bar of SPACE functions. b The search module allows users to search regulatory elements by genomic regions or genes. c Main function of the regulation panel. The upper panel shows the coaccessible peaks of PD-L1 (encoded by CD274). The lower panel shows the potential TFs regulating the LUAD_70975 peak. d The clinical panel allows users to explore the clinical association of a given peak. The five plots show the score distribution across organs, peak score in distinct tumor stages, prognostic value of a peak, tSNE distribution of a given peak, and tSNE of all samples (from left to right). e The immune microenvironment module shows the association between a given peak and 64 immune cells and stromal cells in the tumor microenvironment. f The pathway module allows users to perform gene set enrichment analysis of peak-related genes. g Correlation between BRD4 and LUAD_70975, which can be reproduced in the TF-histone submodule
To validate the computational results of SPACE, we present two cases. First, we used SPACE to explore the regulatory elements (peaks) of PD-L1 (encoded by CD274), an important cancer immunotherapy target.4 The search module identifies 38 nearby peaks within the 100 kb distance to the TSS of CD274 across different cancer types. Among those peaks, the top ranked distal enhancer peak is PRAD_54529 (chr9:5500607-5501108, hg38, Fig. S2a). The enhancer prediction module shows that this peak is located on a super-enhancer (SE_02_28000196) in breast cancer cells (SUM-159), which is in accordance with our recent report.5 The TF submodule shows that 56 TFs, including BRD4 (ENCODE and public ChIP-seq combined), can bind to this regulatory element (Fig. S2c). In SUM-159 breast cancer cells, available data (GSE87424) indicate that BRD4 blockade can inhibit the activity of this peak and therefore downregulate its target gene PD-L1 (Fig. S2d). The importance of this region as well as BRD4 binding was further confirmed by the fact that BRD4 inhibition could greatly reduce the binding of BRD4, MED1, and EP300 to peak PRAD_54529 (Fig. S2e). Knocking out this region in SUM-159 cells can downregulate PD-L1 based on RNA-seq (Fig. S2f). These data confirm that SPACE can predict enhancers of a given gene and their binding TFs.
Second, a recent paper reported the super-enhancer (chr8:128154301-128178044) of the MYC oncogene in lung adenocarcinoma.6 As shown in Fig. S2g, the coaccessible submodule identified a similar enhancer cluster, which is in accordance with the super-enhancer definition. The correlation module shows that all predicted enhancers are highly associated with MYC expression (Fig. S2h, i). This case shows that SPACE prediction is consistent with experimentally validated results. SPACE prediction is a powerful tool for identifying novel functional and clinically relevant regulatory DNA elements of genes of interest.
In summary, SPACE is an all-in-one web server of chromatin accessibility compared with other related resources (Table S1). To the best of our knowledge, SPACE is the first web application supporting ATAC-seq analysis with matched clinical phenotype and transcriptome data. We will continue updating datasets and maintaining web servers in the future. We hope SPACE will be a time-saving tool for cancer biologists to test their hypotheses and a valuable resource for the epigenetics community.
Supplementary information
Acknowledgments
Funding
This work was supported by the National Natural Science Foundation of China (31770935; 81873531; 31970616); the Distinguished Professorship Program of Jiangsu Province to YF; the Distinguished Professorship Program of Jiangsu Province to RM; the National Undergraduate Training Programs for Innovation (201710304030Z); and the National Undergraduate Training Programs for Innovation (201810304026Z).
Competing interests
The authors declare no competing interests.
Contributor Information
Renfang Mao, Email: maorenfang@ntu.edu.cn.
Yihui Fan, Email: fanyihui@ntu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41423-020-0416-9) contains supplementary material.
References
- 1.Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science362, 10.1126/science.aav1898 (2018). [DOI] [PMC free article] [PubMed]
- 2.Zheng R, et al. Cistrome data browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47:D729–D735. doi: 10.1093/nar/gky1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis CA, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 2019;19:133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, et al. A tumor-specific super-enhancer drives immune evasion by guiding synchronous expression of PD-L1 and PD-L2. Cell Rep. 2019;29:3435–3447.e4. doi: 10.1016/j.celrep.2019.10.093. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat. Genet. 2016;48:176–182. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.