Abstract
A drop of seawater contains numerous microspatial niches at the scale relevant to microbial activities. Examples are abiotic niches such as detrital particles that show different sizes and organic contents, and biotic niches resulting from bacteria–phage and bacteria–phytoplankton interactions. A common practice to investigate the impact of microenvironments on bacterial evolution is to separate the microenvironments physically and compare the bacterial inhabitants from each. It remains poorly understood, however, which microenvironment primarily drives bacterioplankton evolution in the pelagic ocean. By applying a dilution cultivation approach to an undisturbed coastal water sample, we isolate a bacterial population affiliated with the globally dominant Roseobacter group. Although varying at just a few thousand nucleotide sites across the whole genomes, members of this clonal population are diverging into two genetically separated subspecies. Genes underlying speciation are not unique to subspecies but instead clustered at the shared regions that represent ~6% of the genomic DNA. They are primarily involved in vitamin synthesis, motility, oxidative defense, carbohydrate, and amino acid utilization, consistent with the known strategies that roseobacters take to interact with phytoplankton and particles. Physiological assays corroborate that one subspecies outcompetes the other in these traits. Our results indicate that the microenvironments in the pelagic ocean represented by phytoplankton and organic particles are likely important niches that drive the cryptic speciation of the Roseobacter population, though microhabitats contributed by other less abundant pelagic hosts cannot be ruled out.
Subject terms: Microbial ecology, Population genetics
Introduction
While waters at the ocean surface are a seemingly well-mixed and diluted matrix, growing evidence has shown that nutrients are not homogeneously distributed at the scale relevant to microbial activities. Marine snow and other large organic particles, for example, can contain high concentrations of organic and inorganic nutrients that exceed those in the bulk seawater by two to four orders of magnitude [1, 2]. Besides these “visible” microscale habitats, the ocean water represents a continuum of organic matter size distribution ranging from the truly dissolved phase, colloids, transparent polymers, to organic gels [3]. These microenvironments often arise from ephemeral nutrient pulses including viral lysis, sloppy feeding, particle sinking, phytoplankton photosynthetic release, and turbulent flow [3–5]. These processes provide transient microscale hotspots at nanometer to millimeter scales, harboring nutrient concentrations up to three orders of magnitude higher than the bulk seawater [6]. Accordingly, many marine bacteria developed novel strategies such as fast motility to respond to these transient processes [7].
Recent studies suggest that biotic interactions among microbial groups such as bacteria–phage [8, 9] and bacteria–phytoplankton interactions [10, 11] are an important source of microscale heterogeneity. Typically, an average milliliter of a nearshore water sample is inhabited by 10,000,000 viruses, 1,000,000 bacteria, 100,000 cyanobacterial cells, 1000 nanoflagellates, 10 dinoflagellates, and 1 diatom [3]. Among the bacterial lineages, members of the Roseobacter group in Alphaproteobacteria are particularly abundant in coastal waters, making up one out of five bacterial cells [12]. An increasing number of phages that infect the Roseobacter group were isolated [13, 14], and they are known to alter the metabolism and shape the population structure of the Roseobacter hosts. For example, one study showed that phage-infected Roseobacter cells contain elevated intracellular metabolites compared to uninfected cells and that the infected cells release labile compounds which are utilized by uninfected ones [15], suggesting that phage infection leads to resource re-partitioning among cells. Another study showed that the distinct phylogenomic clusters of a Roseobacter population isolated from global oceans are not correlated with geographic regions or environmental conditions, but instead are differentiated by subtyping with their co-isolated phages [16], suggesting that phages may play a key role in driving Roseobacter population differentiation and speciation.
Roseobacters are also among the most abundant bacterial lineages associated with three dominant eukaryotic phytoplankton groups in today’s ocean [17–20] including diatoms [21, 22], coccolithophores [23, 24], and dinoflagellates [25, 26]. Much work has been done related to Roseobacter–phytoplankton interactions, which were proposed to occur within phycosphere, zones of concentrated DOM with a few cell diameters surrounding individual phytoplankton cells [10, 27, 28]. For example, in a simple mutualistic laboratory co-culture system, the model Roseobacter strain Ruegeria pomeroyi DSS-3 [29] utilizes the fixed carbon and sulfonate compounds released from the co-cultured diatom and, in return, the bacterium supplies vitamin B12 to the diatom [30]. Another Roseobacter lineage Sulfitobacter prevalently co-occurs with diatoms [21], and strains of this lineage isolated from a diatom species were shown to promote the diatom reproduction by secreting the phytohormone indole-3-acetic acid (IAA), which is synthesized from the diatom-derived or intracellular tryptophan [22]. In addition to these mutualistic interactions, select Roseobacter members act as pathogens to some phytoplankton lineages. For example, a Sulfitobacter member isolated from a coccolithophore species was shown to have algicidal effects on the latter, and the bacterial virulence is enhanced by the dimethylsulfoniopropionate released from the alga [31]. Another study based on a laboratory co-culture of a Roseobacter species Phaeobacter inhibens and a coccolithophore species showed that the bacterium acts initially as a mutualist with the young alga but later becomes a pathogen when the alga gets older, and that the switch of the bacterium’s role depends on the concentration of IAA that the bacterium synthesizes from tryptophan exuded by the algal cells [32]. It is worth mentioning that whether the Roseobacter–phytoplankton interactions indeed occur within phycosphere has not been directly tested due to methodological limitations.
These studies suggest that Roseobacter–phage and Roseobacter–phytoplankton interactions lead to the creation of numerous microscale environments, but whether these niches are equally available to drive evolution of the wild Roseobacter population remains poorly understood. To address this question, we employed a dilution cultivation strategy which provides a better chance to obtain representative Roseobacter members. A single coastal seawater sample was kept from shaking and was not subjected to filtration before bacterial isolation for the preservation of the microenvironments, which differs from the previous studies in which visible niches (e.g., particles and water column) were separated and bacteria from each were collected. Our strategy led to the cultivation of 16 Roseobacter isolates identical at the 16S rRNA gene sequences. Detailed genome-wide single nucleotide polymorphism analyses showed that this population is under ongoing speciation, and functional analyses and physiological assays suggest that the phycosphere and organic particles are the likely niches that drove the ongoing speciation, though microenvironments provided by other less abundant pelagic hosts cannot be excluded. Our procedure also led to the isolation of a gammaproteobacterial Marinobacterium population, whose evolutionary pattern was analyzed and made comparison to that of the Roseobacter population.
Materials and methods
A Roseobacter population and a Marinobacterium population each consisting of 16 isolates were collected from a single 1-L sample of surface seawater at the Southwestern North Pacific coast using a dilution cultivation approach (Fig. S1). Genomes of the 32 strains were sequenced using Illumina, and the genome of the Roseobacter strain xm-d-517 was additionally sequenced using PacBio and assembled into a closed genome with one chromosome and one plasmid.
The main body of the population-level analyses consists of seven parts. The first five parts were done for both populations and the remaining two parts were exclusive to the Roseobacter population. Different parts are interconnected, and the detailed rationale was elaborated in the “Results and discussion”. First, the genetic diversity within each population was approximated by the number of SNPs per Mbp, which was calculated based on the whole-genome alignment produced by progressiveMauve v2.3.1 [33]. Second, a maximum-likelihood phylogenomic tree was constructed for each population using RAxML v8.1.22 [34] based on concatenated single-copy core genes at the amino acid level. The trees were rooted using their most closely related genomes available in Genbank. The two deeply branching clades were named Clade R-I and Clade R-II for the Roseobacter population and named Clade M-I and Clade M-II for the Marinobacterium population. These genome trees were used in the following five parts. Third, the relative frequency of recombination to mutation (ρ/θ) and the relative effect of recombination to mutation (r/m) within each population was determined using ClonalFrameML v1.1 [35]. The software also identifies the recombined DNA segments and further differentiates the segments recombined with external lineages from those within the population. Fourth, whether the population subdivision occurred (i.e., whether independent gene pool exists) and if subdivided, whether the genetic separation is congruent to the phylogenetic separation, was tested using fineSTRUCTURE [36]. Fifth, the core genomic regions underlying population differentiation were identified by comparing SNP density within each clade and between the two clades of each population. Here, the SNP density was measured as the number of SNPs in an overlapping sliding window of 10 kbp over the shared nucleotide sites along the whole-genome alignment. The differentiated genomic regions are expected to show increased SNP density in between-clade comparisons (e.g., Clade R-I versus Clade R-II) because of the fixation of distinct alleles within each clade. Sixth, if the differentiated core genome regions were the result of allelic replacements with divergent lineages, these regions likely left a signature with an unusually large evolutionary rate at the synonymous sites, measured as the number of synonymous substitutions per synonymous site (dS). The genes with unusually large dS values were identified using a recently developed approach [37], which clusters gene families based on the pairwise dS values using the k-means clustering method and subsequently identifies outlier gene families with unusually large dS values. It also allows the inference of the candidate ancestral branches at which recombination occurred with an external lineage (i.e., the allele donor), but identification of the exact branch requires phylogenetic analyses (see the seventh part). The outlier dS analysis also has little information about the identity of the donor, and again phylogenetic analyses help. Seventh, the exact ancestral branch with divergent allele replacements and the potential donors of divergent alleles were identified by comparing the gene trees with the genome tree. Due to the lack of appropriate outgroup lineages, the gene trees were rooted using the midpoint rooting approach, where we postulated that the evolutionary rate at the synonymous sites is constant among closely related lineages.
According to the functional annotations of genes triggering differentiation in the Roseobacter population, a series of assays were performed to establish the potential link between phenotypic and genotypic variation. These assays tended to test swimming motility, sedimentation phenotype, tolerance to H2O2-mediated oxidative stresses, tolerance to NaCl-induced osmotic stress. In addition, the phenotype microarray (PM) technology from BiOLOGTM was used to test the differences in substrate (190 carbon sources) utilization among the representative strains from the two diverged clades. All technical details are provided in the Text S1.
Results and discussion
Genomic features of the Roseobacter population varying at the strain level
The Roseobacter population consisting of 16 strains related to genus Aliiroseovarius was isolated from a single coastal seawater sample (physicochemical data shown in Table S1). Six types of seawater media were used to isolate roseobacters, and five of them recovered strains that contributed to this population. The strains isolated with each medium were named with a corresponding prefix (Table S2). Isolates in the Roseobacter population share identical 16S rRNA gene sequences with the average nucleotide identity (ANI, a measure of genome-wide DNA sequence identity between any two strains) of 99.76 ± 0.18%. After normalizing the genome size (3.33 ± 0.07 Mbp, more genome statistics summarized in Table S3), we showed that the SNP density across the core genome of the Roseobacter population is 4242 per Mbp (Table S4).
The level of genetic diversity in the Roseobacter population described here is far lower than other bacterial populations collected from environmental samples (Table S4). For example, a marine Vibrio population with 20 strains harboring identical 16S rRNA genes has more than 30,000 SNPs/Mbp in their core genomes [38]. In the studies of host-associated bacterial populations, the term “genetically monomorphic population” was coined to describe the lineages containing very few polymorphisms [39]. Examples include a population of typhoid-causing Salmonella enterica with only 446 SNPs/Mbp in 19 strains [40] and a population of plague-causing Yersinia pestis with only 284 SNPs/Mbp in 17 strains [41] (Table S4). The extremely low level of polymorphisms for these host-restricted pathogens is consistent with an evolutionary path dominated by genetic drift [40] or clonal diversification [41, 42]. While the SNP density of the Roseobacter population presented here is not as low as that of the “genetically monomorphic populations,” it is nevertheless the lowest among all studied bacterial and archaeal populations sampled from the ocean, soil, and hot spring (Table S4). This extremely low genetic diversity provides a unique opportunity to look into the early events and mechanisms that drove population differentiation.
Phylogenetic and population structure of the Roseobacter population
The maximum-likelihood phylogeny based on concatenation of all single-copy core genes (the tree shown on the left of Fig. 1a) showed that the Roseobacter population has diverged into two genotypic clusters (hereafter Clade R-I and Clade R-II). Note that these bacteria are not clustered according to the media with which these bacteria were isolated, suggesting that the isolation procedure has no impact on the Roseobacter population structure. For example, Clade R-I consists of three nearly identical strains, each isolated with a distinct medium. Likewise, the remaining 13 more diverse Clade R-II strains are further clustered to four sub-clades, none of which have their members isolated with a single medium. Furthermore, the topological structure of this phylogeny is consistent with the clonal genealogy reconstructed using ClonalFrameML (the tree shown on the left of Fig. 1b), which considers the effect of homologous recombination (HR) on the reconstruction of clonal relationships among individuals in a population [35]. Under the assumption that all recombination events from external sources introduce novel polymorphisms, the model in ClonalFrameML is able to distinguish the substitutions introduced by recombination from the ones caused by point mutations, resulting in an accurate inference of genealogy [35].
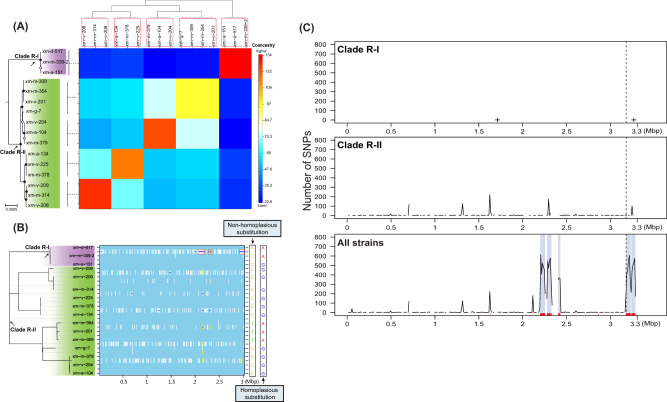
Fig. 1. Differentiation of the Roseobacter population.
a The RAxML maximum-likelihood phylogenomic tree and the fineSTRUCTURE coancestry matrix. The rooted phylogenomic tree is shown on the left (the outgroup not shown). Solid and open circles at the nodes indicate the frequency of the group defined by that node is at least 95 and 80, respectively, in the 100 bootstrapped replicates. The scale bar indicates the number of substitutions per site. The two most deeply branching clades, Clade R-I and Clade R-II, are highlighted in purple and green, respectively. The last common ancestor (LCA) of each clade is marked with an arrow. The coancestry matrix is shown on the right, with warmer colors representing greater percentages of shared ancestry between the strains under comparison. Strains assigned to the same fineSTRUCTURE coancestry population are highlighted with red-dashed boxes, and the dendrogram shows a clustering of the coancestry populations based on the proportion of shared ancestry between coancestry populations using an MCMC model-based clustering method. A vertical bar indicates strains belonging to a monophyletic group, which is connected to the coancestry population through a dashed black line. b The ClonalFrameML inference of recombination events. The genome phylogeny on the left represents a clonal tree after the recombinant DNA segments are removed. On the right is the representation of recombination events along the core genomes of the population for each extant and ancestral branch of the phylogeny. Light blue of the background indicates no substitutions. White vertical bars denote nonhomoplasious substitutions introduced by mutations or replacements of novel alleles from external lineages, whereas the yellow/orange vertical bars refer to homoplasious substitutions usually contributed by intraspecific recombination events. Homoplasious/nonhomoplasious substitutions are each illustrated with an example next to the representation of recombination events. The dark blue horizontal bars represent recombination events, and three long recombined DNA fragments in the Roseobacter population are highlighted with red boxes. c The distribution of SNPs along the genome. The closed genome of strain xm-d-517 is used as the reference genome to count the SNPs within 10-kb sliding windows. The SNP density is counted within Clade R-I and within Clade R-II, respectively, as well as among all strains from these two clades pooled together. The vertical dashed line sets the boundary of the chromosome and the plasmid. The cross symbols in the top plot represent the two SNPs occurring among members of Clade R-I. The rectangular boxes in light blue at the bottom plot indicate the locations of the recombined long DNA segments inferred with ClonalFrameML shown in (b). The red vertical bars over the x-axis represent the 176 core gene families with evidence of divergent allelic replacements based on the outlier dS analysis.
An important parameter characterizing the bacterial population structure is the relative importance of recombination to mutation [43–45]. The frequency of recombination relative to the rate of point mutation (the ρ/θ ratio) was estimated using ClonalFrameML. The results showed that recombination occurs less than once every ten mutation events (ρ/θ = 0.076) in the Roseobacter population. Fraser et al. employed a computer simulation and proposed that ρ/θ of 0.25–0.5 as the threshold delineating a clonal versus a sexual bacterial population [46]. According to this criterion, the Roseobacter population has a clonal population structure. Moreover, ClonalFrameML quantifies the effect of recombination relative to mutation on genetic diversity (the r/m ratio), a statistic that considers ρ/θ, the length of recombined DNA segments, and the allelic divergence [35, 44]. Thus, the r/m ratio estimates the probability that genetic variation at single nucleotide sites is caused by recombination relative to that resulting from mutation. The ClonalFrameML results showed that r/m is ~18 in the Roseobacter population, suggesting that recombination has a much greater impact on genetic diversity than mutation in this population. The very high r/m but very low ρ/θ of this population suggests that recombination events occurred rarely but in potentially long and divergent DNA segments. This hypothesis is supported by the graphical presentation derived from ClonalFrameML (Fig. 1b), which shows that recombination is rare and that three large recombined genomic regions are present in the population.
Subdivision of the Roseobacter population
Phylogenetic separation of closely related lineages is not necessarily driven by genetic isolation of the lineages each with an independent gene pool. It was proposed that phylogenetic trees are useful to infer population structure and genetic separation only when the bacterial population is predominantly clonal with limited effect imposed by recombination [41, 47–51]. We thus tested the hypothesis that the clonal population structure of the Roseobacter population correlates with the population subdivision between Clade R-I and Clade R-II. This can be achieved by explicitly calculating the extent of HR between the two clades using the fineSTRUCTURE coancestry analysis [36]. The algorithm assumes that each individual is a recipient of the DNA from the remaining individuals (i.e., donors) in the population, and finds the individuals that share ancestry across different regions of the genome. This process is performed separately for all individuals in the population. The number of these genomic regions (termed as “chunk count” or “coancestry value”) among all possible donor–recipient pairs was then summarized into a coancestry matrix. Based on this matrix, the software uses a Markov chain Monte Carlo algorithm to group the individuals with similar coancestry patterns into a “fineSTRUCTURE population” [36].
This procedure led to the assignment of the 16 Roseobacter strains to five distinct fineSTRUCTURE populations, one in Clade R-I and four in Clade R-II (strains in red-dashed boxes in the dendrogram at the top of Fig. 1a). As expected, there is a greater proportion of coancestry within each fineSTRUCTURE population than between populations. Interestingly, the proportion of coancestry within the fineSTRUCTURE population corresponding to Clade R-I is very high, whereas that between the Clade R-I population and the other four populations comprising Clade R-II is very low (Fig. 1a). This striking difference suggests that these two lineages were subdivided. Because the Roseobacter population has extremely low recombination frequency, the population subdivision was likely caused by clonal diversification rather than sexual isolation as a result of a recombination barrier. The latter mechanism is used to explain population differentiation in Vibrio cyclitrophicus [38], Sufolobus islandicus [52], Wolbachia [53], Myxococcus xanthus [54], Polynucleobacter [55] and Ruminococcus gnavus [56] among other sexual populations [57]. Another observation is that membership of these five fineSTRUCTURE populations matches that of the five monophyletic groups in the phylogenomic tree, and that the clustering order of the five fineSTRUCTURE populations based on the proportion of shared ancestry among populations (the dendrogram shown at the top of Fig. 1a) accords well with the branching order of the corresponding monophyletic groups shown in the phylogeny (the phylogenomic tree shown on the left of Fig. 1a). This strengthens the hypothesis that phylogenetic trees can be used to infer the population structure of clonal bacteria [51]. As a comparison, the frequently recombined Marinobacterium population did not show the same evolutionary pattern (see Text S2.1).
Genomic regions underlying the Roseobacter population differentiation
We asked which genomic regions of the Roseobacter population were subdivided between Clade R-I and Clade R-II. As population differentiation leads to the fixation of different alleles, genomic regions underlying population differentiation are expected to have a low amount of SNP density when intra-population genomes are aligned, but to show increased SNP density when the inter-population genomes are compared. As illustrated in Fig. 1c, while there are only two SNPs in Clade R-I and more but still a limited number of SNPs in Clade R-II, the SNP density increases sharply when genomes from the two clades were examined together. Furthermore, the SNP density increase is largely restricted to a few chromosomal and plasmid regions, suggesting that these are the genomic regions underlying the population differentiation.
Loci under novel allele replacement overlapped with the genomic regions underlying the Roseobacter population differentiation
As expected, these genomic regions with dense SNPs (shown as rectangular gray shading in Fig. 1c) match well with the three recombined long DNA segments inferred by ClonalFrameML (shown as three red boxes in Fig. 1b). The latter contains 189 protein-coding genes, among which 180 are single-copy gene families. Most polymorphic sites in these genomic regions were subjected to nonhomoplasious substitutions (shown as white vertical bars densely clustered at the three genomic regions; Fig. 1b), whose allelic differences could be explained by a single change (either mutation or recombination with external lineages that are phylogenetically distinct from the population under study [35]) along the phylogeny (on the left of Fig. 1b). As mutations occur more or less randomly across the genome, they are not likely the dominant mechanism leading to these nonhomoplasious substitutions which are clustered in the genomic locations. Thus, these large genomic regions in the Roseobacter population were likely replaced with novel alleles derived from divergent lineages. Notably, one of these genomic regions locates at the plasmid and covers 92% of the plasmid region. When these three regions were excluded from the whole-genome alignment, the ρ/θ decreases slightly from 0.076 to 0.052, but the r/m drops sharply from 18.10 to 1.13 (Table S5). These results suggest that the evolution of the Roseobacter population is profoundly affected by a few recombined DNA segments representing only a tiny fraction (6.67%) of the genomes. Another important prediction by ClonalframeML is that these allele replacements occurred at the ancestral branch giving rise to the last common ancestor (LCA) of Clade R-I (shown as the dark blue horizontal bars within red boxes and the LCA of Clade R-I locating at the same row; Fig. 1b). This indicates that replacements with these divergent alleles drove the differentiation of Clade R-I from Clade R-II.
If recombination with external species is the underlying mechanism, it is expected that allelic replacements by homologous sequences from divergent species leave a strong signature of nucleotide substitution rate at the largely neutral synonymous (silent) sites (dS) in the affected protein-coding genes [58–60]. We have recently developed a population genomic approach that allows for the detection of core genes subject to novel allele replacement [37]. It clusters all possible pairwise dS values over all single-copy core gene families and subsequently identifies the outlier gene family clusters [37] (Fig. S2A). By comparing the pairwise dS values of the affected gene families within and between lineages, this approach [37] further infers the candidate ancestral branches where the allelic replacement events occurred in the species phylogeny (Fig. S2B).
Using k-means clustering (k = 2 based on the majority vote of cluster indices implemented in R package “NbClust” [61], see the details in Text S1; Fig. S2C) of dS for each possible pairwise comparison among the 16 Roseobacter strains across 2846 shared single-copy gene families, we identified two clusters of gene families. One cluster contains 176 families showing unusually large dS for between-clade comparisons and very small dS for within-clade comparisons, whereas the other cluster contains 2670 families showing very small dS for both between-clade and within-clade comparisons (Table S6). This result suggests that genes in the first cluster were likely subject to novel allele replacement either at the LCA of Clade R-I or at the LCA of Clade R-II. Interestingly, 168 of these 176 outlier genes locate in the genomic regions driving population differentiation predicted by the SNP density plot (the “All strains” plot of Fig. 1c) and the ClonalFrameML analysis (the three red boxes in Fig. 1b). In total, the dS clustering method, together with the ClonalFrameML analysis, identified 200 nonredundant core gene families that may be involved in the novel allele replacement and likely drive the genetic separation of Clade R-I from Clade R-II.
Accessory genomes may not play a leading role in driving population differentiation
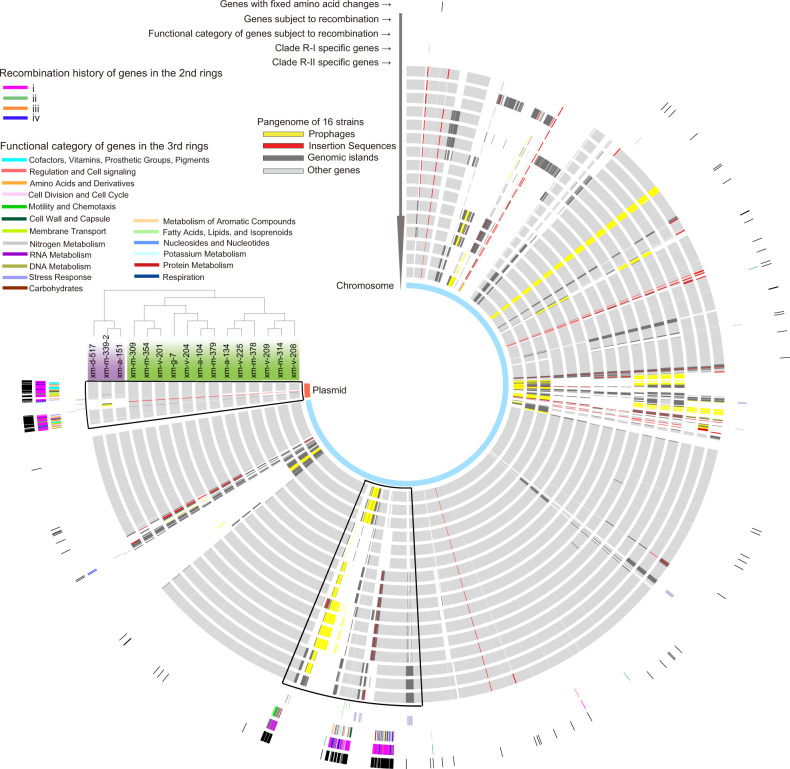
The above analyses focused on the genes in the core genomes (i.e., 2898 gene families shared by all 16 strains in which 2846 are single-copy families) that drive differentiation between Clade R-I and Clade R-II in the Roseobacter population. Here, we further explored the 736 gene families in the accessory genomes that are shared by a subset of the 16 strains. Among the accessory genes, 44 and 9 families were universally and exclusively found in Clade R-I and Clade R-II, respectively. Most (37/53) of these clade-specific genes encode proteins with unknown functions (Table S7). A few encode mobile genetic elements (MGEs). Among the remaining, genes specific to Clade R-I involve in regulation, restriction modification, oligopeptide transportation and opine metabolism, and genes specific to Clade R-II are limited to regulation. Next, both core and accessory genes were mapped to the population’s pangenome (Fig. 2), which was constructed using the closed genome of xm-d-517 (Fig. S3) as a backbone. More than half of the genes specific to Clade R-I co-localize with the 200 core genes that were largely affected by recombination (Fig. 2), consistent with the finding that the major allelic replacements of the 200 core gene loci occurred at the LCA of Clade R-I. Furthermore, mapping of MGEs showed that most of the 200 core gene families co-localize with MGEs including those being part of the core genomes (e.g., the plasmid) and part of the accessory genomes (e.g., some of the genomic islands, insertion sequences, and prophages) (Fig. 2). Taken together, our analyses showed that some MGEs from both the core and accessory genomes may be associated with the divergent allele replacement events, but the accessory genomes rarely carry functionally important clade-specific genes and thus may have a limited role in driving the differentiation of the Roseobacter population.
Fig. 2. The pangenome of the Roseobacter population.
From outer to inner rings: (1) core genes with fixed amino acid changes between Clade R-I (taxa shaded in purple) and Clade R-II (taxa shaded in green); (2) core genes (n = 200) subjected to recombination with external lineages. These families are classified into four groups each with a distinct color. Each group represents a distinct history of recombination events illustrated in Fig. S9; (3) SEED subsystem functional category assignment for each of the 200 core genes, with each category shown in a distinct color. Unassigned genes are not represented; (4–5) genes universally and exclusively found in Clade R-I (purple) and Clade R-II (green), respectively; (6–21) genomes of the 16 Roseobacter strains, with the order following the display of these strains in their phylogeny attached to the circos plot. The genome of xm-d-517 is closed, which is used as a skeleton to build the pangenome plot of these 16 strains. The core genes subject to recombination with external lineages are clustered in two adjacent chromosomal regions (framed in one box) and one plasmid region (framed in another box). Mobile genetic elements including prophages (yellow), insertion sequences (red) and genomic islands (dark gray) are mapped to each genome; (22) Chromosome and plasmid are indicated in light blue and light red, respectively.
Genome-wide nonsynonymous changes are concentrated in the core genomic regions driving population differentiation
Most of the 200 core genes that triggered population differentiation left genetic signatures at synonymous (silent) sites, manifested as unusually large dS values in these genes, but they are also expected to accumulate differences at nonsynonymous (amino acid changing) sites to enable functional changes of these genes. Indeed, among the 194 (out of 200) single-copy core gene families subjected to recombination with external lineages, 179 genes harbor the substitution at the amino acid level, which is in sharp contrast to only 309 genes showing amino acid substitutions in the remaining 2652 genes in core genomes (Table S8; χ2 test, p < 0.001). Furthermore, among the 2746 amino acid variants across all 2846 single-copy gene families, 2156 (78.51%) occur in those 194 single-copy core families (Table S8; χ2 test, p < 0.001). Among the 2156 amino acid variants found in those 194 families, 1832 (84.97%) are biallelic variants each universally and exclusively present in either Clade R-I or Clade R-II (Fig. 2). Hence, clade-specific allele fixation at nonsynonymous sites is highly concentrated in the genomic regions driving population differentiation, suggesting that important functional changes may have occurred in some of the encoded proteins at these loci.
Phycosphere is a likely niche that drove Roseobacter population differentiation
The 200 core gene families are involved in a variety of cellular functions, including organic substrate utilization (monosaccharides, organic acids, amino acids, polyamines), vitamin B7 (biotin) biosynthesis, molybdenum and tungstate acquisition, resistance to oxidative stress, lipopolysaccharide synthesis, polar flagella assembly, DNA repair, RNA processing and modification, cell division, and cell cycle (Table S9). Many of these functions match well with the known strategies that Roseobacter and other marine bacteria use to interact with eukaryotic phytoplankton, including motility and chemotaxis as a prerequisite to establish symbiosis with phytoplankton [62], degradation of extracellular reactive oxygen species commonly produced by phytoplankton and enriched in phycosphere [63, 64], and provision of biotin to phytoplankton in exchange of organic substrates [17, 65, 66]. Our results suggest that the phycosphere, a microscale layer rich in organic matter surrounding the algal cells [10, 27, 28], is a potential niche that drove the Roseobacter population differentiation. However, since only 5–8% of the known marine phytoplankton species are biotin auxotrophs [67, 68], our interpretation is limited. Instead, some of these functions might be equally important when roseobacters interact with the abundant organic particles or other less abundant hosts in the pelagic environments.
Roseobacter population differentiation at the physiological level
If ancestral allelic replacements at the 200 loci triggered population differentiation, it is expected that these genotypic variations may lead to phenotypic variations and that at least some of the phenotypic differences were transmitted to their descendants in today’s ocean. We thus performed a few physiological assays to phenotypically diagnose two representative strains (xm-d-517 and xm-m-339-2) from Clade R-I and two (xm-m-314 and xm-v-204) from Clade R-II.
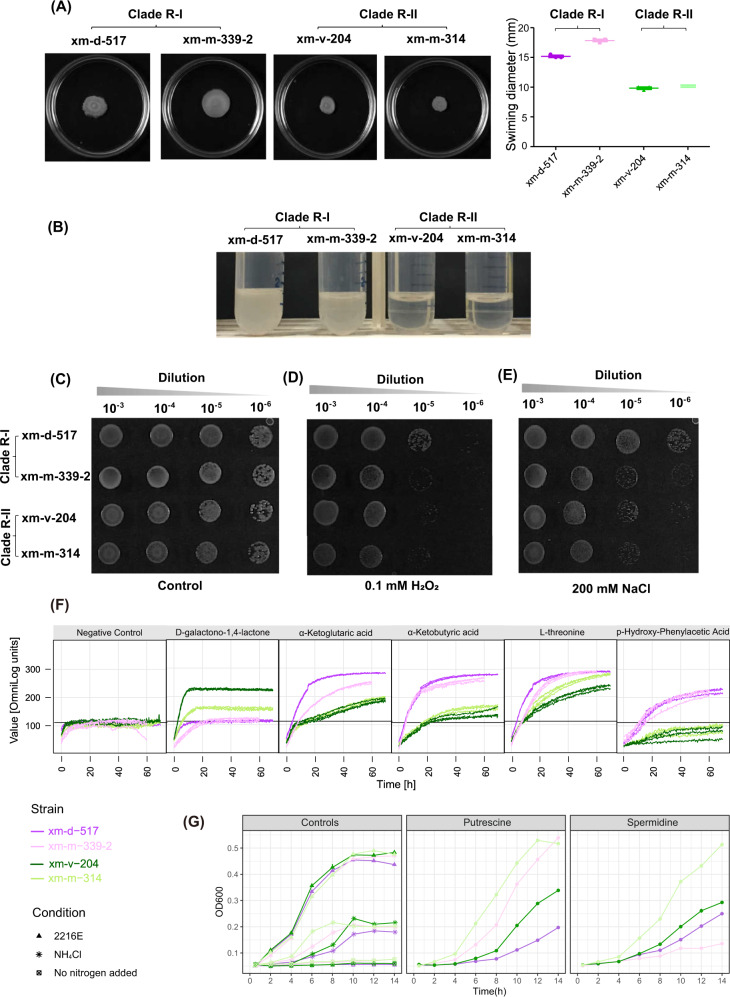
First, motility assays were motivated by the prediction of the genes (from xm-d-517_02347 to xm-d-517_02354) involved in polar flagella assembly within the 200 core genes (Table S9). After growing in the semi-solid agar plate (0.18%, w/v) for 11 days, the Clade R-I members showed significantly greater swimming zones than the Clade R-II members (Fig. 3a), indicating stronger swimming motility of the former. Consistent with this observation, the sedimentation experiment showed that while the Clade R-II members settled to the bottom following 24 h incubation in liquid culture, the Clade R-I members failed to sediment (Fig. 3b). The distinct sedimentation phenotype indicates greater transcription levels of the flagellar genes in Clade R-I members according to a recent report [69]. As being motile is a prerequisite for roseobacters to establish symbiosis with phytoplankton [62], stronger motility of the Clade R-I members increases their chance to meet and interact with phytoplankton [10].
Fig. 3. Phenotypic differentiation of the Clade R-I (represented by xm-d-517 and xm-m-339-2) and Clade R-II (represented by xm-v-204 and xm-m-314) of the Roseobacter population.
a Swimming motility assay on semi-solid agar plates. 3 μl of a cell suspension from sub-culture was spotted at the center of a semi-solid marine broth 2216 plate. After incubation for 11 days at 28 °C, the images were photographed and shown in the left panel. Results are representative of those from three individual assays with identical results. In the right panel, data shown was determined by measuring the swimming halo diameter. b Sedimentation of Clade R-I and Clade R-II members. The Clade R-II members show a clear sedimentation phenotype in marine broth 2216 medium after 24 h of incubation at room temperature without shaking, whereas the Clade R-I members did not settle under the same conditions. The growth of the Clade R-I and Clade R-II members under oxidative stress (d) and osmotic stress (e), respectively. Tenfold serial dilutions of the strains were spotted onto marine broth 2216 plates, which were used as the control (c). Plates were imaged after two days. f The respiration curves of five substrates that are utilized significantly differently as carbon sources by Clade R-I and Clade R-II based on the analysis of phenotype microarray (PM) microplates. Results are those of three replicated analyses. The curves for all carbon sources in both microplates (PM01 and PM02) are shown in Figs. S4 and S5. (g) Growth experiments used to test whether polyamines (putrescine and spermidine) can be utilized as a sole nitrogen source, in which three replicates were performed for each strain. Left: the negative control without any nitrogen source, the positive control when NH4Cl is used as a sole nitrogen source, and another control inoculated in rich medium (DifcoTM Marine broth 2216). Middle and right: the growth curves of the four strains with putrescine and spermidine as a sole nitrogen source, respectively.
Next, three genes were predicted to be involved in oxidative stress response within the 200 core genes. They encode a catalase-peroxidase (xm-d-517_03124) for degradation of hydrogen peroxide (H2O2) and other reactive oxygen species [70], a hydrogen peroxide-inducible genes activator (oxyR; xm-d-517_03125) that activates the former gene [71], and a regulator (soxR; xm-d-517_02264) that activates the transcription of a complex oxidative stress regulon in response to superoxide-generating agents [72]. The assay results showed that members in Clade R-I, especially xm-d-517, have greater tolerance to H2O2-mediated oxidative stress than Clade R-II members (Fig. 3c vs. d), potentially increasing the chance of Clade R-I members to survive at phycosphere where H2O2 is enriched compared to the bulk seawater [63, 64].
Third, the gene encoding a choline dehydrogenase (xm-d-517_02345) is included in the 200 core genes. This gene is a part of the choline–glycine betaine pathway, which may increase a bacterium’s osmotic tolerance [73]. Our assay results showed that Clade R-I members, especially strain xm-d-517, show stronger tolerance to NaCl-induced osmotic stress than Clade R-II members (Fig. 3c vs. 3e), though whether phycosphere has a distinct osmotic pressure from the bulk seawater has rarely been discussed. Note that choline can be an excellent nitrogen source to members of the Roseobacter group [74]. Therefore, an alternate explanation for the observed divergent choline dehydrogenases between the two clades is that Clade R-I and Clade R-II members may show differential efficiency in utilizing choline as a nitrogen source.
Lastly, as ~40 of the 200 core genes were predicted to take up and/or catabolize organic compounds including carbohydrates, amino acids, and polyamines among others (Table S9), we employed the PM technology [75] to systematically investigate the substrate utilization differences between the two clades. As expected, most of the tested 190 carbon sources were not differentially utilized because of the extremely high genetic similarity among the members of the Roseobacter population. Five substrates including D-galactono-1,4-lactone (Fig. S4-C02), α-Ketoglutaric acid (Fig. S4-D06), α-Ketobutyric acid (Fig. S4-D07), and l-threonine (Fig. S4-G04), and p-Hydroxy-Phenylacetic acid (Fig. S4-H02), however, indeed differentiated the two clades (Fig. 3f), with D-galactono-1,4-lactone supporting higher growth of the Clade R-II members and the other four more favorably utilized by the Clade R-I members (Table S10). Through phenotype-to-genotype mapping with the opm package [76], we showed that the utilization of l-threonine may be linked to the ilvA gene (xm-d-517_02199 encoding l-threonine dehydratase which catalyzes the conversion of l-threonine to α-Ketobutyric acid and ammonium), which is a part of the 200 core genes (Table S9).
A few other important substrates including putrescine (see the PM results in Fig. S5-H08) are also differentially utilized by the four strains when they are used as a sole carbon source, but the pattern disagrees with the phylogenetic divide of these strains. As putrescine and spermidine are two important types of polyamines, which are prevalent in the marine environments and may serve as both nitrogen and carbon sources for the Roseobacter group [77], their utilization was tested with additional growth assays (see Table S11 and Text S2.2 for details). When used as a sole carbon source, both polyamines showed different utilization among the strains but inconsistent with the phylogenetic divide (Fig. S6). When utilized as a sole nitrogen source, however, spermidine supported higher growth of the Clade R-II members than that of the Clade R-I members (one-way ANOVA, p < 0.001; Fig. 3g), though putrescine did not differentiate the two clades. While the above phenotypic differences may be ascribed to the genotypic differences of the related core genes due to allelic replacements at these loci, direct evidence supporting such link is not available.
History and pattern of novel allele replacements in the Roseobacter population
We showed that Clade R-I members outcompeted Clade R-II members in most of the assayed physiological traits, and these traits are exclusively encoded in the core genes that drove Roseobacter population differentiation. This observation led us to hypothesize that the LCA shared by Clade R-I and Clade R-II may not be able to efficiently exploit the phycosphere; instead, it was the LCA of Clade R-I that was replaced with novel alleles at these core gene loci, and these events enabled the Clade R-I members to more efficiently explore the phycosphere niche and eventually drove their genetic separation from the parental population which is now represented by Clade R-II. Alternatively, the LCA shared by Clade R-I and R-II may interact with one phytoplankton taxon frequently, and the allelic replacements at the LCA of Clade R-I allowed it to efficiently explore a new phytoplankton taxon, which may release more nutrients but impose a higher oxidative stress. In this case, adaptation to different groups of phytoplankton may drive the genetic divergence of the two clades. Addressing these questions requires additional analyses, because the bioinformatics analyses presented so far provided limited information regarding the history of the recombination events affecting these 200 core gene families. For example, the phylogenomic tree shows that Clade R-I is subtended by, rather than embedded in, Clade R-II (Fig. 1a left), thus it cannot tell that Clade R-I was the derived subpopulation. Likewise, the fineSTRUCTURE coancestry matrix shows that Clade R-I and Clade R-II each have a largely distinct ancestor and thus are genetically separated (Fig. 1a right), but it does not have information regarding which clade evolved earlier. Although the ClonalFrameML analysis supported that the novel allele replacements occurred at the LCA of Clade R-I (Fig. 1b), it is not clear which donor lineages contributed to the novel alleles and whether multiple recombination events occurred with small DNA segments or few events occurred with long segments.
These questions can be addressed by comparing the topology of the gene trees of the 200 core genes subjected to divergent allele replacements (see gene tree examples in Fig. S7) with that of the species tree. A vast majority of the 200 gene trees showed that Aliiroseovarius crassostreae is the most closely related lineage to the Roseobacter population (Fig. S8A), suggesting that A. crassostreae or some missing lineage closely related to it is the potential donor that contributed to the novel alleles at these core loci. However, the lack of appropriate outgroups prevented a reliable inference of the evolutionary relationship between Clade R-I, Clade R-II, and A. crassostreae (Fig. S8B, C, see Text S2.3). We therefore employed an alternative approach to deduce the gene tree topology. This approach was based on the between-clade neutral genetic distance (measured as between-clade dS values), and the root was set between the two clades with the greatest distance (see Text S2.3). Next, the comparison of topology was performed between the inferred gene trees and the species tree to extrapolate the recombination history of these genes (Fig. S9; Text S2.3).
The recombination history analysis showed that the 200 core genes drove speciation by replacing novel alleles from other Roseobacter lineages related to A. crassostreae. It identified the LCA of Clade R-I as the primary recipient of the novel alleles. As these 200 core genes are clustered into two adjacent chromosomal regions and a plasmid region, the allelic replacements at the LCA of Clade R-I likely proceeded by a few recombination events involving long DNA segments. This is also supported by the very low ρ/θ (relative rare recombination events) but a high r/m ratio (long DNA segments involving recombination) calculated from the core genome alignments (Table S5). Our approach further inferred that the ultimate allele donor lineages for 152 of the 200 genes (sum of the genes following Fig. S9-i and S9-ii) are phylogenetically distinct from those for the remaining 48 genes (sum of the genes following Fig. S9-iii and S9-iv). One explanation is that the new allelic replacements at the initial recombination events involving long DNA segments were not adaptive or even deleterious at some loci, and fine-tuning at these loci occurred by recombining with a different external lineage.
Conclusion
Our finding that population differentiation occurred in a Roseobacter population with an exceedingly low SNP density is surprising (Table S4) when compared with reported free-living prokaryotic populations showing evidence of speciation but much greater SNP density (Table S4). Furthermore, in many populations studied previously, accessory genomes change much faster than the core genomes and the former plays a leading role in driving population differentiation [54, 78–84], but speciation of this Roseobacter population was predominantly driven by allelic replacements at three core genomic regions which together make ~6% of the genomic DNA. It suggests that ecological differentiation of the Roseobacter population proceeded by adjusting existing functions at the SNP level rather than gaining completely novel capabilities. In addition, the rare recombination (ρ/θ = 0.076) and extremely low genomic diversity within Clade R-I suggest that the differentiation in the Roseobacter population may be a result of genome-wide selective sweep (see Text S2.4). To our knowledge, this is likely the first evidence that genome-wide selective sweep drives bacterial speciation in a natural environment. As a comparison, we analyzed a sympatric population affiliated with gammaproteobacterial Marinobacterium recovered from the same sample (Table S12). Although members of the Marinobacterium population are more diverse than members of the Roseobacter population at the 16S rRNA gene, the whole-genome ANI, and the SNP density levels, the former shows no evidence of speciation (Figs. S10 and S11), likely owing to its high recombination rate and the reduced genome (Fig. S12) lacking motility and chemotaxis genes (Fig. S13) needed to build symbiosis with phytoplankton or explore other microenvironments (see Text S2.1).
There has been ample evidence showing that members of the Roseobacter group are among the most active bacteria that participate in bacteria–phage and bacteria–phytoplankton interactions in the pelagic ocean [14, 21]. It is also becoming increasingly clear that these trophic interactions largely drive the oceanic carbon and nutrient cycles [10, 85], but how the microscale niches resulting from these interactions drive the microbial genotype formation and evolution has been understudied. Convincing evidence is available that phages are an important driver in the differentiation of a Roseobacter population related to Ruegeria mobilis [16]. However, that population harbors a huge amount of intraspecific diversity that exceeds our Roseobacter population by a factor of 15 (62,535 versus 4242 SNPs per Mb), and they were sampled from global oceans that vary considerably in environmental parameters including nutrients, temperature, salinity, and dissolved oxygen among others. Therefore, a possibility that fine-scale genetic differentiation within the population defined by phage subtyping cannot be precluded. In recent years, Roseobacter members have been used as model bacteria to study bacteria–phytoplankton interactions [22, 30–32, 86]. While interactions with phytoplankton have been implicated as one of the most important forces driving Roseobacter evolution [17], evolutionary biology evidence has never been available. Our comprehensive analyses presented here favored a role of Roseobacter–phytoplankton interaction over that of Roseobacter–phage interaction in driving the evolution of a pelagic Roseobacter population, though other types of trophic interaction or organic particle utilization cannot be ruled out. Further studies are needed to find more direct evidence for these competing hypotheses.
Supplementary information
Acknowledgements
We thank Ying Sun for sharing her bioinformatics pipeline, Xiaoyuan Feng for his helpful discussion, Kan Zhu for his assistance in the access to the OmniLOG instrument, and Leon Zou (BiOLOG Inc.) for his helpful suggestions on the Biolog experiments. This work was supported by the National Key R&D Program of China (2018YFC0309800), the National Natural Science Foundation of China (41776129), the National Key R&D Program of China (2016YFA0601400 to YZ), the Shenzhen City Knowledge Innovation Plan (JCYJ20160530174441706 to AY), the Hong Kong Research Grants Council General Research Fund (14163917), the Hong Kong Environment and Conservation Fund (15/2016), the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), and the Direct Grant of CUHK (4053257 & 3132809).
Data availability
Genomic sequences of the Roseobacter and Marinobacterium population are available at the NCBI GenBank database under the accession number WBXQ00000000-WBYV00000000.
Code availability
The scripts used for the population structure analyses, recombination history inference, SNP density plot and allelic replacements inference have been deposited in the online repository (https://github.com/Xiaojun928/Population-genomics).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaojun Wang, Yao Zhang, Minglei Ren
Supplementary information
The online version of this article (10.1038/s41396-020-00743-7) contains supplementary material, which is available to authorized users.
References
- 1.Prgzelin BB, Alldredge AL. Primary production of marine snow during and after an upwelling event. Limnol Oceanogr. 1983;28:1156–67. [Google Scholar]
- 2.Shanks AL, Trent JD. Marine snow: microscale nutrient patches. Limnol Oceanogr. 1979;24:850–4. [Google Scholar]
- 3.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–91. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 4.Moran MA. The global ocean microbiome. Science. 2015;350:aac8455. doi: 10.1126/science.aac8455. [DOI] [PubMed] [Google Scholar]
- 5.Stocker R. Marine microbes see a sea of gradients. Science. 2012;338:628–33. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 6.Stocker R, Seymour JR, Samadani A. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA. 2008;105:4209–14. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stocker R, Seymour JR. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev. 2012;76:792–812. doi: 10.1128/MMBR.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenwasser S, Ziv C, Creveld SGvan, Vardi A. Virocell metabolism: metabolic innovations during host–virus interactions in the ocean. Trends Microbiol. 2016;24:821–32. doi: 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 10.Seymour JR, Amin SA, Raina J-B, Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat Microbiol. 2017;2:1–12. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 11.Smriga S, Fernandez VI, Mitchell JG, Stocker R. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci USA. 2016;113:1576–81. doi: 10.1073/pnas.1512307113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S, et al. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73:4559–69. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bischoff V, Bunk B, Meier-Kolthoff JP, Spröer C, Poehlein A, Dogs M, et al. Cobaviruses—a new globally distributed phage group infecting Rhodobacteraceae in marine ecosystems. ISME J. 2019;13:1404–21. doi: 10.1038/s41396-019-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan Y, Chen F. Bacteriophages that infect marine roseobacters: genomics and ecology. Environ Microbiol. 2019;21:1885–95. doi: 10.1111/1462-2920.14504. [DOI] [PubMed] [Google Scholar]
- 15.Ankrah NYD, May AL, Middleton JL, Jones DR, Hadden MK, Gooding JR, et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014;8:1089–100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenschein EC, Nielsen KF, D’Alvise P, Porsby CH, Melchiorsen J, Heilmann J, et al. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 2017;11:569–83. doi: 10.1038/ismej.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo H, Moran MA. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78:573–87. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–98. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 19.Ramanan R, Kim B-H, Cho D-H, Oh H-M, Kim H-S. Algae–bacteria interactions: evolution, ecology and emerging applications. Biotechnol Adv. 2016;34:14–29. doi: 10.1016/j.biotechadv.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–11. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 21.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–84. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 23.Green DH, Echavarri-Bravo V, Brennan D, Hart MC. Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. BioMed Res Int. https://www.hindawi.com/journals/bmri/2015/194540/. Accessed 28 May 2020. [DOI] [PMC free article] [PubMed]
- 24.González JM, Simó R, Massana R, Covert JS, Casamayor EO, Pedrós-Alió C, et al. Bacterial cmmunity structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–46. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park BS, Guo R, Lim W-A, Ki J-S. Pyrosequencing reveals specific associations of bacterial clades Roseobacter and Flavobacterium with the harmful dinoflagellate Cochlodinium polykrikoides growing in culture. Mar Ecol. 2017;38:maec.12474. [Google Scholar]
- 26.Li S, Chen M, Chen Y, Tong J, Wang L, Xu Y, et al. Epibiotic bacterial community composition in red-tide dinoflagellate Akashiwo sanguinea culture under various growth conditions. FEMS Microbiol Ecol. 2019;95:fiz057. doi: 10.1093/femsec/fiz057. [DOI] [PubMed] [Google Scholar]
- 27.Bell W, Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull. 1972;143:265–77. [Google Scholar]
- 28.Cole JJ. Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst. 1982;13:291–314. [Google Scholar]
- 29.Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–3. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 30.Durham BP, Dearth SP, Sharma S, Amin SA, Smith CB, Campagna SR, et al. Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ Microbiol. 2017;19:3500–13. doi: 10.1111/1462-2920.13834. [DOI] [PubMed] [Google Scholar]
- 31.Barak-Gavish N, Frada MJ, Ku C, Lee PA, DiTullio GR, Malitsky S, et al. Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci Adv. 2018;4:eaau5716. doi: 10.1126/sciadv.aau5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, et al. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife. 2016;5:e17473. doi: 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling AE, Mau B, Perna NT. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Luo H. Homologous recombination in core genomes facilitates marine bacterial adaptation. Appl Environ Microbiol. 2018;84:e02545–17. doi: 10.1128/AEM.02545-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 40.Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–93. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–3. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Achtman M. Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos Trans R Soc Lond B Biol Sci. 2012;367:860–7. doi: 10.1098/rstb.2011.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Didelot X, Maiden MCJ. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–22. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 45.Hanage WP. Not so simple after all: bacteria, their population genetics, and recombination. Cold Spring Harb Perspect Biol. 2016;8:a018069. doi: 10.1101/cshperspect.a018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–80. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holt KE, Baker S, Weill F-X, Holmes EC, Kitchen A, Yu J, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44:1056–9. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–21. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhi X-Y, Zhao W, Li W-J, Zhao G-P. Prokaryotic systematics in the genomics era. Antonie Van Leeuwenhoek. 2012;101:21–34. doi: 10.1007/s10482-011-9667-x. [DOI] [PubMed] [Google Scholar]
- 51.Yahara K, Furuta Y, Oshima K, Yoshida M, Azuma T, Hattori M, et al. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol Biol Evol. 2013;30:1454–64. doi: 10.1093/molbev/mst055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML, et al. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 2012;10:e1001265. [DOI] [PMC free article] [PubMed]
- 53.Ellegaard KM, Klasson L, Näslund K, Bourtzis K, Andersson SGE. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 2013;9:e1003381. doi: 10.1371/journal.pgen.1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, Velicer GJ, et al. A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J. 2016;10:2468–77. doi: 10.1038/ismej.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoetzinger M, Hahn MW. Genomic divergence and cohesion in a species of pelagic freshwater bacteria. BMC Genom. 2017;18:794. doi: 10.1186/s12864-017-4199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arevalo P, VanInsberghe D, Elsherbini J, Gore J, Polz MF. A reverse ecology approach based on a biological definition of microbial populations. Cell. 2019;178:820–34. doi: 10.1016/j.cell.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 57.Bobay L-M, Ochman H. Biological species are universal across life’s domains. Genome Biol Evol. 2017;9:491–501. doi: 10.1093/gbe/evx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engel P, Stepanauskas R, Moran NA. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 2014;10:e1004596. doi: 10.1371/journal.pgen.1004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes AL, French JO. Homologous recombination and the pattern of nucleotide substitution in Ehrlichia ruminantium. Gene. 2007;387:31–7. doi: 10.1016/j.gene.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Hughes AL, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J Bacteriol. 2005;187:2698–704. doi: 10.1128/JB.187.8.2698-2704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: an R package for determining the relevant number of clusters in a data set. J Stat Softw. 2014;61:1–36. [Google Scholar]
- 62.Raina J-B, Fernandez V, Lambert B, Stocker R, Seymour JR. The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol. 2019;17:284–94. doi: 10.1038/s41579-019-0182-9. [DOI] [PubMed] [Google Scholar]
- 63.Hünken M, Harder J, Kirst GO. Epiphytic bacteria on the Antarctic ice diatom Amphiprora kufferathii Manguin cleave hydrogen peroxide produced during algal photosynthesis. Plant Biol. 2008;10:519–26. doi: 10.1111/j.1438-8677.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- 64.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper MB, Kazamia E, Helliwell KE, Kudahl UJ, Sayer A, Wheeler GL, et al. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 2019;13:334–45. doi: 10.1038/s41396-018-0274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang YZ, Koch F, Gobler CJ. Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc Natl Acad Sci USA. 2010;107:20756–61. doi: 10.1073/pnas.1009566107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Helliwell KE. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol. 2017;216:62–8. doi: 10.1111/nph.14669. [DOI] [PubMed] [Google Scholar]
- 69.Gao R, Krysciak D, Petersen K, Utpatel C, Knapp A, Schmeisser C, et al. Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated Burkholderia glumae PG1 strain. Appl Environ Microbiol. 2015;81:7993–8007. doi: 10.1128/AEM.01043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 71.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–8. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 72.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–84. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landfald B, Strøm AR. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J Bacteriol. 1986;165:849–55. doi: 10.1128/jb.165.3.849-855.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lidbury I, Kimberley G, Scanlan DJ, Murrell JC, Chen Y. Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Mol Microbiol. 2015;17:5048–62. doi: 10.1111/1462-2920.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bochner BR, Gadzinski P, Panomitros E. Phenotype microArrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–55. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vaas LAI, Sikorski J, Hofner B, Fiebig A, Buddruhs N, Klenk H-P, et al. opm: an R package for analysing OmniLog(R) phenotype microarray data. Bioinformatics. 2013;29:1823–4. doi: 10.1093/bioinformatics/btt291. [DOI] [PubMed] [Google Scholar]
- 77.Mou X, Vila-Costa M, Sun S, Zhao W, Sharma S, Moran MA. Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ Microbiol Rep. 2011;3:798–806. doi: 10.1111/j.1758-2229.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 78.Porter SS, Chang PL, Conow CA, Dunham JP, Friesen ML. Association mapping reveals novel serpentine adaptation gene clusters in a population of symbiotic. Mesorhizobium ISME J. 2017;11:248–62. doi: 10.1038/ismej.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andam CP, Gogarten JP. Biased gene transfer in microbial evolution. Nat Rev Microbiol. 2011;9:543–55. doi: 10.1038/nrmicro2593. [DOI] [PubMed] [Google Scholar]
- 80.Boucher Y, Cordero OX, Takemura A. Endemicity within global Vibrio cholerae populations. mBio. 2011;2:1–8. doi: 10.1128/mBio.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–9. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polz MF, Alm EJ, Hanage WP. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013;29:170–5. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cordero OX, Polz MF. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol. 2014;12:263–73. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 84.Hoetzinger M, Schmidt J, Jezberová J, Koll U, Hahn MW. Microdiversification of a pelagic Polynucleobacter species Is mainly driven by acquisition of genomic islands from a partially interspecific gene pool. Appl Environ Microbiol. 2017;83:e02266–16. doi: 10.1128/AEM.02266-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, et al. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA. 2016;113:3143–51. doi: 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Christie-Oleza JA, Sousoni D, Lloyd M, Armengaud J, Scanlan DJ. Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat Microbiol. 2017;2:17100. doi: 10.1038/nmicrobiol.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences of the Roseobacter and Marinobacterium population are available at the NCBI GenBank database under the accession number WBXQ00000000-WBYV00000000.
The scripts used for the population structure analyses, recombination history inference, SNP density plot and allelic replacements inference have been deposited in the online repository (https://github.com/Xiaojun928/Population-genomics).