Abstract
The killing of tumor cells by CD8+ T cells is suppressed by the tumor microenvironment, and increased expression of inhibitory receptors, including programed cell death protein-1 (PD-1), is associated with tumor-mediated suppression of T cells. To discover cellular defects triggered by tumor exposure and associated PD-1 signaling, we established an ex vivo imaging approach to investigate the response of antigen-specific, activated effector CD8+ tumor infiltrating lymphocytes (TILs) after interaction with target tumor cells. Although TIL–tumor cell couples readily formed, couple stability deteriorated within minutes. This was associated with impaired F-actin clearing from the center of the cellular interface, reduced Ca2+ signaling, increased TIL locomotion, and impaired tumor cell killing. The interaction of CD8+ T lymphocytes with tumor cell spheroids in vitro induced a similar phenotype, supporting a critical role of direct T cell–tumor cell contact. Diminished engagement of PD-1 within the tumor, but not acute ex vivo blockade, partially restored cell couple maintenance and killing. PD-1 thus contributes to the suppression of TIL function by inducing a state of impaired subcellular organization.
Introduction
Cancer cells are commonly recognized by the immune system, and immune cells constitute a large part of the tumor mass. Cytotoxic T lymphocytes (CTLs) are capable of directly killing cancer cells; however, their killing ability is widely suppressed after they have entered the tumor, where they are known as tumor-infiltrating lymphocytes (TILs). A number of key contributors to tumor-mediated immune suppression in vivo have been characterized. Most prominently, tumor-reactive T cells increase their cell surface expression of inhibitory receptors, including cytotoxic T lymphocyte–associated antigen 4 (CTLA-4), programmed cell death protein-1 (PD-1), lymphocyte activation gene 3 (LAG3), T cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibitory motif domains (TIGIT), and T cell molecule with immunoglobulin and mucin domain 3 (TIM3) (1, 2). Monoclonal antibody–mediated blockade of CLTA-4 and PD-1 has yielded substantial clinical success in enhancing the antitumor immune response (3). Additional mechanisms of tumor-mediated immune suppression include the recruitment of tolerogenic immune cells, notably CD4+ regulatory T cells (Tregs) into the tumor (4, 5), and the expression of suppressive soluble mediators, such as adenosine and prostaglandin E2 (PGE2) (6-8). Whereas it is clear that tumor cell killing is diminished due to the immunosuppressive tumor microenvironment, it is still uncertain as to which cellular steps in target tumor cell killing are impaired in TILs and how such impairment is controlled by the established mediators of tumor-mediated immune suppression.
PD-1 is a critical mediator of tumor-mediated immune suppression and it is increased in abundance on the surface of T cells in response to continuous antigen exposure. In persistent viral infections, PD-1 signaling maintains an exhausted phenotype among CD8+ T cells (9). PD-1 engagement has multiple effects on T cell activation. These include activation of phosphatase and tensin homolog (PTEN) (10), recruitment of the phosphatase sarcoma homology 2 domain phosphatase 2 (SHP-2) to the interface between CTLs and their target tumor cells (11, 12), suppression of the sustained activation of Akt and Ras pathways with consequences for cell cycle regulation (13), inhibition of glycolysis and the promotion of lipolysis and oxidative metabolism (14), and the increased abundance of the proapoptotic protein Bim (15). PD-1 engagement inhibits the T cell stop signal upon contact with an antigen presenting cell (APC) (16) and the formation of stable cell couples (17). However, it remains unresolved as to whether and how PD-1 signaling disrupts the cellular events required for CTL-mediated target cell killing.
The killing of target cells by CTLs requires carefully orchestrated steps of cellular reorganization, a process known as polarization. CTLs must bind to the target cells, use actin polymerization to stabilize the cellular interface (known as the immune synapse), relocate the microtubule organizing center (MTOC) from behind the nucleus to the center of the cellular interface, and finally release the contents of cytolytic granules directed toward the target cell (18, 19). The increase in the T cell cytoplasmic calcium ion (Ca2+) concentration, as mediated by store-operated calcium entry using the endoplasmic reticulum (ER) calcium sensor stromal interaction molecule (STIM) and the plasma membrane calcium channel Orai, is a key signaling step required for cytolytic granule release (20). The increase in intracellular Ca2+ concentration in TILs becomes diminished, likely driven by defects in proximal T cell receptor (TCR) signal transduction. Demonstrating the importance of Ca2+ signaling for CTL function and tumor control, loss of or mutations in STIM and Orai impair cytolytic killing and immune control of tumors, and the activation of Ca2+ channels in CD8+ T cells can restrain tumor growth (21-25). The increase in the T cell cytoplasmic Ca2+ concentration and actin dynamics are bi-directionally linked (26).
To investigate how T cell polarization and related signaling are altered when TILs interact with their target tumor cells, we adapted a well-established model of antigen-driven tumor recognition to live-cell imaging approaches ex vivo. A renal carcinoma cell line (Renca) expressing the influenza virus haemagglutinin (HA) as a neo-antigen (RencaHA) is recognized by CD8+ T cells from Clone 4 TCR transgenic mice (27). When adoptively transferred into a RencaHA-tumor bearing BALB/c mouse, naïve Clone 4 T cells are primed within the tumor-draining lymph nodes, infiltrate the tumor, and acquire a suppressed phenotype that is characterized by diminished cytokine secretion, decreased killing ability, and enhanced inhibitory receptor expression (28). Here, we adoptively transferred in vitro–activated Clone 4 CD8+ T cells into RencaHA tumor–bearing mice. These transduced Clone 4 T cells homed to the tumor and acquired a suppressed phenotype. They were re-isolated for an ex vivo investigation of their interaction with RencaHA tumor cells. Clone 4 TILs displayed impaired cell couple maintenance with their tumor targets, which was associated with impaired F-actin distribution at the target cell interface and reduced Ca2+ signaling, resulting in impaired tumor cell killing. PD-1 expression on Clone 4 TILs was enhanced. Direct interaction of Clone 4 T cells with RencaHA tumor cell spheroids in vitro induced a similar phenotype. Blocking PD-1 engagement in vivo in the tumor microenvironment, but not acutely in vitro, enhanced tumor clearance and improved the maintenance of TIL cells. Thus, we have discovered a cellular mechanism of tumor-mediated immune suppression: Impaired maintenance of T cell polarization reduced the cytotoxicity of TILs and a polarization-impaired state was induced by the inhibitory receptor PD-1.
Results
The cytolytic ability of CD8+ T cells is diminished as a result of tumor infiltration
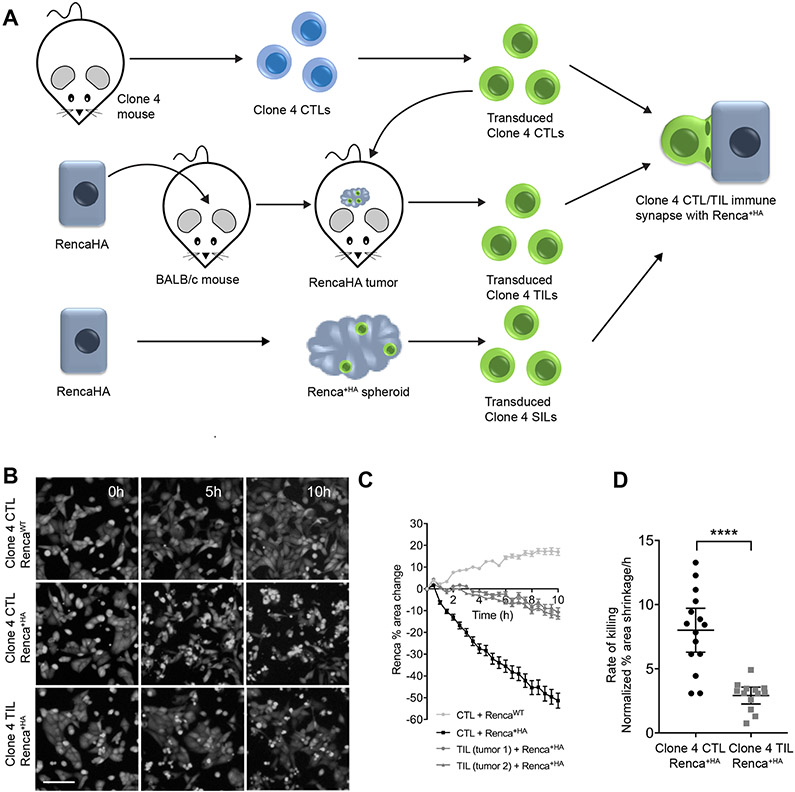
To investigate the killing of tumor target cells by CD8+ TILs, we adapted a mouse model of neo-antigen CD8+ T cell tumor recognition to enable ex vivo, live-cell imaging approaches (Fig. 1A). RencaHA renal carcinoma cells express the haemagglutinin (HA) protein from influenza A/PR/8/H1N1 as a neo-antigen. The Clone 4 TCR recognizes the dominant H2-Kd–restricted HA peptide 518-526 (IYSTVASSL). In all experiments, CD8+ T cells from Clone 4 TCR transgenic mice were activated in vitro with HA peptide–pulsed splenocytes and retrovirally transduced to express GFP-tagged proteins of interest. Cells were FACS sorted to ensure that sensor abundance was close to endogenous protein amounts (29, 30). Activated, transduced Clone 4 CD8+ T cells were adoptively transferred into BALB/c mice bearing established subcutaneous RencaHA tumors, and after 96 hours the GFP-positive TILs were isolated from the tumors by FACS sorting.
Fig. 1. Tumor exposure impairs the killing ability of CD8+ T cells.
(A) Experimental schematic for imaging the interaction of HA peptide–pulsed Renca tumor cells (Renca+HA) with retrovirally transduced, HA-specific, in vitro–activated CTLs in comparison to the same cells after adoptive transfer into mice with established Renca+HA cells tumors or co-culture with RencaHA spheroids. (B to D) Fluorescence microscopy analysis of the killing of CTV-labelled Renca or Renca+HA cells in vitro by Clone 4 CTLs or TILs. (B) Images are representative of 13 experiments. Scale bar, 100 μm. (C) The percentage changes in the area of Renca cells are means ± SEM from a minimum of three fields of view/time and are representative of seven experiments. (D) Average Renca+HA cell death rates are means ± 95% confidence interval from seven independent experiments. ****P < 0.0001 by Student’s t test with Welch’s correction.
To explore the cytolytic capability of tumor-specific CTLs and TILs, we developed an imaging-based ex vivo killing assay. Renca cells plated in a glass-bottom imaging plate were pulsed with HA peptide (Renca+HA) and were then overlaid with Clone 4 CTLs or TILs. We found that CTLs efficiently lysed target cells, decreasing the imaging area covered by Renca+HA cells at a rate of 8 ± 3 %/hour (Fig. 1, B to D). Similarly, Clone 4 CTLs specifically lysed BALB/c splenocytes pulsed with HA peptide in vivo (fig. S1). Non-HA peptide–pulsed Renca cells continued to proliferate, which confirmed that lysis was dependent on cognate antigen recognition. In contrast, Clone 4 TILs were poorly cytolytic, as expected (39, 45, 56). The area covered by Renca+HA cells was cleared by Clone 4 TILs at a significantly reduced rate of 3 ± 1 %/h (Fig. 1, B to D).
MTOC polarization in TILs and cell couple maintenance is impaired
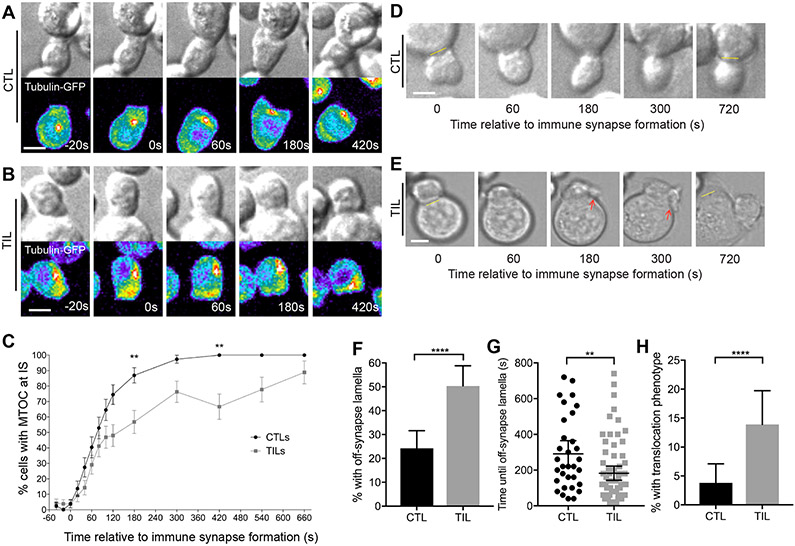
In CTLs, relocation of the MTOC from behind the nucleus to the target cell interface after cell coupling is critical for lytic granule release and cytolytic activity (31). To visualize CD8+ T cell MTOC polarization toward the Renca target cell interface, tubulin-GFP–expressing, HA-specific Clone 4 CTLs and TILs were imaged during their interaction with Renca+HA cells by spinning disk confocal microscopy. Whereas all CTLs relocated their MTOC to the cellular interface within 7 min, fewer TILs quickly relocated their MTOC towards the tumor cell contact site (Fig. 2, A to C, fig. S2 and Movie S1).
Fig. 2. Compared to CTLs, TILs have delayed MTOC polarization and diminished cell couple stability.
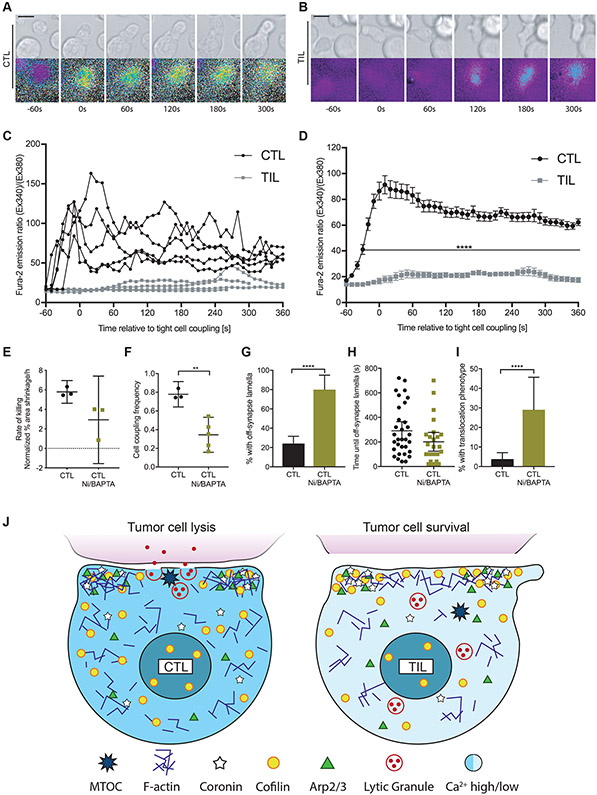
(A to C) Microscopy analysis of tubulin-GFP–expressing, HA-specific CTLs (A) and TILs (B) interacting with Renca+HA cells over time. (A and B) DIC (top) and false-colored fluorescence intensity (bottom) images are representative of at least three independent experiments. (C) The percentage of cells with their MTOC localized at the immune synapse (IS) are means ± SEM of 51 CTLs and 55 TILs from all experiments. (D to H) Microscopy analysis of HA-specific CTLs and TILs interacting with Renca+HA cells over time. (D and E) DIC images with the original position of tight cell couple formation with Renca+HA cells (yellow lines) and off-synapse lamellae (red arrows) are representative of at least six independent experiments. The percentages of cells with off-synapse lamellae (F), time of the first appearance of off-synapse lamella (G), and the percentage of cells that translocated away from the contact site (H) are means ± 95% confidence interval of 138 CTLs and 137 TILs from all experiments. Scale bars, 5 μm. *P < 0.05, **P < 0.01 (G), **P < 0.0036 (C, to account for Bonferroni correction), ****P < 0.0001 by proportions z-test (C, F and H), or Mann Whitney u-test (G).
We also investigated Clone 4 CD8+ T cell–target cell coupling and morphology and found that TILs formed tumor cell couples less frequently than did CTLs: Whereas the initial contact of Clone 4 TILs with Renca+HA target cells led to the formation of tight cell couples in 31 ± 6% of contacts, 78 ± 3% of Clone 4 CTL–tumor cell contacts led to the formation of tight couples. Clone 4 CTLs formed symmetrical, stable interfaces with Renca+HA target cells throughout 12 min of observation (Fig. 2D). Initial immune synapses between Clone 4 TILs and Renca+HA target cells were also symmetrical, but polarization was diminished starting as rapidly as seconds after tight cell coupling (Fig. 2E). To quantify this difference in Clone 4 CD8+ T cell polarization, we determined the occurrence of lamellae pointing away from the cellular interface (Fig. 2E, red arrows). T cells continuously extend large lamellae, but whereas directing such lamellae towards the interface is expected to be stabilizing, directing them away from the interface (“off-synapse lamellae”) may weaken the interface (32, 33). Off-synapse lamellae occurred in a higher proportion of TILs than of CTLs (Fig. 2F) and were observed earlier during cell coupling (Fig. 2G). Furthermore, TILs remained at the initial site of target cell binding less efficiently: 14 ± 3% of TILs translocated across the target cell surface by at least one interface diameter from the initial site of target cell binding, whereas only 4 ± 2% of CTLs exhibited translocation (Fig. 2H), as was expected based on previous studies (34, 35). Thus, our data suggest that although HA-specific TILs effectively establish cell couples with peptide-pulsed Renca target cells, they do not maintain cell couples well.
Peripheral F-actin ring formation is destabilized in TILs compared to that in CTLs
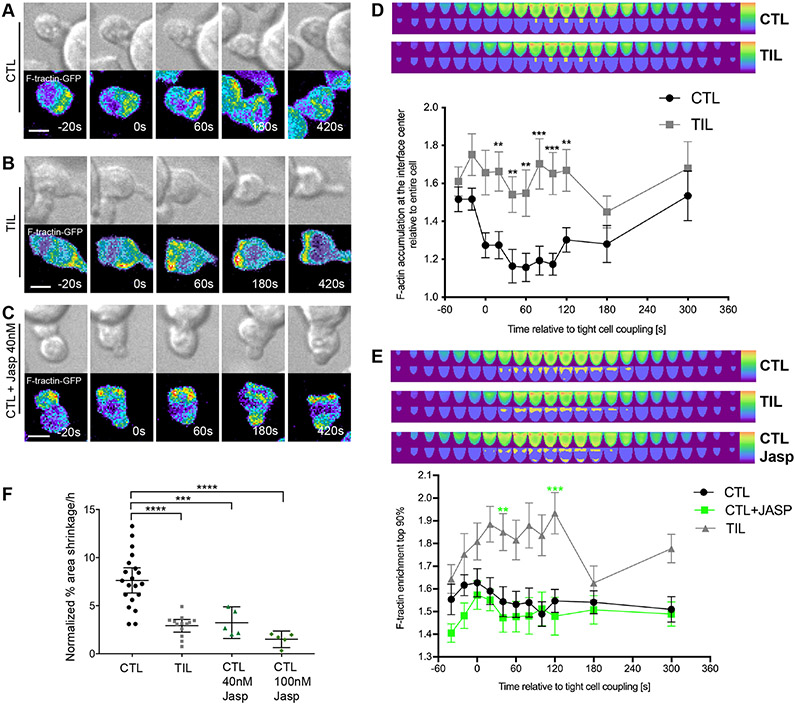
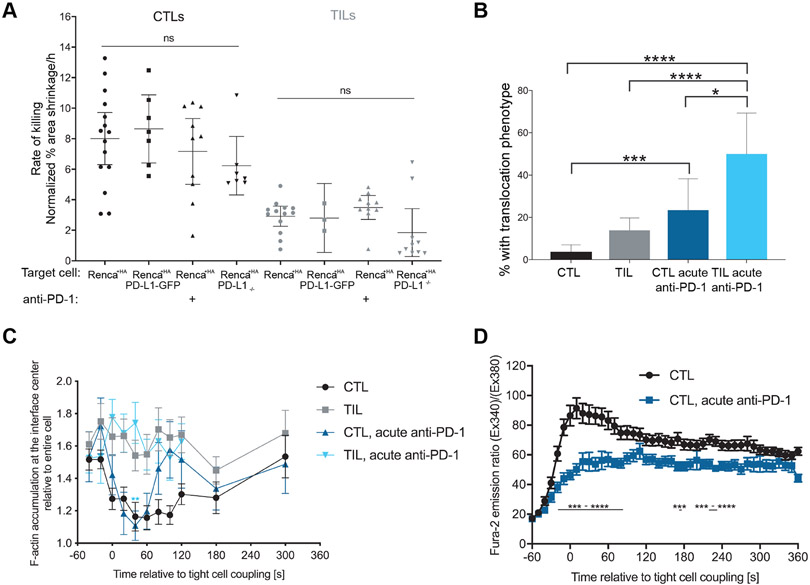
The immune synapse is stabilized by an F-actin ring around its periphery. In cytolytic killing, clearance of F-actin from the interface center can be associated with effective secretion of cytolytic granules (36), but not always (37, 38). To determine whether impaired F-actin distributions could underpin the impaired maintenance of TIL–target cell couples, we visualized F-actin with an F-actin–binding peptide conjugated to GFP: F-tractin-GFP (39). Clone 4 CTLs and TILs expressing F-tractin-GFP were imaged (Fig. 3, A to C), and the data were automatically shape-normalized and computationally analyzed (Fig. 3, D and E, fig. S3A and Movie S2). To investigate F-actin clearance from the interface center, we measured the ratio of F-actin accumulation at the interface center relative to the entire cell (Fig. 3D). At the time of the first contact between Clone 4 T cells and Renca+HA target cells, F-actin was comparably enriched in CTLs and TILs. However, F-actin enrichment at the interface center rapidly decreased in Clone 4CTLs to 1.3-fold or less over the next 2 min, indicative of central clearing. In contrast, F-actin enrichment remained greater than 1.5-fold in Clone 4 TILs. Addressing F-actin distributions cell-wide, we measured enrichment in the 10% of the cell volume with the greatest amounts of F-tractin, which largely corresponded to the cellular interface with an emphasis on its edge (Fig. 3D). Enrichment of F-actin was increased in antigen-specific TILs relative to that in CTLs (Fig. 3E). However, TILs displayed substantially reduced F-actin amounts overall, as determined by phalloidin staining (fig. S3, B and C). Thus, the increased relative interface enrichment in Clone 4 TILs was unlikely to correspond to greater F-actin densities, which suggests that the dominant F-actin defect in Clone 4 TILs was impaired F-actin clearance from the interface center. This defect just preceded the time of occurrence of off-interface lamellae (Fig. 2G) and the time at which MTOC polarization began to fail (Fig. 2C).
Fig. 3. Impaired F-actin distributions at the immune synapse are associated with poor TIL killing ability.
(A to E) Microscopy analysis of F-tractin-GFP–expressing Clone 4 CTLs, with and without treatment with 40 nM Jasplakinolide, and TILs interacting with Renca+HA cells over time. (A to C) DIC (top) and false-colored fluorescence intensity (bottom) images are representative of at least two independent experiments. (D and E) Computational analysis of GFP fluorescence distribution is displayed in horizontal slices perpendicular to the immune synapse averaged over all cells and times with the location of the central cylinder (D) and the 10% of the cell volume with the most intense F-actin accumulation (E) highlighted in yellow. Quantification of F-actin enrichment in TILs and CTLs (D) or in TILs and CTLs treated with and without Jasplakinolide (Jasp) (E) in the highlighted volumes are means ± SEM of 60 CTLs, 51 TILs, or 40 Jasp-treated CTLs from at least three independent experiments. (F) Fluorescence microscopy analysis of the killing of Renca+HA cells in vitro by HA-specific CTLs treated with or without Jasp and by TILs. Average Renca+HA cell death rate data are means ± SEM from at least four independent experiments. Scale bars, 5 μm. **P < 0.0045 (D and E, to account for Bonferroni correction), ***P < 0.001, and ****P < 0.0001 by Student’s t-test (D), one-way ANOVA (E), or pair-wise Student’s t-test with Welch’s correction (F).
To investigate whether impaired cell couple maintenance could result in loss of cytolytic effector function, Clone 4 CTLs were treated with a low concentration of Jasplakinolide, a macrocyclic peptide that promotes actin filament nucleation. At this concentration of Jasplakinolide, cell coupling is not substantially impaired but F-actin turnover is slowed (40, 41). Interface F-actin accumulation was slightly diminished after treatment with Jasplakinolide, because the 10% cell volume with the greatest F-tractin amounts were not exclusively focused on the interface anymore, but included regions at the T cell distal pole (Fig. 3E). Treatment with Jasplakinolide significantly reduced the rate of target cell lysis by CTLs to that displayed by TILs (Fig. 3F). Treatment of Clone 4 CTLs with a higher concentration of Jasplakinolide that impairs cell coupling (41) further reduced killing (Fig. 3F). The similar phenotypes of Jasplakinolide-treated Clone 4 CTLs and untreated TILs suggest that tumor-induced disruption of actin remodeling may suppress the lytic activity of TILs.
Interface recruitment of negative regulators of F-actin is more extensive in TILs than in CTLs
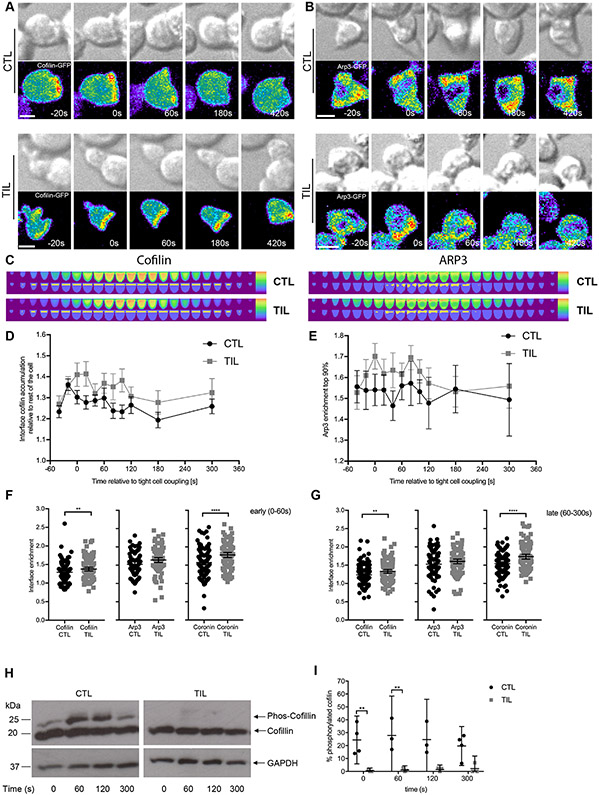
T cell actin remodeling is a dynamic process, which is promoted by the Arp2/3 complex. Coronin 1A and cofilin inhibit the Arp2/3 complex and promote the severing and depolymerization of F-actin, respectively. Actin regulators need to localize to regions of T cell F-actin turnover to be effective (30). To investigate mechanisms of altered F-actin dynamics, we imaged Clone 4 CTLs and TILs that were transduced to express cofilin-GFP, Arp3-GFP, or Coronin 1A-GFP (Fig. 4, A to G; fig. S4, A to G; and Movies S3 to S5). Although we found no difference in the relative amounts of Arp3 recruited to the interface when we compared Clone 4 CTLs and TILs, the amounts of cofilin and Coronin1A that were recruited were greater in Clone 4 TILs within seconds of tight cell coupling than in Clone 4 CTLs (Fig. 4F). These differences in cofilin and Coronin 1A interface enrichment persisted after prolonged tight cell coupling (Fig. 4G). The distribution of the cofilin-activating phosphatase chronophin (42) was similar to that of cofilin itself (fig. S4, H to K, and Movie S6). To establish whether the increase in amount of cofilin recruited to the interface was associated with enhanced cofilin activation, we determined the relative amounts of active non-phosphorylated cofilin by phos-tag Western blotting analysis of Clone 4 CTL and TIL lysates activated on plates coated with antibodies against CD3. At all times, the percentage of active, unphosphorylated cofilin was increased within TILs when compared to that in CTLs (Fig. 4, H and I, and fig. S5A). However, the overall abundance of cofilin was reduced in Clone 4 TILs when compared to that in CTLs, as determined by Western blotting analysis (fig. S5B). The increased interface retention of cofilin and Coronin 1A in TILs after cell coupling suggests that impaired F-actin stability in TILs could underpin the diminished F-actin translocation to the edge of the interface (Fig. 3D) and the accompanying defect in cell couple maintenance (Fig. 2, D and E) that we observed earlier.
Fig. 4. Cofilin and coronin, but not Arp2/3, are more extensively enriched at the cell contact interface in TILs.
(A to G) Microscopy analysis of cofilin-GFP–expressing (A) or Arp3-GFP–expressing (B) HA-specific CTLs and TILs interacting with Renca+HA cells over time. DIC (top) and false-colored fluorescence intensity (bottom) images are representative of at least two independent experiments. (C) Computational analysis of GFP fluorescence distribution is displayed in horizontal slices perpendicular to the immune synapse averaged over all cells and time points with the interface region cylinder (cofilin) and the 10% of the cell volume with the most intense Arp3 accumulation highlighted. (D to G) Quantification of interface GFP enrichment at individual times (D and E) and at early (F) and late (G) times in the highlighted volumes are means ± 95% confidence interval of 90 CTLs and 58 TILs (D) or 56 CTLs and 51 TILs (E ) from all experiments with the addition of coronin 1A data (fig. S4, D to G) in (F) and (G). (H and I) Western blotting analysis of phosphorylated cofilin (Phos-Cofilin) in the lysates of HA-specific CTLs and TILs after activation with antibodies against CD3 for the indicated times. Blots (H) are representative of at least three independent experiments (see also fig. S5A). (I) The relative amounts of Phos-Cofilin are expressed as a percentage of that of total cofilin and are means ± 95% confidence interval as normalized to the abundance of GAPDH from all experiments. Scale bars, 5 μm. *P < 0.05, **P < 0.01, and ****P < 0.0001 by Student’s t-test (D to G) or two-way ANOVA (I).
Activation-induced cytoplasmic Ca2+ influx is impaired in TILs compared to that in CTLs
To identify signaling defects associated with TIL polarization impairment, we also investigated the T cell cytoplasmic Ca2+ concentration in Clone 4 T cells after activation by Renca+HA cells with the Ca2+-sensitive, ratiometric fluorescent dye Fura-2 (Fig. 5 A and B, and Movie S7). In Clone 4 CTLs, the cytoplasmic Ca2+ concentration increased rapidly after initial target cell contact, often after formation of a first small membrane contact. In contrast, after activation, the cytoplasmic Ca2+ concentration increase in Clone 4 TILs was delayed sometimes up to minutes and was reduced in magnitude (Fig. 5, C and D), as expected (21, 24, 25). To determine whether the Ca2+ signaling defect could be linked to impaired killing and cell couple maintenance, we treated HA-specific CTLs with Ni2+ to block plasma membrane calcium channels and 1,2-Bis(2-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid tetrakis acetoxymethyl ester (BAPTA) to buffer cytoplasmic Ca2+. Renca+HA target cells exhibited 5.8 ± 0.3% area shrinkage/hour without treatment in our imaging-based killing assay and 2.9 ± 1.0% after treatment with Ni2+ and BAPTA (Fig. 5E). The frequency of tight cell couple formation between Clone 4 TILs and Renca+HA target cells was reduced from 78 ± 3% without treatment to 34±7% after treatment with Ni2+ and BAPTA (Fig. 5F). Whereas 80 ± 7% of CTLs treated with Ni2+ and BAPTA produced off-synapse lamellae, only 24 ± 4% of untreated CTLs exhibited the same behavior (Fig. 5G) although the onset time did not differ substantially (Fig. 5H). Similarly, 29 ± 8% of CTLs treated with Ni2+ and BAPTA translocated over the target cell surface by at least a full interface diameter, but only 4±2% of untreated CTLs translocated over the same distance (Fig. 5I). These data suggest that diminished Ca2+ signaling could contribute to impaired cell couple maintenance in TILs (Fig. 5J).
Fig. 5. Compared to CTLs, TILs display a reduced cytoplasmic Ca2+ influx.
(A to D) Microscopy analysis of Fura-2–loaded Clone 4 CTLs and TILs interacting with Renca+HA cells over time. (A and B) DIC (top) and false-colored fluorescence images (bottom) are representative of at least two independent experiments. (C and D) Ratios of Fura-2 emission upon excitation at 340 nm over 380 nm are from five randomly chosen single CTLs and TILs (C)or are means ± SEM from 50 CTLs and 14 TILs from at least two independent experiments (D). (E) Fluorescence microscopy analysis of the killing of Renca+HA cells in vitro by HA-specific CTLs treated with or without NiCl2 and BAPTA-AM, as indicated. Average Renca+HA cell death rate data are means ± SEM from three independent experiments. (F to I) Microscopy analysis of HA-specific CTLs treated with or without NiCl2 and BAPTA-AM interacting with Renca+HA cells over time. (F) The cell coupling frequencies are means ± 95% confidence interval of at least three independent experiments. The percentage of Clone 4 CTLs with off-synapse lamellae (G), the time of the first appearance of off-synapse lamellae (H), and the percentage of cells that translocated away from the initial contact site (I) are means ± 95% confidence interval of 138 CTLs (from Fig. 2, F to H) and 31 CTLs after treatment from three independent experiments. (J) Graphic summary of TIL defects. Cell shapes, cytoplasmic Ca2+ concentrations, and the distributions of lytic vesicles and the indicated cytoskeletal components and regulators of actin dynamics are given schematically for CTLs (left) and TILs (right) at 1 to 3 min of tight cell coupling. Scale bars, 5 μm. **P < 0.01 and ****P < 0.0001 by Student’s t-test (D and F), Mann Whitney u-test (H), or proportion’s z-test (G and I).
Direct interaction between CTLs and tumor cell spheroids induces a polarization-impaired state
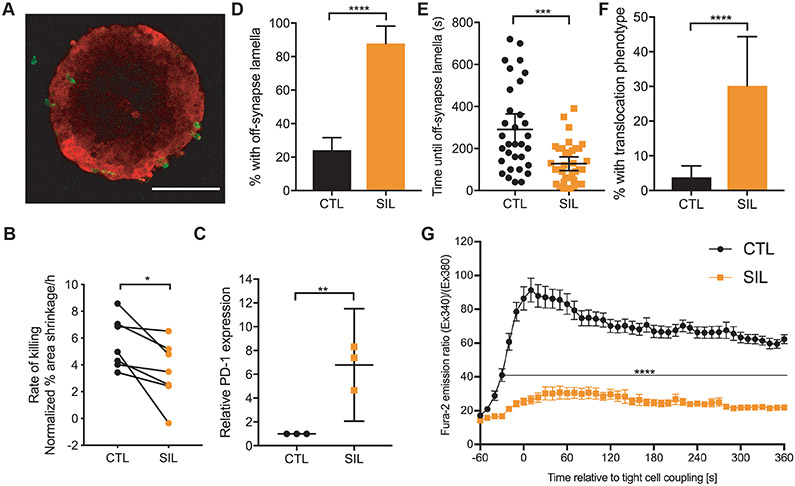
To determine whether infiltration of CTLs into three-dimensional (3D) tumor cell structures was sufficient to impair the polarization of CD8+ T cells, we grew Renca cells as spheroids in Matrigel. Overnight interaction of Clone 4 CTLs with spheroids incubated with HA peptide (Fig. 1A) resulted in Clone 4 cell infiltration to generate spheroid-infiltrating lymphocytes (SILs) (Fig. 6A). After we re-isolated these cells from the spheroids, we found that the rate of killing of Renca+HA target cells was reduced from 5.6 ± 0.7% area shrinkage/hour after co-culture with activated CTLs to 3.5 ± 0.9% after co-culture with SILs (Fig. 6B). PD-1 abundance was increased in SILs when compared to that in CTLs (Fig. 6C). Additionally, the frequency of the formation of tight cell couples after initial SIL contact with Renca+HA target cells was reduced when compared to that of Clone 4 CTLs from 78 ± 3% to 47 ± 4% (P = 0.007), and the maintenance of SIL couples with Renca+HA cells was impaired (Fig. 6, D to F). Finally, the cytoplasmic Ca2+ concentration was substantially reduced after activation of SILs compared to that in CTLs (Fig. 6G). Thus, infiltration of Clone 4 CTLs into Renca+HA cell spheroids induced a polarization-impaired state that shared critical features with that of functionally impaired TILs.
Fig. 6. Co-culture of CTLs with Renca tumor cell spheroids induces a polarization-impaired state.
(A) Two-photon microscopy analysis of the infiltration of Clone 4 CTLs into spheroids overnight. The 2D section through the center of a spheroid of mCherry-Renca cells (red) with calcein-labeled, HA-specific SILs (green) is representative of three independent experiments. (B) Fluorescence microscopy analysis of the killing of Renca+HA cells in vitro by HA-specific CTLs and SILs. Average Renca+HA cell death rate data are means ± SEM from seven independent experiments. (C) Flow cytometric analysis of the relative expression of PD-1 on HA-specific CTLs and SILs. Data are normalized medians ± 95% confidence interval from three independent experiments. (D to F) Microscopy analysis of HA-specific CTLs and SILs interacting with Renca+HA cells over time. The percentage of cells with off-synapse lamellae (D), time of the first occurrence of off-synapse lamellae (E), and the percentage of cells that translocated away from the contact site (F) are means ± 95% confidence interval of 138 CTLs (from Fig. 2, F to H) and 43 SILs from three independent experiments. (G) Fluorescence microscopy analysis of Fura-2–loaded HA-specific CTLs and SILs interacting with HA peptide–pulsed Renca+HA cells over time. Normalized Fura-2 emission data are means ± SEM from 50 CTLs (from Fig. 5D) and 40 SILs from three independent experiments. Scale bar, 100 μm. *P < 0.05, ***P < 0.001, and ****P < 0.0001 by paired Student’s t-test (B), Student’s t-test with Welch’s correction (C), proportion’s z-test (D and F), Mann Whitney u-test (E), or Student’s t-test (G).
Blocking PD-1 in vivo enhances TIL cytotoxicity and tumor rejection
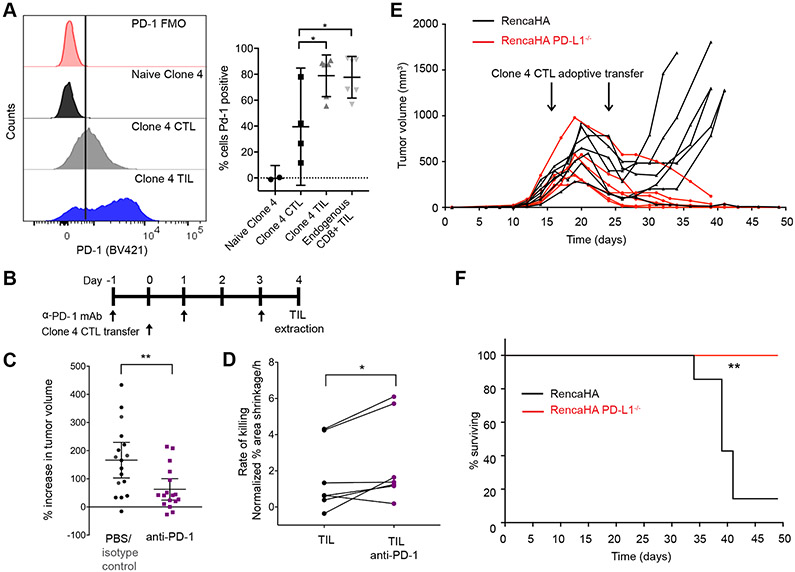
To identify potential in vivo regulators of the diminished maintenance of TIL cytoskeletal polarization, we investigated the role of PD-1. The expression of PD-1 limits anti-tumor immunity (1, 3), and we confirmed that the abundance of PD-1 was increased on TILs and endogenous CD8+ T cells from RencaHA tumors when compared to that on activated CTLs (Fig. 7A). When RencaHA tumor–bearing mice were treated with a blocking monoclonal antibody (mAb) against PD-1 for 6 days before and after the adoptive transfer of Clone 4 CTLs (Fig. 7B), tumor growth was significantly reduced after antibody treatment when compared to that in control treated mice (Fig. 7C), as was expected based on previous findings (3). In addition, killing of Renca+HA tumor cells ex vivo by Clone 4 TILs isolated from anti-PD-1–treated mice was substantially enhanced compared to that by Clone 4 TILs isolated from untreated control mice (Fig. 7D).
Fig. 7. Loss of PD-1 engagement in vivo improves TIL killing ability.
(A) Flow cytometric analysis of the cell surface expression of PD-1 on naïve and primed Clone 4 CTLs, Clone 4 TILs, and endogenous CD8+ TILs. Left: Histograms are representative of at least two independent experiments. Right: Data are means ± 95% confidence interval. (B and C) Analysis of subcutaneous RencaHA tumor growth in mice after treatment with antibody against PD-1 and adoptive transfer of Clone 4 CTLs, as indicated. The percentage increase in tumor volume within 5 days of the initiation of antibody treatment for 17 mice/condition with means ± 95% confidence interval are from five independent experiments. (D) Fluorescence microscopy analysis of the killing of Renca+HA cells in vitro by HA-specific TILs from untreated control mice or mice treated with antibodies against PD-1. Average Renca+HA cell death rate data with means ± 95% confidence interval are from six independent experiments. (E and F) Analysis of subcutaneous RencaHA and RencaHA PD-L1−/− tumor growth in mice after adoptive transfer of HA-specific CTLs at the indicated times. Tumor volume (E) and survival (F) data of 7 RencaHA and 6 RencaHA PD-L1−/− tumor-bearing mice are pooled from two independent experiments. *P < 0.05 and **P < 0.01 by one-way ANOVA (A), Mann Whitney u-test (C), paired Student’s t-test (D), or Mantel-Cox test (F).
PD-1 inhibitory signaling is activated after engagement by its ligand programmed death-ligand 1 (PD-L1), which is expressed on tumor cells, tumor-associated immune cells, or both (1). To investigate the functional relevance of tumor-expressed PD-L1, we generated a RencaHA-PD-L1−/− cell line (fig. S6A). Loss of PD-L1 expression induced spontaneous tumor regression in 4 out of 10 inoculated mice and increased survival (fig. S6, B and C). In combination with the adoptive transfer of HA-specific Clone 4 CTLs, all mice bearing RencaHA-PD-L1−/− tumors underwent complete tumor clearance, compared to only 1 of 7 control mice (Fig. 7, E and F). Thus, these data suggest that PD-L1 on RencaHA cells contributes to the suppression of adoptively transferred Clone 4 CTLs. Together, the experiments with PD-1 blocking antibodies and PD-L1-deficient tumor cells indicate that PD-1 is a critical element of in vivo tumor immune suppression in the RencaHA model.
PD-1 promotes the impaired maintenance of TIL polarization in vivo
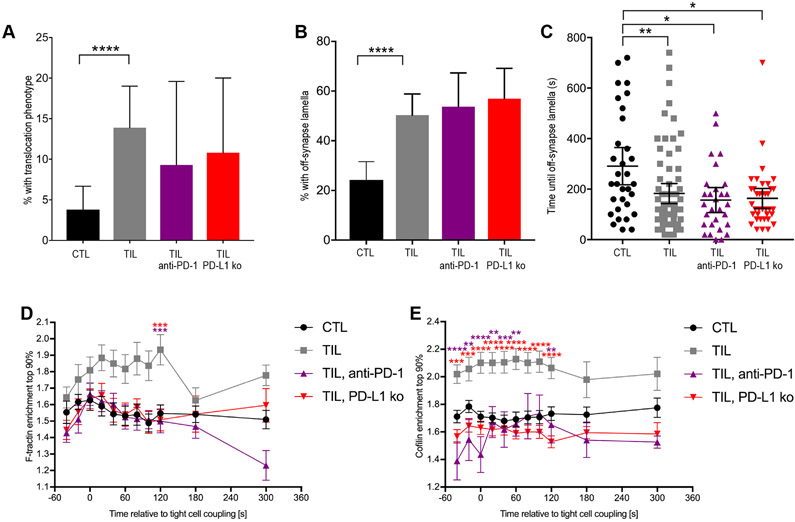
To investigate whether blockade of PD-1 engagement within the tumor microenvironment restored TIL polarization, we studied TILs isolated from RencaHA tumor–bearing mice treated with antibody against PD-1 or from RencaHA-PD-L1−/− tumor-bearing mice and found that 9 ± 5% and 11 ± 5%, respectively, displayed translocation of at least one interface diameter over the target cell surface after immune synapse formation as compared to 14 ± 3% of TILs treated with buffer alone (fig. S7A). The production of off-synapse lamellae was not reduced by PD-1 blockade (fig. S7, B and C). In TILs from mice treated with antibodies against PD-1 and in TILs from RencaHA-PD-L1−/− tumor-bearing mice, F-actin clearing from the interface center was significantly enhanced at multiple times and resembled that of Clone 4 CTLs (Fig. 8A and fig. S7D). Similarly, cofilin accumulation at the interface was not enhanced in Clone 4 TILs from mice treated with antibody against PD-1 or in Clone 4 TILs isolated from mice with RencaHA-PD-L1−/− tumors (Fig. 8, B and C, and fig. S7E). Together, these data suggest that PD-1 engagement impairs cytoskeletal rearrangement in TILs in vivo and thus may contribute to their diminished cytolytic function.
Fig. 8. Loss of PD-1 engagement in vivo partially rescues cytoskeletal polarization.
(A) Fluorescence microscopy analysis of F-tractin-GFP–expressing HA-specific CTLs and TILs from the indicated mice interacting with Renca+HA cells. Data showing the accumulation of F-actin at the center of the immune synapse are means ± SEM of 60 CTLs, 51 TILs (from Fig. 3D), 42 TILs from mice treated with antibody against PD-1, and 43 TILs from RencaHA PD-L1−/− tumors from two independent experiments. (B to D) Fluorescence microscopy analysis of cofilin-GFP–expressing HA-specific CTLs and TILs from the indicated mice interacting with HA peptide–pulsed Renca+HA cells. Data showing the accumulation of cofilin-GFP at the interface at individual time points (B) and at early (C) and late (D) times are means ± SEM (B) or are means ± 95% confidence interval (C and D) of 90 CTLs, 58 TILs (from Fig. 4D), 32 TILs from mice treated with antibody against PD-1, and 55 TILs from RencaHA PD-L1−/− tumors from at least two independent experiments. *P < 0.05, **P < 0.01 (D), **P < 0.0045 (A, to account for Bonferroni correction) by one-way ANOVA.
Acute in vitro blockade of PD-1 does not restore cytoskeletal polarization or killing
To further understand the role of PD-1, we investigated the effects of acute PD-1 blockade performed only during in vitro killing and imaging assays. When HA-specific Clone 4 TILs or CTLs were treated with the same antibody against PD-1 as that used in vivo or when RencaHA-PD-L1−/− target cells were used, target cell killing was not enhanced (Fig. 9A). Similarly, the lytic activity of HA-specific TILs or CTLs was not inhibited by RencaHA target cells that overexpressed PD-L1 (Fig. 9A and fig. S8A). These data suggest that PD-1 does not regulate cytolysis by TILs and CTLs in vitro in an acute manner.
Fig. 9. Acute in vitro blockade of PD-1 does not restore killing or cytoskeletal polarization.
(A) Fluorescence microscopy analysis of the killing of Renca+HA, Renca+HA PD-L1−/−, and Renca +HA PD-L1-GFP cells in vitro by HA-specific CTLs and TILs in the presence or absence of antibody against PD-1, as indicated. Average Renca+HA cell death rates with means ± 95% confidence interval are from at least four independent experiments. (B) Microscopy analysis of HA-specific CTLs and TILs interacting with HA peptide–pulsed Renca+HA cells in the presence or absence of antibody against PD-1, as indicated. The percentage of cells that translocated away from the contact site are means ± 95% confidence interval of 138 CTLs, 137 TILs (from Fig. 2, F to H), 35 CTLs treated with anti-PD-1, and 24 TILs treated with anti-PD-1 from two independent experiments. (C) Fluorescence microscopy analysis of F-tractin-GFP–expressing HA-specific CTLs and TILs interacting with HA peptide–pulsed Renca+HA cells in the presence or absence of antibody against PD-1, as indicated. Data showing the accumulation of F-tractin-GFP data at the center of the immune synapse are means ± SEM of 60 CTLs, 51 TILs (from Fig. 3D), 41 CTLs+anti-PD-1, and 28 C TILs+anti-PD-1 from two independent experiments. (D) Fluorescence microscopy analysis of Fura-2–loaded, HA-specific CTLs with Renca+HA cells in the presence or absence of antibody against PD-1, as indicated. Normalized Fura-2 emission data are means ± SEM of 50 CTLs (from Fig. 5D) and 52 CTLs treated with anti-PD-1 from two independent experiments. *P < 0.05, **P < 0.0045 (to account for Bonferroni correction), ***P < 0.001, and ****P < 0.0001; by Kruskal-Wallis test with comparison to ‘Renca+HA’ (A, CTL), Kruskal-Wallis test and one-way ANOVA because it is difficult to satisfy all assumptions for either test (A, TIL) proportion’s z-test (B), one-way ANOVA (C), or Student’s t-test (D).
When we investigated the effect of acute PD-1 blockade on F-actin distribution and Ca2+ influx in HA-specific CTLs and TILs, we found that it did not enhance these critical elements of cellular polarization (Fig. 9, B to D, and fig. S8, B to D). On the contrary, acute treatment with antibody against PD-1 increased the percentage of Clone 4 CTL and TIL cell couples with Renca+HA target cells that translocated from their initial target cell binding site by at least one interface diameter increased from 4±2% to 24±7% for CTLs and from 14±3% to 50±9% for TILs (Fig. 9B). However, acute PD-1 blockade did not affect F-actin central clearing (Fig. 9C) nor enrichment in the 10% of the cell volume with the highest F-tractin amounts (fig. S8D). Acute PD-1 blockade also did not enhance Ca2+ signaling in Clone 4 CTLs, but indeed impaired it around the time of tight cell coupling (Fig. 9D). Whereas PD-1 blockade in vivo thus restored a substantial fraction of the impaired cell couple maintenance of TILs, acute in vitro blockade could not do so. On the contrary, PD-1 blockade impaired some elements of CTL signaling and cell polarity consistent with in vitro data in a model of persistent viral infection (17).
Discussion
The mechanisms underpinning the defective killing ability of TILs are still uncertain. A number of alterations in TIL activation have been described previously, including reduced actin polymerization (43-45), reduced Ca2+ signaling among other proximal defects (24, 46, 47), increased expression of the inhibitory signaling mediators phosphatase sarcoma homology 2 domain phosphatase 1 (SHP-1) and cytokine-inducible SH2-containing protein (Cish) (46, 48), and decreased functional avidity of the TCR (49). In human TILs activated by super-antigen–incubated B cell lines, MTOC recruitment to the cellular interface and F-actin clearance from its center are impaired in a Galectin-3–dependent manner (50). Metabolic defects in the glucose-poor tumor microenvironment are likely to occur because the enhancement of glycolysis in TILs leads to improved tumor killing (51, 52). By establishing an experimental system for the rapid ex vivo imaging of the antigen-driven interaction of TILs with their target tumor cells, we further described the cellular mechanism of immune suppression by tumor cells: Prolonged PD-1 signaling in vivo impaired Ca2+-sensitive cytoskeletal polarization and diminished the lytic activity of TILs. We recapitulated key features of TIL suppression using the interaction of only CTLs and tumor cell spheroids. Time is a critical element of this mechanism because, first, the polarization defect affected cell couple maintenance more than cell couple formation and, second, PD-1 blockade was only effective over days within the tumor microenvironment, but not acutely in vitro.
Whereas TILs and SILs formed cell couples with their target tumor cells, often, within seconds of cell coupling, TILs and SILs increasingly developed lamellae pointing away from the target cell interface. Recruitment of the Arp2/3 complex to the interface periphery, the principal means by which T cells generate F-actin (53), was similar between TILs and CTLs. In contrast, recruitment of cofilin and Coronin 1A, key negative regulators of F-actin, was enhanced in TILs compared to that in CTLs. This is expected due to slower actin turnover by inhibition of the Arp2/3 complex by Coronin 1A and destabilization of F-actin filaments by cofilin. Reduced F-actin turnover is consistent with the decreased ability of TILs to move F-actin to the interface edge while clearing the center, which is considered to be critical for effective killing of target cells (36). Four elements of tumor immune suppression have already been linked to cofilin. The prostaglandin PGE2 is an established element of Clone 4 tumor immunosuppression (54) and it inhibits actin polymerization during phagocytosis through the activation of cofilin (55). Similarly, adenosine receptors, which in the tumor microenvironment mediate immune suppression by adenosine (6, 7), inhibit cofilin inactivation in cardiomyocytes (56). Cofilin activation can also be inhibited by oxidation in T cells and during mesenchymal cell motility (57, 58). The low-oxygen environment of solid tumors (59) is consistent with enhanced cofilin activity. Finally, reduced amounts of intracellular ATP induce cofilin activation by chronophin in neurons (60), a process that is equally conceivable in the energy-starved tumor microenvironment. Consistent with our suggestion that coronin 1A impairs F-actin dynamics in TILs, localized coronin 1A inhibition in CD4+ T cells enables the efficient formation of a signaling-enhancing, lamellal F-actin network (40).
We observed that diminished PD-1 engagement in vivo partially restored the ability of TILs to maintain a polarized cell couple with and kill their target tumor cells. In contrast, acute blockade of PD-1 during the interaction of TILs with target Renca tumor cells in vitro did not rescue either impaired TIL polarization or target cell killing. These data suggest that extended in vivo engagement of PD-1 induces a ”polarization-impaired state” that may require hours to days to form and revert. In support of this hypothesis, co-culture of primary T cells with various tumor cell types for 24 hours diminishes the ability of the T cells to form cell couples with allogeneic antigen–pulsed B cells (44), which is partially controlled by PD-1 (43). Similarly, in a model of persistent lymphocytic choriomeningitis virus (LCMV) infection, T cell mobility, an indicator of polarization, was impaired in a PD-1–dependent manner (17). Over time, T cells can switch between suppressed and active states. In vitro culture of TILs in IL-2 for more than 6 hours can overcome their diminished ability to kill target tumor cells (61), and resting of TILs for >24 hours in vitro restores impaired tetramer binding to the TCR (49). In the LCMV model, restoration of T cell motility in vivo became apparent after 1 hour of anti-PD-1 treatment and increased over the next 16 hours (17).
PD-1 could also exert indirect effects within the tumor microenvironment through enhancing the lineage stability of FoxP3+ Tregs (62), suppressing IFN-γ–dependent innate and adaptive immunity (63), and enabling the recruitment of novel peripheral T cell clonotypes to the tumor (64, 65). In addition, engagement of the costimulatory receptor CD28 is required for the restoration of T cell function by PD-1 blockade in vivo (66, 67). Whereas HA-specific Clone 4 TILs may have access to the CD28 ligands CD80 and CD86 on the surface of tumor-infiltrating professional APCs, they had no such opportunity during their interaction with Renca+HA target cells in vitro. At this time, it is still unresolved as to whether the extended time of PD-1 engagement or indirect effects of PD-1 within the tumor is more important in the establishment of the polarization-impaired state of TILs. The need for extended PD-1 engagement within the context of the tumor microenvironment makes it difficult to link the established signaling roles of PD-1, which have largely been investigated acutely and in vitro, to the in vivo inhibition of TIL polarization and function by PD-1. Nevertheless, transcriptional changes in TILs are conceivable, because sustained engagement of another receptor, the TCR by antigen, can substantially alter the transcriptional program of T cells (68), as was also shown in the context of cancer (69). In this context, differential expression of cytoskeletal components as determined here may contribute to the TIL polarization defect even though the dependence of this effect on PD-1 still needs to be determined. Metabolic changes are also conceivable, because enhanced tumor cell killing by blockade of PD-1 is associated with increased glycolysis in TILs (52). Future investigations of the mechanisms of PD-1 function should take into account the difference between the sustained in vivo and acute in vitro roles that we have observed.
Materials and Methods
Cells and media
Complete medium consisted of RPMI-1640 with L-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone defined, US source, GE healthcare), 50 μM β-mercaptoethanol (Gibco), and PenStrep (Gibco) at 100 U/mL penicillin and 100 μg/mL streptomycin. RencaWT cells were maintained in complete medium. RencaHA cells were maintained in complete medium supplemented with 100 μg/ml geneticin. Please note that ”Renca+HA” refers to either RencaWT or RencaHA cells incubated with a large excess of exogenous agonist peptide. ”RencaWT” refers to RencaWT cells that were not incubated with peptide. Clone 4 T cells were maintained in complete medium supplemented with 50 U/mL rh-IL-2 (NIH/NCI BRB preclinical repository; referred to as “IL-2 medium”). Phoenix cells were maintained in DMEM with 4. 5g/L D-glucose, L-glutamine and sodium pyruvate (Gibco), supplemented with 10% FBS (Hyclone define, US source, GE healthcare), MEM non-essential amino acids (Gibco), PenStrep (Gibco), 300 μg/mL Hygromycin (Invitrogen), and 1 μg/mL Diphtheria toxin (Sigma). Renca WT, RencaHA, and Phoenix cell lines were tested for the absence of mycoplasma.
Mice
Clone 4 TCR transgenic mice (RRID: IMSR_JAX:005307) were bred and maintained at the University of Bristol under specific pathogen–free conditions. All Clone 4 mice were culled for experimental use between 6 and 12 weeks of age. BALB/c mice were purchased from Charles River Laboratories at 6 weeks of age and then maintained at the University of Bristol under specific pathogen–free conditions. All mice were culled using schedule one methods and any experimental procedures were conducted in accordance with protocols approved by the UK Home Office.
Clone 4 CD8+ T cell isolation, stimulation, and retroviral transduction
The procedures used for the isolation and stimulation of TCR transgenic T cells were extensively detailed previously (29, 70, 71). Briefly, red blood cells were removed from dissociated spleens using ACK lysis buffer (Gibco) and splenocytes were plated at 5 x 106 cells per well, in 1 mL of complete medium, in a 24-well plate. KdHA peptide (IYSTVASSL) was added to a final concentration of 1 μg/mL, and the cells were incubated overnight. Primed cultures were washed five times in PBS to remove unbound peptide and then retrovirally transduced as extensively detailed previously (29, 70, 71). Briefly, T cells were transduced to express GFP-tagged proteins using recombinant Moloney murine leukemia virus (MMLV). Plasmids for the retroviral expression of F-tractin-GFP (71), tubulin-GFP (72), cofilin-GFP (30), coronin 1A-GFP (30) and chronophin-GFP (71) were described previously. T cells were resuspended in 2 mL of retrovirus-enriched supernatant from transfected Phoenix-E cells. Protamine sulphate was added to a final concentration of 8 μg/mL. Plates were centrifuged for 2 hours at 200g at 32°C. After transduction, the cells were re-plated at 4 x 106 cells per well, in 2 mL of IL-2 medium, in a 24-well plate. Cells were cultured for a further two days. CTLs were sorted and imaged or were adoptively transferred into RencaHA tumor–bearing mice, on day four.
RencaHA tumor growth in BALB/c mice and adoptive transfer of Clone 4 CD8+ T cells
To induce tumor growth, 6- to 8-week old female BALB/c mice were injected subcutaneously in the scruff of the neck with 1 x 106 RencaHA cells suspended in PBS. The mice were monitored regularly to assess tumor growth. Tumor size was measured using a caliper and calculated using the following equation: volume (V) = L2 x W/2, where length (L) was the longest measurement and width (W) was measured perpendicularly to length. Mice were culled if tumors exceeded 15 mm in any one direction or any humane end points were reached. GFP-positive Clone 4 CTLs were resuspended in PBS at 2 x 107 cells/mL, and 200 μL of cells were injected into the tail vein of a tumor-bearing mouse. Unless otherwise specified, only mice bearing tumors greater than 500 μm3 in volume were used, and tumors were harvested 96 hours after transfer.
In vivo anti–PD-1 immunotherapy
Mice received three doses of either monoclonal antibody against PD-1 (RMP1-14, Bio X cell, RRID: AB_10949053) or rat IgG2a isotype control (2A3, Bio X cell, RRID: AB_1107769) on days one, three, and five of treatment by tail vein injection. Each dose consisted of 250 μg of mAb in 200 μL of PBS. Adoptive transfer of CTLs occurred on day two, and the mice were culled on day six. Tumor measurements were taken on days one and six before culling.
Isolation of TILs
Tumors were collected into 3.2 mL of RPMI-1640, roughly chopped with sterile scissors, and tumor cell dissociation enzymes (Miltenyi Biotec) were added according to the manufacturer’s instructions. Tumors were incubated at 37°C for 45 min and briefly vortexed every 10 min to enhance dissociation. The tumor suspension was then passed through a 40-μm sieve, and red blood cells were removed with ACK lysis buffer. Cells were resuspended in 600 μL of MACs buffer (PBS , 2 mM EDTA, 0.5% BSA) with 65 μL of MACs microbeads (Miltenyi) and incubated at room temperature for 30 min. CD8a (Ly-2) microbeads were used to enrich for CD8+ T cells, whereas CD45 microbeads were used to enrich for all lymphocytes. Positive selection of the magnetically labelled cell populations was performed using MACS LS-columns (Miltenyi Biotec). Collected cells were resuspended in 500 μL of imaging buffer for immediate sorting or 500 μL of FACS buffer for antibody staining.
Generation of SILs
RencaWT mCherry cells were resuspended at a concentration of 1 x 105 cells/ml, mixed with Matrigel (Corning) at 4°C, seeded in a 24-well plate at a final concentration of 500 cells/blob, and left to solidify for 10 min at 37°C. Cell medium (2 ml) was added to each well and incubated at 37°C for 11 days. Each blob was washed twice in PBS and incubated for 30 min with 1 ml of Cell Recovery Solution (Corning). Spheroids were collected in a 15-ml Falcon tube and pulsed with KdHA peptide at a final concentration of 2 μg/mL for 1 hour. Pulsed spheroids were re-embedded in Matrigel together with 5 x 105 primed Clone 4 CTLs per blob. The following day, blobs were washed twice in PBS and incubated with 1 ml of Cell Recovery Solution (Corning). Spheroids were collected, washed through a 70-μm sieve, and then disaggregated to retrieve SILs in 500 μL of imaging buffer for immediate FACS sorting.
Imaging of Clone 4 CTLs, TILs, and SILs
After retroviral transduction or TIL/SIL extraction, T cell cultures were sorted to isolate GFP-positive T cells and resuspended in ‘imaging buffer’ (10% FBS in PBS with 1 mM CaCl2 and 0.5 mM MgCl2). As target cells, 1 x 106 RencaWT cells were pulsed with KdHA peptide at a final concentration of 2 μg/mL for 1 hour (to generate Renca+HA cells). Imaging was performed in glass-bottomed, 384-well optical imaging plates (Brooks life science systems) at 37°C with a Perkin Elmer UltraVIEW ERS 6FE confocal system attached to a Leica DM I6000 inverted epifluorescence microscope and a Yokogawa CSU22 spinning disk. A 40x oil-immersion lens (NA=1.25) with 2x2 camera binning (x and y pixel size of 0.4 μm) was used for all imaging experiments, unless otherwise stated. Every 20 s for 15 min, a z-stack of 21 GFP images (1 μm z-spacing) was acquired, as well as a single, mid-plane differential interference contrast (DIC) image. Some imaging experiments involved prior treatment of T cells with 40 nM Jasplakinolide, 10 μg/mL anti-PD-1 mAb (RMP1-14, Bio X cell, RRID: AB_10949053), or 5 mM NiCl2 with 20 μM BAPTA-AM (Molecular Probes). In all cases, reagents were added at the desired concentration for 30 min before imaging, and the cells were incubated at 37°C, for BAPTA-AM at 30°C. All reagents, with the exception of BAPTA-AM, were also present throughout the imaging process. To image the increase in the cytoplasmic Ca2+ concentration, Clone 4 T cells were incubated with 2 μM Fura-2 AM (Molecular Probes) for 30 min at room temperature in imaging buffer and washed twice thereafter. Because of limiting cell numbers, such washing was not feasible for Clone 4 TILs. Clone 4 T cells were activated with Renca+HA cells as described earlier and then every 10 s for 15 min, one bright-field image, one fluorescence image with excitation at 340 nm, and one fluorescence image with excitation at 380 nm were acquired at 37°C with a 40x oil objective (NA=1.25) on a Leica DM IRBE-based wide filed system equipped with Sutter DG5 illumination and a Photometrics Coolsnap HQ2 camera. For analysis, field-averaged background fluorescence was subtracted from the fluorescence data and the ratio of the Fura-2 images upon excitation at 340 nm versus 380 nm was calculated and multiplied by 100 to fit into the 8-bit display scale. Average ratio within a circular region of interest of the dimensions of the T cell was determined over time for each T cell.
Analysis of live-cell imaging data
Image analysis protocols have previously been described (29, 70, 71). Using Metamorph image analysis software (Molecular Devices), we identified CTLs forming conjugates with Renca target cells using the DIC reference images. Tight cell couple formation was defined as the first time point at which a maximally spread immune synapse formed, or two frames after initial cell contact, whichever occurred first. To assess CTL and TIL morphology, every DIC frame after tight cell couple formation was assessed for the presence of off-synapse lamellae, defined as transient membrane protrusions pointing away from the immune synapse, followed by retraction. For each cell couple, the initial position of the immune synapse on the Renca+HA target cell was compared to the position in the final frame. If the T cell had migrated by a distance greater than the diameter of the immune synapse, this was classed as a translocation event. To analyze protein spatiotemporal distribution, we used an improved version of the method described by Roybal et al. (30) as implemented in version 2.8 of the open source CellOrganizer software (http://CellOrganizer.org) (70, 73, 74). The first improvement is referred to as synapse tracking. The previous method required annotation of the cell couple synapse position for each time point, whereas the modified method requires only annotation at the initial time point. The fluorescence image for the initial frame is segmented by thresholding using the method described by Otsu (75), and an ellipse is fitted to the segmented object containing the manually annotated synapse position (the T cell). A window of 61 by 61 around the synapse position is chosen, and a circle containing the target cell is found by Hough transformation of the DIC image (76). The coordinates of the endpoints of the synapse are found as the intersection points of the T cell ellipse and the target cell circle. This process is repeated for preceding or subsequent frames with the T cell location information in the previous frame used to find the T cell in the current frame. The coordinates of the two synapse endpoints are saved across time. The second improvement is referred to as alignment refinement. To correct for inadvertent rotation during the initial alignment of all of the cells within the population to be analyzed, we implemented two modifications. The first step is to infer a rotational angle around the z-axis with a regression model using as features the intensities of evenly distributed coordinates in the cylinder originating from the synapse center. The second step is to decide whether to flip the cell by 180 degrees around the y-axis using a binary classifier with the total intensity of the first and second half of all slices as features. The regressor and classifier were trained with images that had been manually annotated. Finally, we fit a line through the synapse region in the central slice and perform a fine correction of the rotation around the z-axis using the angle of that line. As a first measurement of protein enrichment, we defined the 10% of the cell volume with the highest fluorescence intensity across the average of the entire population of analyzed cells and determined enrichment in this volume relative to the entire cell, as established previously (30). Additional regions of interest were defined by geometrical criteria. For the synapse region in cofilin-GFP–expressing T cells, we used a cylinder normal to and one voxel distant from the synapse plane, with its axis centered on the synapse region, a radius of 8 voxels, and a height of 4 voxels. For the region defining the interface center in F-tractin-GFP–expressing cells, we used a cylinder similar to the one in the cofilin analysis, but with a radius of 8/3 voxels. The computational analysis of cofilin-GFP distributions required additional adaptations because cofilin-GFP accumulated prominently in the nucleus. Enrichment in the 10% of the cell volume with the highest fluorescence intensity did not reliably reflect interface enrichment. Because the T cell nucleus often closely approaches the cellular interface, a cylindrical interface measurement region captured more of the interface accumulation but still not exclusively so. Therefore, we also sorted time points into those with >135% fluorescence intensity in regions of interface accumulation above cellular background and without accumulation, computationally determined enrichment in the 10% of the cell volume with the highest fluorescence intensity at only the time points with accumulation, and multiplied the enrichment intensity by the fraction of cell couples with cofilin accumulation.
Determination of F-actin amounts by Phalloidin staining
T cells were stained with Alexa Fluor 488–conjugated phalloidin as previously described (41) and cover slips were imaged as 3D image stacks on a Leica TCS SP8 AOBS laser scanning microscope using a 40x oil NA=1.3 objective. Using ImageJ, z-stacks of phalloidin-stained T cells were maximum-projected, and these images were then 2D median–filtered using a filter radius of 1, enabling noise reduction and edge-preservation. Otsu global thresholding (75) was used to binarize the images. Because this algorithm systematically overestimated the threshold, each threshold value was multiplied by 0.2 to provide a consistent offset. Objects were detected as regions of contiguous foreground pixels (8-way connectivity); as such, holes were filled, and a watershed algorithm was used to split merged objects. Cell areas and mean fluorescent intensities were measured and cell areas were converted to volumes assuming spherical geometry.
T cell killing assays
Renca+HA or RencaWT cells were stained with 10 μM cell trace violet (CTV) and plated at a density of 1 x 104 cells per well of a 384-well plate (Perkin Elmer) in 100 μL of complete medium. Cells were incubated at 37°C for a minimum of 5 hours to allow adherence and spreading. Before imaging, complete medium was exchanged for imaging buffer (50 μL/well). Clone 4 CTLs, TILs, or SILs were resuspended in imaging buffer at a density of 2 x 105 cells/mL. The imaging plate was then mounted on a Leica DMI6000 inverted epifluorescence widefield microscope at 37°C with a humidified CO2 enrichment attachment, a motorized stage, and adaptive focus control enabled by image analysis software (Leica LAS-X). CTV stained Renca cells were imaged, and the positions for two or three fields of view were saved per well. 50 μL of the T cell culture (giving a total of 1x104 cells/well) were added per well. Images of CTV-stained Renca cells were acquired every 30 min for 10 hours, using adaptive focus control to prevent drift. Using Metamorph image analysis software, a mask was generated using thresholding for light objects and adjusted to match the fluorescent signal. The same thresholding criteria were then used for all conditions within that experiment. The area of the field covered by CTV-stained Renca+HA cells was measured, the average percentage change in cell area was calculated for each time point, and a gradient describing the rate of cell area change was calculated. This was corrected for a control condition in which no T cells were added, providing a normalized value for the rate of cell area change across all experiments.
Generation of RencaHA-PD-L1−/− and PD-L1-GFP overexpressing cell lines
The methods we used closely follow those previously described (77). We designed sgRNAs using three programs: Benchling, Broad institute, and MIT CRISPR design. The six top common hits were selected. DNA oligonucleotides encoding these sgRNAs were inserted into the pSpCas9(BB)-2A-GFP vector (Addgene). RencaHA cells were transfected with these vectors using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 hours, FACS was used to separate single GFP-positive cells into the wells of a 96-well plate. Colonies were expanded and screened for PD-L1 loss by flow cytometry. Several clones were further screened to compare MHC class I expression, HA expression, and in vitro proliferative ability to the original RencaHA cell line, and the most similar clone (B10) was selected. The following primers encoding gRNA sequences were used for the CRISPR/Cas9-mediated knockout of PD-L1: pSpCas9PDL1 b F (CACCGTCCAAAGGACTTGTACGTGG) and pSpCas9PDL1 b R (AAACCCACGTACAAGTCCTTTGGAC). The cDNA encoding PD-L1 was cloned into a pSRαGFP vector to create a PD-L1–GFP fusion construct. RencaWT cells were then transfected with this construct using Lipofectamine 2000 and the cells were selected in medium containing geneticin (200 μg/ml). Cells were FACS-sorted for GFP-positive cells and analyzed by staining with antibodies against PD-L1. Flow cytometry was used to assess the expression and localization of GFP-conjugated PD-L1.
Phos-tag Western blotting to assess cofilin phosphorylation
Round-bottomed, 96-well plates were coated overnight with 10 μg/mL anti-CD3 antibody (145-2C11, BioLegend, RRID: AB_2632707) in PBS at 4°C. Clone 4 CTLs or TILs were resuspended in complete medium at a concentration of 1.5 x 106 cells/mL and incubated at 37°C. CTLs or TILs (100 μl) were added to the well together with 100μL of complete medium. Cells were centrifuged in the plate for 30 s at 250g to ensure uniform contact with the anti-CD3–coated plastic. The plate was immediately incubated at 37°C for 1, 2, or 5 min. After incubation, the cells were lysed with 100 μL of ice-cold RIPA buffer (Cell Signaling). Phos-tag reagent (Wako) was added to 15% SDS-PAGE gels according to the manufacturer’s protocols. Standard ECL protocols were used for immunodetection. The antibodies used were anti-cofilin (D3F9, Cell Signaling, RRID: AB_10622000), anti-GAPDH (14C10, Cell Signaling, RRID: AB_10693448), and horseradish peroxidase (HRP)-conjugated anti-IgG (Cell Signaling).
Analysis of cell surface receptor staining by flow cytometry
Cells to be fixed were stained with Zombie live/dead fixable exclusion dyes (BioLegend). Cells were incubated with Fc receptor blocking antibody (anti-CD16/CD32, clone 93, eBioscience). Surface staining was conducted in PBS containing 0.5% FBS. Cells were fixed with 4% paraformaldehyde (PFA) using standard protocols. Antibodies used for cell surface staining included: anti-CD279/PD-1 BV785 (29F.1A12, Biolegend, RRID: AB_2563680); anti-CD274/PD-L1 BV421 (MIH5, BD Bioscience, cat no 564714); anti-CD90.1/Thy1.1 PerCP Cy5.5 (OX-7, Biolegend, RRID: AB_961437); anti-Vβ8.1/Vβ8.2 TCR FITC (KJ16-133, eBioscience, RRID: AB_465261). SIL PD-1 expression was normalized to that of CTLs.
In vivo killing assay
BALB/c mice received tail vein injections of 4 x 106 primed Clone 4 CTLs or with PBS alone. On the second day after adoptive transfer, BALB/c splenocytes were pulsed with or without KdHA peptide at a final concentration of 2 μg/mL for 1 hour and then washed and resuspended in PBS. KdHA-pulsed splenocytes were then stained with 5 μM CellTrace violet (CTV) (high), whereas unpulsed splenocytes were stained with 0.5 μM CTV (low). Splenocytes were washed and resuspended in PBS. Immediately before injection, high- and low-stained splenocytes were mixed in equal numbers, and 5x106 cells were injected into the BALB/c mice. After 24 hours, the splenocytes were analyzed by flow cytometry and gated on live/dead dye exclusion and CTV staining to compare numbers of CTVhigh KdHA-pulsed splenocytes and CTVlow control splenocytes.
Statistical analysis
Data were checked for conformity to the Normal Distribution using the SAS procedure UNIVARIATE, including the Anderson-Darling and Shapiro-Wilk tests and graphical methods. To achieve conformity to the Normal Distribution, subcellular distribution enrichment ratios had to be transformed to logarithms. Data from independent experiments were pooled if the 95% confidence intervals of the independent experiments overlapped. To confirm the validity of such pooling of data from independent experiments, sources of variation were analyzed in more detail in three representative data sets (fig. S9). Specifically, our analysis asked how significant differences were between experimental repeats within a treatment, such as CTL experimental repeat 1 versus 2 versus however many repeats there were, as opposed to differences between treatments, CTLs versus TILs. First, each data set was tested for auto-correlation between repeated measurements on the same cell. First-order auto-correlation (P < 0.05) was detected in all sets of repeated measurements by the Durbin-Watson statistic (SAS procedure AUTOREG). Accordingly, further analysis was conducted by the method of General Linear Models with an AR(1) covariance matrix in SAS procedure MIXED using REML estimation. The results showed that, whereas the difference between treatments was always significant, experiments within treatments did not differ significantly. We applied this analysis to three representative experiments. The SAS/STAT14.3 package of the SAS Proprietary Software, version 9.4 was used. In the determination of time until the first off-interface lamella (Fig. 2G), differences between CTLs and TILs were significant with P = 0.0035; differences between experimental repeats had a P value of 0.06. In the determination of cofilin interface enrichment (fig. S4B), differences between CTLs and TILs were significant with P = 0.0015; differences between experimental repeats had a P value of 0.67. In the determination of the increase in the cytoplasmic Ca2+ concentration, differences between CTLs and TILs were significant with P < 0.0001; differences between experimental repeats had a P value of 0.99. Differences between experimental conditions were then analyzed at individual times. The Bonferroni correction for multiple comparisons was applied so that, for example across 11 time points analyzed, only differences with P < 0.0045 were considered to be significant and are reported as such. Applying this stringent correction increased the rate of false negatives. Our quantification of protein distributions, however, involved repeated measurements that are not necessarily independent, so the over-conservatism of the Bonferroni adjustment provides a compensatory safeguard. Imaging data grouped over time ranges were not affected by this problem. The frequency of occurrence of off-synapse lamellae or cellular translocation was analyzed using the z transformation for proportions. All other data types were analyzed by Student’s t tests, unpaired, unpaired with Welch’s correction for unequal variance, multiple t-tests with two-stage set-up method of Benjamini, Krieger and Yekutielli, ANOVA, or by nonparametric tests, Mann Whitney u-tests, or Kruskal-Wallis test, as appropriate and indicated in the figure legends. In few cases where it was difficult to satisfy all assumptions for the statistical tests under consideration, P values were calculated using the most appropriate tests, and the test resulting in the highest P value was reported as the most conservative approach. Standard tests were executed with GraphPad Prism version 7.0e.
Supplementary Material
Fig. S1. Clone 4 CTLs efficiently kill KdHA-pulsed splenocytes in vivo.
Fig. S2. Diminished MTOC polarization of Clone 4 TILs toward the T cell–target cell interface.
Fig. S3. F-actin distribution is impaired in TILs.
Fig. S4. Greater enrichment of cofilin, coronin 1A, and chronophin in TILs than in CTLs.
Fig. S5. Increased cofilin phosphorylation, but reduced cofilin abundance, in TILs
Fig. S6. Characterization of the RencaHA PD-L1−/− cell line and corresponding tumors.
Fig. S7. Loss of PD-1 engagement in vivo rescues cytoskeletal polarization.
Fig. S8. Acute in vitro blockade of PD-1 does not restore cytoskeletal polarization.
Fig. S9. Data pooling.
Movie S1. Representative interactions of a tubulin-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S2. Representative interactions of a F-tractin-GFP–expressing Clone 4 CTL, TIL, and Jasplakinolide-treated CTL with Renca+HA target cells.
Movie S3. Representative interactions of a cofilin-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S4. Representative interaction of a coronin 1A-GFP–expressing Clone 4 CTL with a Renca+HA target cell.
Movie S5. Representative interactions of an Arp3-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S6. Representative interaction of a chronophin-GFP–expressing Clone 4 CTL with a Renca+HA target cell.
Movie S7. Representative interactions of a Fura-2–loaded Clone 4 CTL and TIL with a Renca+HA target cell.
Acknowledgments:
We acknowledge the University of Bristol Flow Cytometry and Wolfson BioImaging facilities for providing equipment and technical support.
Funding: This work was supported by grants from the Wellcome Trust (102387/Z/13/Z/ to R.A. and 201254/Z/16/Z to G.E.), the Biotechnology and Biological Sciences Research Council (BB/P011578/1 – 15 NFSBIO to C.W.), the European Research Council (PCIG11-GA-2012-321554 to C.W.), the U.S. National Science Foundation (MCB1616492 to R.F.M.), the U.S. National Institutes of Health (P41 GM103712 to R.F.M.), and AstraZeneca, UK (to C.W.).
Footnotes
Competing interests: S.J.D. is a full-time employee and shareholder at AstraZeneca. The other authors declare that they have no competing interests.
Data and materials availability: The microscopy movies collected in this study and the models created from them are available at https://doi.org/10.1184/R1/11994639 and http://murphylab.cbd.cmu.edu/data/CTL2020/. The methods used for image analysis and modeling are available as part of the CellOrganizer package (http://www.cellorganizer.org). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Schildberg FA, Klein SR, Freeman GJ, Sharpe AH, Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity 44, 955–972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson AC, Joller N, Kuchroo VK, Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M, Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine 369, 122–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY, Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med 210, 2435–2466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng MW, Ngiow SF, von Scheidt B, McLaughlin N, Sparwasser T, Smyth MJ, Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res 70, 7800–7809 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Beavis PA, Milenkovski N, Henderson MA, John LB, Allard B, Loi S, Kershaw MH, Stagg J, Darcy PK, Adenosine Receptor 2A Blockade Increases the Efficacy of Anti-PD-1 through Enhanced Antitumor T-cell Responses. Cancer immunology research 3, 506–517 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Young A, Ngiow SF, Barkauskas DS, Sult E, Hay C, Blake SJ, Huang Q, Liu J, Takeda K, Teng MWL, Sachsenmeier K, Smyth MJ, Co-inhibition of CD73 and A2AR Adenosine Signaling Improves Anti-tumor Immune Responses. Cancer cell 30, 391–403 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, Sahai E, Reis e Sousa C, Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell 162, 1257–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R, Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Patsoukis N, Li L, Sari D, Petkova V, Boussiotis VA, PD-1 increases PTEN phosphatase activity while decreasing PTEN protein stability by inhibiting casein kinase 2. Mol Cell Biol 33, 3091–3098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T, Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J Exp Med 209, 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL, SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol 173, 945–954 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA, Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal 5, ra46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, Li L, Boussiotis VA, PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nature communications 6, 6692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons RM, Liu X, Pulko V, Harrington SM, Krco CJ, Kwon ED, Dong H, B7-H1 limits the entry of effector CD8(+) T cells to the memory pool by upregulating Bim. Oncoimmunology 1, 1061–1073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, Azuma M, Krummel MF, Bluestone JA, Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat Immunol 10, 1185–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinselmeyer BH, Heydari S, Sacristan C, Nayak D, Cammer M, Herz J, Cheng X, Davis SJ, Dustin ML, McGavern DB, PD-1 promotes immune exhaustion by inducing antiviral T cell motility paralysis. J Exp Med 210, 757–774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritter AT, Asano Y, Stinchcombe JC, Dieckmann NM, Chen BC, Gawden-Bone C, van Engelenburg S, Legant W, Gao L, Davidson MW, Betzig E, Lippincott-Schwartz J, Griffiths GM, Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity 42, 864–876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wulfing C, Purtic B, Klem J, Schatzle JD, Stepwise cytoskeletal polarization as a series of checkpoints in innate but not adaptive cytolytic killing. Proc Natl Acad Sci U S A 100, 7767–7772 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pores-Fernando AT, Zweifach A, Calcium influx and signaling in cytotoxic T-lymphocyte lytic granule exocytosis. Immunol Rev 231, 160–173 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Kim KD, Bae S, Capece T, Nedelkovska H, de Rubio RG, Smrcka AV, Jun CD, Jung W, Park B, Kim TI, Kim M, Targeted calcium influx boosts cytotoxic T lymphocyte function in the tumour microenvironment. Nature communications 8, 15365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidinger C, Shaw PJ, Feske S, STIM1 and STIM2-mediated Ca(2+) influx regulates antitumour immunity by CD8(+) T cells. EMBO Mol Med 5, 1311–1321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feske S, ORAI1 and STIM1 deficiency in human and mice: roles of store-operated Ca2+ entry in the immune system and beyond. Immunol Rev 231, 189–209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SF, Fouquet S, Chapon M, Salmon H, Regnier F, Labroquere K, Badoual C, Damotte D, Validire P, Maubec E, Delongchamps NB, Cazes A, Gibault L, Garcette M, Dieu-Nosjean MC, Zerbib M, Avril MF, Prevost-Blondel A, Randriamampita C, Trautmann A, Bercovici N, Early T cell signalling is reversibly altered in PD-1+ T lymphocytes infiltrating human tumors. PLoS One 6, e17621 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Cintron EJ, Monu NR, Frey AB, Tumor-induced disruption of proximal TCR-mediated signal transduction in tumor-infiltrating CD8+ lymphocytes inactivates antitumor effector phase. J Immunol 185, 7133–7140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babich A, Burkhardt JK, Coordinate control of cytoskeletal remodeling and calcium mobilization during T-cell activation. Immunol Rev 256, 80–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkinson SR, Williams NA, Morgan DJ, The role of intercellular adhesion molecule-1/LFA-1 interactions in the generation of tumor-specific CD8+ T cell responses. J Immunol 174, 3401–3407 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Janicki CN, Jenkinson SR, Williams NA, Morgan DJ, Loss of CTL function among high-avidity tumor-specific CD8+ T cells following tumor infiltration. Cancer Res 68, 2993–3000 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Singleton KL, Roybal KT, Sun Y, Fu G, Gascoigne NR, van Oers NS, Wülfing C, Spatiotemporal patterning during T cell activation is highly diverse. Sci Signal 2, ra15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roybal KT, Buck TE, Ruan X, Cho BH, Clark DJ, Ambler R, Tunbridge HM, Zhang J, Verkade P, Wulfing C, Murphy RF, Computational spatiotemporal analysis identifies WAVE2 and cofilin as joint regulators of costimulation-mediated T cell actin dynamics. Sci Signal 9, rs3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupfer A, Singer SJ, Cell biology of cytotoxic and helper T cell functions: immunofluorescence microscopic studies of single cells and cell couples. Annu. Rev. Immunol 7, 309–337 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Sinai P, Dozmorov IM, Song R, Schwartzberg PL, Wakeland EK, Wulfing C, T/B-cell interactions are more transient in response to weak stimuli in SLE-prone mice. Eur J Immunol 44, 3522–3531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims TN, Soos TJ, Xenias HS, Dubin-Thaler B, Hofman JM, Waite JC, Cameron TO, Thomas VK, Varma R, Wiggins CH, Sheetz MP, Littman DR, Dustin ML, Opposing effects of PKCtheta and WASp on symmetry breaking and relocation of the immunological synapse. Cell 129, 773–785 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Mrass P, Takano H, Ng LG, Daxini S, Lasaro MO, Iparraguirre A, Cavanagh LL, von Andrian UH, Ertl HC, Haydon PG, Weninger W, Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J Exp Med 203, 2749–2761 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, Amigorena S, In vivo imaging of cytotoxic T cell infiltration and elimination of a solid tumor. J Exp Med 204, 345–356 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritter AT, Kapnick SM, Murugesan S, Schwartzberg PL, Griffiths GM, Lippincott-Schwartz J, Cortical actin recovery at the immunological synapse leads to termination of lytic granule secretion in cytotoxic T lymphocytes. Proc Natl Acad Sci U S A 114, E6585–E6594 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rak GD, Mace EM, Banerjee PP, Svitkina T, Orange JS, Natural killer cell lytic granule secretion occurs through a pervasive actin network at the immune synapse. PLoS Biol 9, e1001151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown AC, Oddos S, Dobbie IM, Alakoskela JM, Parton RM, Eissmann P, Neil MA, Dunsby C, French PM, Davis I, Davis DM, Remodelling of cortical actin where lytic granules dock at natural killer cell immune synapses revealed by super-resolution microscopy. PLoS Biol 9, e1001152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi J, Wu XS, Crites T, Hammer JA 3rd, Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol Biol Cell 23, 834–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Britton GJ, Ambler R, Clark DJ, Hill EV, Tunbridge HM, McNally KE, Burton BR, Butterweck P, Sabatos-Peyton C, Hampton-O'Neil LA, Verkade P, Wulfing C, Wraith DC, PKCtheta links proximal T cell and Notch signaling through localized regulation of the actin cytoskeleton. eLife 6, 20003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roybal KT, Mace EM, Clark DJ, Leard AD, Herman A, Verkade P, Orange JS, Wulfing C, Modest Interference with Actin Dynamics in Primary T Cell Activation by Antigen Presenting Cells Preferentially Affects Lamellal Signaling. PLoS One 10, e0133231 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gohla A, Birkenfeld J, Bokoch GM, Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol 7, 21–29 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Ramsay AG, Clear AJ, Fatah R, Gribben JG, Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood 120, 1412–1421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsay AG, Clear AJ, Kelly G, Fatah R, Matthews J, Macdougall F, Lister TA, Lee AM, Calaminici M, Gribben JG, Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: implications for the tumor microenvironment and immunotherapy. Blood 114, 4713–4720 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koneru M, Monu N, Schaer D, Barletta J, Frey AB, Defective adhesion in tumor infiltrating CD8+ T cells. J Immunol 176, 6103–6111 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Monu N, Frey AB, Suppression of proximal T cell receptor signaling and lytic function in CD8+ tumor-infiltrating T cells. Cancer Res 67, 11447–11454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heim L, Friedrich J, Engelhardt M, Trufa DI, Geppert CI, Rieker RJ, Sirbu H, Finotto S, NFATc1 Promotes Antitumoral Effector Functions and Memory CD8(+) T-cell Differentiation during Non-Small Cell Lung Cancer Development. Cancer Res 78, 3619–3633 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Palmer DC, Guittard GC, Franco Z, Crompton JG, Eil RL, Patel SJ, Ji Y, Van Panhuys N, Klebanoff CA, Sukumar M, Clever D, Chichura A, Roychoudhuri R, Varma R, Wang E, Gattinoni L, Marincola FM, Balagopalan L, Samelson LE, Restifo NP, Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J Exp Med 212, 2095–2113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blohm U, Roth E, Brommer K, Dumrese T, Rosenthal FM, Pircher H, Lack of effector cell function and altered tetramer binding of tumor-infiltrating lymphocytes. J Immunol 169, 5522–5530 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Petit AE, Demotte N, Scheid B, Wildmann C, Bigirimana R, Gordon-Alonso M, Carrasco J, Valitutti S, Godelaine D, van der Bruggen P, A major secretory defect of tumour-infiltrating T lymphocytes due to galectin impairing LFA-1-mediated synapse completion. Nature communications 7, 12242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, Kaech SM, Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, Tonc E, Schreiber RD, Pearce EJ, Pearce EL, Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nolz JC, Gomez TS, Zhu P, Li S, Medeiros RB, Shimizu Y, Burkhardt JK, Freedman BD, Billadeau DD, The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr Biol 16, 24–34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basingab FS, Ahmadi M, Morgan DJ, IFNgamma-Dependent Interactions between ICAM-1 and LFA-1 Counteract Prostaglandin E2-Mediated Inhibition of Antitumor CTL Responses. Cancer immunology research 4, 400–411 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Serezani CH, Kane S, Medeiros AI, Cornett AM, Kim SH, Marques MM, Lee SP, Lewis C, Bourdonnay E, Ballinger MN, White ES, Peters-Golden M, PTEN directly activates the actin depolymerization factor cofilin-1 during PGE2-mediated inhibition of phagocytosis of fungi. Sci Signal 5, ra12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeidan A, Gan XT, Thomas A, Karmazyn M, Prevention of RhoA activation and cofilin-mediated actin polymerization mediates the antihypertrophic effect of adenosine receptor agonists in angiotensin II- and endothelin-1-treated cardiomyocytes. Molecular and cellular biochemistry 385, 239–248 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Klemke M, Wabnitz GH, Funke F, Funk B, Kirchgessner H, Samstag Y, Oxidation of cofilin mediates T cell hyporesponsiveness under oxidative stress conditions. Immunity 29, 404–413 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Cameron JM, Gabrielsen M, Chim YH, Munro J, McGhee EJ, Sumpton D, Eaton P, Anderson KI, Yin H, Olson MF, Polarized cell motility induces hydrogen peroxide to inhibit cofilin via cysteine oxidation. Curr Biol 25, 1520–1525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erapaneedi R, Belousov VV, Schafers M, Kiefer F, A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J 35, 102–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang TY, Minamide LS, Bamburg JR, Bokoch GM, Chronophin mediates an ATP-sensing mechanism for cofilin dephosphorylation and neuronal cofilin-actin rod formation. Dev Cell 15, 691–703 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radoja S, Saio M, Schaer D, Koneru M, Vukmanovic S, Frey AB, CD8(+) tumor-infiltrating T cells are deficient in perforin-mediated cytolytic activity due to defective microtubule-organizing center mobilization and lytic granule exocytosis. J Immunol 167, 5042–5051 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Stathopoulou C, Gangaplara A, Mallett G, Flomerfelt FA, Liniany LP, Knight D, Samsel LA, Berlinguer-Palmini R, Yim JJ, Felizardo TC, Eckhaus MA, Edgington-Mitchell L, Martinez-Fabregas J, Zhu J, Fowler DH, van Kasteren SI, Laurence A, Bogyo M, Watts C, Shevach EM, Amarnath S, PD-1 Inhibitory Receptor Downregulates Asparaginyl Endopeptidase and Maintains Foxp3 Transcription Factor Stability in Induced Regulatory T Cells. Immunity 49, 247–263 e247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, Chang H, Lovitch SB, Horak C, Weber JS, Weirather JL, Wolchok JD, Postow MA, Pavlick AC, Chesney J, Hodi FS, MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med 10, (2018). [DOI] [PubMed] [Google Scholar]
- 64.Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, Curtis C, Bucktrout SL, Davis MM, Chang ALS, Chang HY, Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, Chiang EY, Iftikhar H, O'Gorman WE, Au-Yeung A, Takahashi C, Goldstein LD, Poon C, Keerthivasan S, de Almeida Nagata DE, Du X, Lee HM, Banta KL, Mariathasan S, Das Thakur M, Huseni MA, Ballinger M, Estay I, Caplazi P, Modrusan Z, Delamarre L, Mellman I, Bourgon R, Grogan JL, Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579, 274–278 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD, T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL, Sharpe AH, Freeman GJ, Blazar BR, Turka LA, Owonikoko TK, Pillai RN, Ramalingam SS, Araki K, Ahmed R, Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson PO, Manzo BA, Sundstedt A, Minaee S, Symonds A, Khalid S, Rodriguez-Cabezas ME, Nicolson K, Li S, Wraith DC, Wang P, Persistent antigenic stimulation alters the transcription program in T cells, resulting in antigen-specific tolerance. Eur J Immunol 36, 1374–1385 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, Lauer P, Brockstedt DG, Knoblaugh SE, Hammerling GJ, Schell TD, Garbi N, Greenberg PD, Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ambler R, Ruan X, Murphy RF, Wulfing C, Systems Imaging of the Immune Synapse. Methods Mol Biol 1584, 409–421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roybal KT, Mace EM, Mantell JM, Verkade P, Orange JS, Wulfing C, Early Signaling in Primary T Cells Activated by Antigen Presenting Cells Is Associated with a Deep and Transient Lamellal Actin Network. PLoS One 10, e0133299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tskvitaria-Fuller I, Rozelle AL, Yin HL, Wülfing C, Regulation of sustained actin dynamics by the TCR and costimulation as a mechanism of receptor localization. J. Immunol 171, 2287–2295 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Murphy RF, CellOrganizer: Image-derived models of subcellular organization and protein distribution. Methods in cell biology 110, 179–193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buck TE, Li J, Rohde GK, Murphy RF, Toward the virtual cell: automated approaches to building models of subcellular organization "learned" from microscopy images. Bioessays 34, 791–799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Otsu N, A threshold selection method from gray-level histograms. . IEEE Trans Systems Man Cybernetics 9, 62–66 (1979). [Google Scholar]
- 76.Duda RO, Hart PE, Use of the Hough transformation to detect lines and curves in pictures. SRI INTERNATIONAL MENLO PARK CA ARTIFICIAL INTELLIGENCE CENTER SRI-TN-36, (1971). [Google Scholar]
- 77.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system. Nature protocols 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Clone 4 CTLs efficiently kill KdHA-pulsed splenocytes in vivo.
Fig. S2. Diminished MTOC polarization of Clone 4 TILs toward the T cell–target cell interface.
Fig. S3. F-actin distribution is impaired in TILs.
Fig. S4. Greater enrichment of cofilin, coronin 1A, and chronophin in TILs than in CTLs.
Fig. S5. Increased cofilin phosphorylation, but reduced cofilin abundance, in TILs
Fig. S6. Characterization of the RencaHA PD-L1−/− cell line and corresponding tumors.
Fig. S7. Loss of PD-1 engagement in vivo rescues cytoskeletal polarization.
Fig. S8. Acute in vitro blockade of PD-1 does not restore cytoskeletal polarization.
Fig. S9. Data pooling.
Movie S1. Representative interactions of a tubulin-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S2. Representative interactions of a F-tractin-GFP–expressing Clone 4 CTL, TIL, and Jasplakinolide-treated CTL with Renca+HA target cells.
Movie S3. Representative interactions of a cofilin-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S4. Representative interaction of a coronin 1A-GFP–expressing Clone 4 CTL with a Renca+HA target cell.
Movie S5. Representative interactions of an Arp3-GFP–expressing Clone 4 CTL and TIL with Renca+HA target cells.
Movie S6. Representative interaction of a chronophin-GFP–expressing Clone 4 CTL with a Renca+HA target cell.
Movie S7. Representative interactions of a Fura-2–loaded Clone 4 CTL and TIL with a Renca+HA target cell.