Abstract
Organismal interactions within microbial consortia and their responses to harmful intruders remain largely understudied. An important step toward the goal of understanding functional ecological interactions and their evolutionary selection is the study of increasingly complex microbial interaction systems. Here, we discovered a tripartite biosystem consisting of the fungus Aspergillus nidulans, the unicellular green alga Chlamydomonas reinhardtii, and the algicidal bacterium Streptomyces iranensis. Genetic analyses and MALDI-IMS demonstrate that the bacterium secretes the algicidal compound azalomycin F upon contact with C. reinhardtii. In co-culture, A. nidulans attracts the motile alga C. reinhardtii, which becomes embedded and surrounded by fungal mycelium and is shielded from the algicide. The filamentous fungus Sordaria macrospora was susceptible to azalomycin F and failed to protect C. reinhardtii despite chemotactically attracting the alga. Because S. macrospora was susceptible to azalomycin F, this data imply that for protection the fungus needs to be resistant. Formation of the lichen-like association between C. reinhardtii and A. nidulans increased algal growth. The protection depends on the increased amounts of membrane lipids provided by resistant fungi, thereby generating a protective shelter against the bacterial toxin. Our findings reveal a strategy whereby algae survive lethal environmental algicides through cooperation with fungi.
Subject terms: Microbial ecology, Microbiome, Microbial ecology, Antibiotics, Fungal ecology
Introduction
Algae and fungi have coexisted and coevolved for at least 400 million years, as deduced from fossil records showing macroalgae parasitized by fungi [1], as well as lichens [2, 3]. During the course of evolution, terrestrial algae and fungi have formed lichens but some have lost this ability [4]. Tight associations between algae and fungi, in particular in lichens, provide examples of mutualism that has been maintained over hundreds of millions of years up to the present [4]. However, the benefits for the algal partners remain a matter of debate as the mycobiont often gains a fitness advantage at the expense of the photobiont. Furthermore, the mycobiont can control the growth and metabolism of the photobiont, which is referred to as “helotism” [5], a type of “algal slavery” [6] with reference to the master–servant relationship of ancient sparta.
Microorganisms are often subject to rapidly changing biotic and abiotic conditions in their natural habitats. To cope with these environmental changes, they often produce natural products that are low molecular mass metabolites [7, 8]. They are often dispensable for growth, produced at a certain stage in the life cycle of the producing organism and can be of pharmaceutical value [7]. In many cases, these natural products are weapons to defend habitats against competing microorganisms. Metagenomic analysis has revealed that lichens are colonized by various other microorganisms and form “microhabitats” themselves [9, 10]. Among the potentially beneficial colonizers, there are streptomycetes equipped with an antibiotic arsenal [11–14]. How this multipartner interaction remains stable through mutualistic exchange and how lichens cope with potentially toxin-producing colonizers is largely unknown.
Co-existance of filamentous fungi, streptomycetes and other producers of natural products often triggers the production of these effector chemicals [15]. For instance, Streptomyces rapamycinicus and its closest relative S. iranensis (Supplementary Fig. 1) are capable of eliciting the production of the archetypical lichen metabolites orsellinic acid and lecanoric acid in the fungal partner A. nidulans. However these metabolites do not show activity against S. rapamycinicus, the closest relative to S. iranensis [16–18]. Since A. nidulans interacts with the green alga C. reinhardtii [19] and lichens are colonized by potentially harmful streptomycetes [11, 12, 14], we hypothesized that S. iranensis might interfere with this algal–fungal association. Here, we have found a tripartite interaction of the model green alga C. reinhardtii with the established bipartite A. nidulans-S. iranensis co-culture system [17]. We found that both partners have distinct effects on the alga: (i) the bacterium kills the alga using the natural product azalomycin F, (ii) the alga is protected by fungi resistant against azalomycin F like A. nidulans, and (iii) the presence of A. nidulans increases the growth rate and overall biomass of C. reinhardtii. We show that the algal–fungal association itself is a defense mechanism against toxins.
Materials and methods
Microbial strains and plasmids
Microbial strains and plasmids used in this study are listed in Supplementary Table 1.
Streptomyces iranensis mutants
For deleting genes of S. iranensis the method described by Netzker et al. [20] was essentially followed. Oligonucleotides applied for gene deletion are listed in Supplementary Table 2.
Production and purification of azalomycin F from S. iranensis
To obtain samples of the azalomycin F complex from S. iranensis, 50 mL of TSBY [21] were inoculated with 5 × 108 spores and the bacteria were incubated for 4 days at 28 °C at 180 rpm. The entire culture was centrifuged to separate the biomass from the culture supernatant. After lyophilisation of the biomass, the content of the pellet was extracted twice with 50 mL methanol at 60 °C. Both extraction solutions were combined and evaporated to dryness under reduced pressure and dissolved in 2 mL 75% (v/v) aqueous methanol (MeOH(aq)). The extract was subsequently loaded onto an N-vinylpyrrolidone-divinylbenzene copolymer resin SPE column (Macherey-Nagel, Düren, Germany), washed with 50% (v/v) MeOH(aq) and eluted with 70% (v/v) MeOH(aq) to obtain purified azalomycin F complex. The eluent was reduced to dryness, dissolved in pure MeOH, and 5 μL were loaded onto an ultrahigh-performance liquid chromatography-mass spectrometry for analysis. The chromatographic system consisted of an UltiMate 3000 binary rapid-separation liquid chromatograph with photodiode array detector (Thermo Fisher Scientific, Dreieich, Germany). The MS was performed on an LTQ XL linear ion trap mass spectrometer (Thermo Fisher Scientific, Dreieich, Germany) equipped with an electrospray ion source. The sample was analyzed on a 150 mm by 4.6 mm Accucore reversed-phase (RP)-MS column with a particle size of 2.6 μm (Thermo Fisher Scientific, Dreieich, Germany) at a flow rate of 1 mL/min with the following gradient: 0.1% (v/v) HCOOH-MeCN/0.1% (v/v) HCOOH–H2O 0/100, increased to 80/20 in 15 min and then to 100/0 in the following 20 min, held at 100/0 for 2 min, and reversed to 0/100 in 2 min, with detection range at 190–400 nm. For the azalomycin F-deficient mutants, cultivation, extraction, and analysis of azalomycin F are described in the Supplementary Methods Section 1.3.
Sample preparation for matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-IMS)
MALDI-IMS was carried out on indium tin oxide (ITO) covered glass slides (Bruker Daltonics, Bremen, Germany). Slides were sterilized over a Bunsen burner and 3 mL of 1% (w/v) TAP [22] agar were spread onto the slide for S. iranensis monoculture. For co-cultivation, 1.5 mL 1% (w/v) TAP agar was spread onto one half of the ITO glass slide and allowed to solidify. The other half was covered with TAP agar, containing submerged C. reinhardtii. For this purpose, a C. reinhardtii culture was centrifuged at 3500 × g for 5 min and the supernatant was discarded. The pellet was resuspended in liquid TAP agar with a temperature of about 35 °C to yield an OD750 of 2. This suspension was then added to the other side of the slide and allowed to solidify. Finally, 15 µL S. iranensis mycelium, prewashed in PBS, was set between both agar sides. The slides were incubated at 26 °C and a light intensity of 30 µE m−2 s−1 in petri dishes for 5 days and subsequently dried in a hybridization oven at 37 °C for 48 h. For MALDI-IMS analysis, see Supplementary Methods 1.5.
Chemotaxis assay
Chemotaxis was measured by counting C. reinhardtii cells that migrated into glass capillaries loaded with a potential attractant [23, 24]. Three microliters capillaries (Drummond Microcaps®, Drummond Scientific, Broomall, PA, USA) were loaded with TAP supplemented with 10 g/L glucose, 0.5 g/L MgCl2∙7H2O, 1 mL/L trace elements, 10 mL/L 0.5 M arginine, 3 mL/L 3 mM FeSO4 and 3 mL/L 0.1% (w/v) p-aminobenzoic acid (PABA) as control. Equally supplemented supernatant of an A. nidulans culture grown in TAP medium served as experimental sample and as a negative control PBS was used. The capillaries were placed upright into a 2 mL reaction tube. For each sample, three technical replicates were tested. 100 µL of a C. reinhardtii suspension with an OD750 of 4 were placed at the bottom of each 2 mL reaction tube. When attracted, the C. reinhardtii cells had to swim against gravity. Incubation was carried out at 26 °C in the dark for 5 h. After incubation, the capillaries were cleaned with a paper towel to remove attached cells, and the content was transferred to 27 µL of PBS + 4% (v/v) paraformaldehyde. C. reinhardtii cells which had migrated into the capillaries were counted using a Neubauer counting chamber.
Chlorophyll quantification assay
The assay was performed in order to assess the chlorophyll content within a C. reinhardtii suspension and to estimate the viability of the cells 24 h after treatment with azalomycin F. Each well was inoculated with 100 µL TAP supplemented with glucose, MgSO4, PABA, FeSO4, trace elements, and arginine (see Supplementary Information 1.1) and containing 104 conidia of A. nidulans. The 96-well plates were incubated at 37 °C for 16–18 h in a plate shaker with 1000 rpm. Then, 100 µL of a C. reinhardtii suspension with an OD750 of 2 was added to each well and fluorescence was measured (excitation 480 nm, emission 684 nm) on a Tecan Infinite M200 pro microplate reader (Tecan Trading AG, Männedorf, Switzerland). To allow C. reinhardtii to migrate into the mycelium, the plate was incubated for 5 h at 26 °C, 120 rpm and 30 µE m−2 s−1. Then, 200 ng azalomycin F were added and the fluorescence was measured again to determine the values for the start of the treatment. Incubation was continued for 24 h before measuring fluorescence using an excitation wavelength of 480 nm and measuring emission at 684 nm.
Co-cultivation chamber experiments
Co-cultivation chambers were assembled as previously reported by Paul et al. [25]. The cultivation chambers were separated by a polyvinylidene fluoride (PVDF) membrane (Merck, Darmstadt, Germany) with a pore size of 0.1 µm. Each chamber was filled with 250 mL TAP supplemented with glucose, MgSO4, PABA, FeSO4, trace elements, and arginine (see Supplementary Information 1.1). C. reinhardtii was inoculated to obtain a final OD750 of 0.1 and A. nidulans mycelium was inoculated from a Miracloth-filtered overnight culture. The chambers were incubated at 26 °C, 80 rpm and 30 µE m−2 s−1. OD750 measurement was conducted on SERVA Nanophotometer Pearl (SERVA Heidelberg, Germany). After 24 h, 12.5 mL of 20% (w/v) glucose, 0.5 g/L MgSO4∙7H2O, 750 µL of 0.1% (w/v) PABA, 750 µL of 10 mM FeSO4, and 2.5 mL of 0.5 M L-arginine were added to each half of the co-cultivation chamber. A. nidulans dry mass was determined by filtering the mycelium after the experiment and drying it for 7 days at 60 °C.
Protection of C. reinhardtii by external polar lipids
Phosphatidylinositol sodium salt from soybean was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cardiolipin (CL), 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), 16:0-i15:0 phosphatidylcholine (PC), d18:1/18:0 C18 glucosyl(β)ceramide (GC), and extract of polar lipids of Saccharomyces cerevisiae (YE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). DPPG, CL, and GC were dissolved in 95:5 methanol:water to obtain a 0.5 mg/mL stock solution. PC was dissolved in 95:5 methanol:water to obtain a 4 mg/mL stock solution. YE was purchased as a 25 mg/mL solution in chloroform. For application of the lipids to C. reinhardtii, the solvent was evaporated under a gentle stream of nitrogen. The lipids were dissolved in a culture of C. reinhardtii with an OD750 of 2. The final concentrations of the lipids were chosen similar to Xu et al. [26]. To dissolve the lipids in TAP, the cultures were incubated in a ThermoMixer® (Eppendorf, Hamburg, Germany) at 30 °C and rapid shaking for 30 min. Afterwards, the cultures were incubated in 24 well plates at 26 °C, 120 rpm and 30 µE m−2 s−1. Autofluorescence was measured after 24 h of incubation as described for the chlorophyll quantification assay.
Results
Streptomyces iranensis specifically releases algicidal azalomycin F in presence of C. reinhardtii
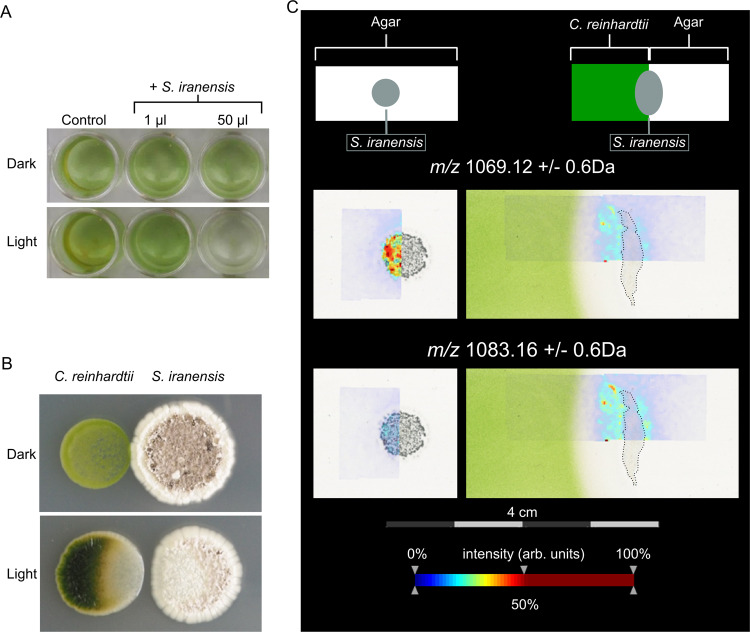
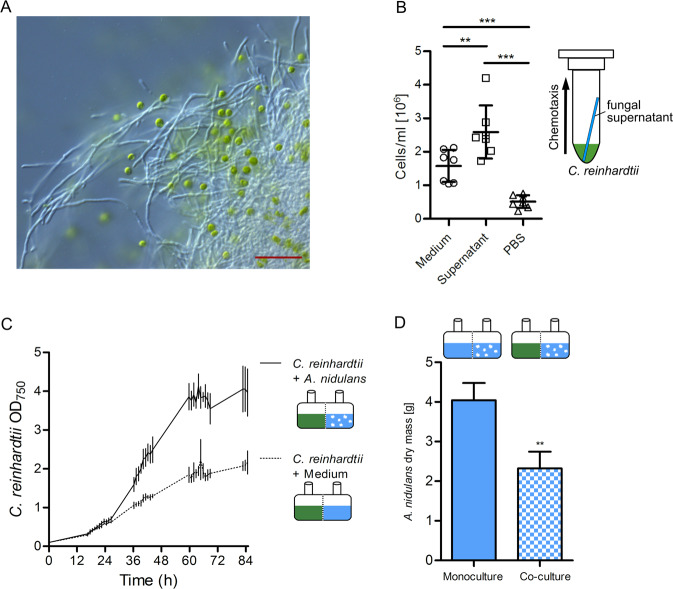
A. nidulans and the green alga C. reinhardtii quickly assemble in co-culture to form an algal–fungal association [19]. We conducted co-cultivation experiments of C. reinhardtii and S. iranensis to identify possible effects of the bacterium on the alga. When co-cultured with S. iranensis under light, C. reinhardtii lost its green color in liquid medium (Fig. 1a) as well as on solid agar (Fig. 1b). Because decolorization is indicative of cell death of C. reinhardtii [27], S. iranensis likely produces one or several algicidal agents.
Fig. 1. S. iranensis actively secretes azalomycin F showing light dependent algicidal activity.
Co-cultures of S. iranensis and C. reinhardtii (a, b). Co-cultivation in light and dark with and without S. iranensis in TAP medium (a) and on agar (b). Loss of green coloration indicates killing of C. reinhardtii. c MALDI-IMS analysis. Top: schematic position of the S. iranensis colony, sterile TAP agar and TAP agar containing C. reinhardtii on the ITO-slide. MALDI-IMS images indicate color-coded abundance of ion m/z = 1069.12 ± 0.6 Da (azalomycin F3a, [M + H]+) and ion m/z = 1083.16 ± 0.6 Da (azalomycin F4a, [M + H]+). Color code: blue = low abundance, red = high abundance of ion. MALDI-IMS shows that both azalomycins are released only in co-cultivation of S. iranensis with C. reinhardtii which was inoculated into the left half of the agar.
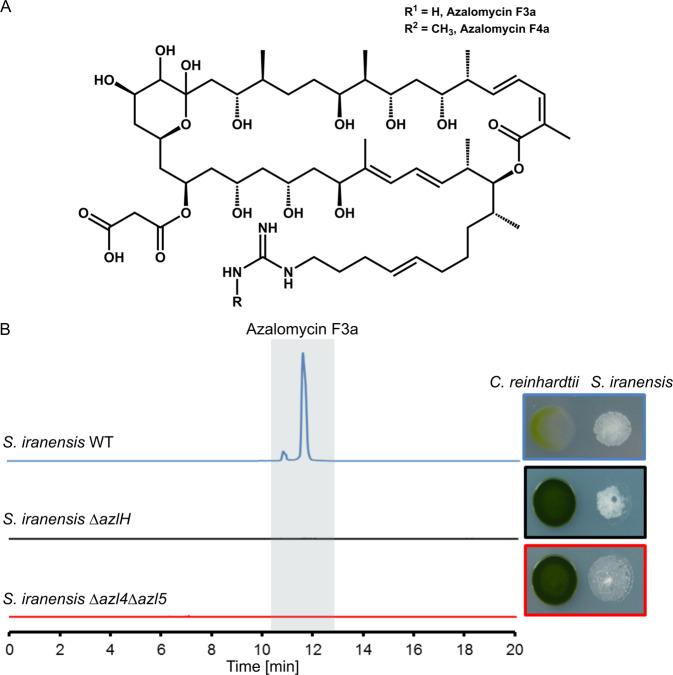
Since C. reinhardtii was decolorized over a certain distance on solid agar (Fig. 1b, c), we detected and visualized compounds that are located in the killing zone by MALDI-IMS. Two ions with m/z = 1069.12 and m/z = 1083.16 were detected (Fig. 1c). AntiSMASH analysis of the S. iranensis genome suggested that these two compounds could be azalomycin F3a and F4a [21, 28]. The azalomycin F complex is formed by a group of macrolactones, which were first isolated from Streptomyces hygroscopicus var. azalomyceticus. Azalomycin F consists of a 34-membered macrolactone ring and a guanidine-containing side chain (Fig. 2a, [29]) and is active against fungi as well as Gram-positive bacteria [30, 31]. A comparison of the polyketide synthase (PKS) genes (Supplementary Fig. 2) and the genes coding for tailoring enzymes (Supplementary Table 3) of the published azalomycin F biosynthetic gene cluster with the gene cluster found in S. iranensis supports the assumption that the two found ions indeed correspond to the azalomycin F complex. To substantiate this finding, azalomycin F-deficient mutants of S. iranensis were generated (Fig. 2, Supplementary Fig. 3). In the ∆azl4∆azl5 mutant strain, the azalomycin F biosynthesis genes azl4 and azl5 were deleted. Azl4 catalyzes the activation of 4-guanidinobutyric acid by addition of coenzyme A to the molecule. Azl5 consecutively loads 4-guanidinobutyryl-CoA onto the azalomycin F PKS [32]. In the second mutant strain ∆azlH, the azlH gene, which encodes a part of the PKS [30, 32], was deleted. Neither of the mutants ∆azl4∆azl5 and ∆azlH produced detectable amounts of azalomycin F and both failed to decolorize C. reinhardtii (Fig. 2b). MALDI-IMS showed that azalomycin F was exclusively present inside the bacterial colony in monoculture of S. iranensis, while it was released in co-culture of the bacterium with C. reinhardtii (Fig. 1c). We conclude that azalomycin F is the sole algicidal agent that is present under our culturing conditions and that S. iranensis produces and releases it in the presence of C. reinhardtii.
Fig. 2. Structure of azalomycin F and its production by S. iranensis.
a Azalomycin F complex consists of azalomycins F3a and F4a. b MS analysis in positive mode of S. iranensis wild type (WT), S. iranensis mutants ∆azl4∆azl5 and ∆azlH. The mutants failed to produce azalomycin F and to decolorize C. reinhardtii in co-culture (pictures on the right side). Extracted Ion Chromatogram: m/z = 1068.65–1068.80.
Azalomycin F-induced algal decolorization is light dependent
We observed that C. reinhardtii was not decolorized when it was co-cultivated with S. iranensis in the dark (Fig. 1a, b). This was not due to lack of release of azalomycin F because MALDI-IMS showed that azalomycin F was still released by S. iranensis in the presence of C. reinhardtii in the dark (Supplementary Fig. 4). To provide evidence for a light-dependent algicidal activity of azalomycin F, C. reinhardtii cells were stained with SYTOX Blue after treatment with the algicide under light and in the dark. SYTOX Blue is a non-permeating dye that exhibits fluorescence upon binding to nucleic acids. Therefore, blue fluorescence is only detectable when cell membranes are leaky in damaged or dead cells. A positive SYTOX Blue signal was only detected when C. reinhardtii was treated with azalomycin F under illumination (Supplementary Fig. 5). Azalomycin F-treated cells cultured in the dark maintained their red autofluorescence, indicating that C. reinhardtii was not killed by azalomycin F in the dark. To analyze whether the effect of azalomycin F in light is connected to photosynthesis, we investigated the C. reinhardtii mutants cc4147 FuD7 (psbA deletion) and cc4385 (psbD deletion) (Supplementary Fig. 6). psbA and psbD code for the D1 and D2 reaction center proteins of photosystem II, respectively [33, 34]. As shown in Supplementary Fig. 6, azalomycin F also had a lethal effect on both mutant strains indicating that photosynthesis is not required for its lethal activity. To investigate the spectrum of algae affected by azalomycin F, we expanded our studies to microalgae belonging to different classes. Both Euglena gracilis and Haematococcus pluvialis were susceptible to azalomycin F as indicated by decolorization, a positive SYTOX blue signal and rounding up of the cells (Supplementary Fig. 7). Thus, a wide range of microalgae are susceptible to azalomycin F.
Association of Aspergillus nidulans and Chlamydomonas reinhardtii increased algal survival in presence of azalomycin F
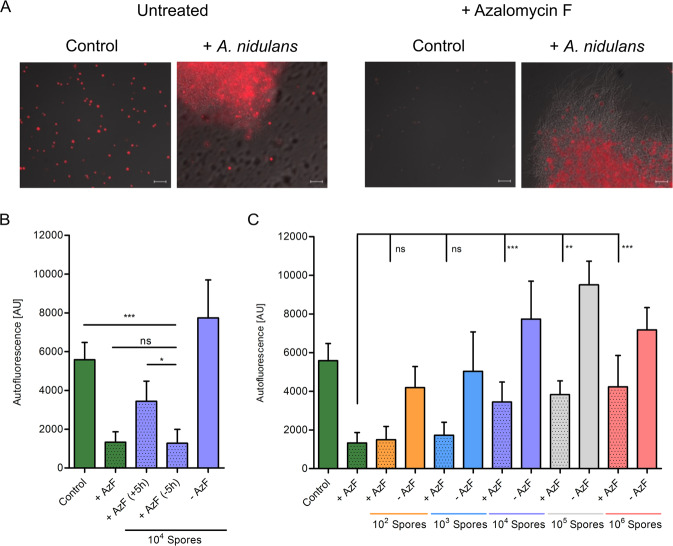
We observed C. reinhardtii cells that still appeared green and alive in a tripartite interaction with A. nidulans and S. iranensis (Supplementary Fig. 8). These data imply that presence of A. nidulans reduces the algicidal activity of S. iranensis. We therefore analyzed whether A. nidulans protects C. reinhardtii against purified azalomycin F, since the fungus proved to be more resistant to azalomycin F than the alga (Supplementary Fig. 9). For this purpose, we co-cultivated A. nidulans with C. reinhardtii and allowed the alga to swim into the mycelia before azalomycin F was added to the co-culture. Viability of C. reinhardtii was measured by chlorophyll autofluorescence (Fig. 3a). As shown in Fig. 3b, C. reinhardtii cells that were allowed to migrate into fungal mycelia were less affected by azalomycin F compared with C. reinhardtii cells that were not given the time to do so. Increasing initial fungal spore inocula were applied prior to addition of C. reinhardtii, azalomycin F treatment and autofluorescence measurement (Fig. 3c). A higher number of spores should lead to a higher fungal biomass and increased fungal biomass correlated with better protection, as demonstrated by titrating out the effect of azalomycin F in the presence of 104 spores and above (Fig. 3c). In order to estimate the specificity of this protection, we tested further fungal species. Supplementary Fig. 10 shows that S. cerevisiae attracts and protects C. reinhardtii from azalomycin F as well. A tight association, as seen with A. nidulans, was not observed (Supplementary Fig. 11). Sordaria macrospora, an important fungus to study sexual development [35], however, failed to protect C. reinhardtii, although the fungus attracted the alga and formed associations comparable to A. nidulans (Supplementary Figs. 10, 11). Our data thus suggest that the protection is associated with self-resistance, since only fungi that are resistant against azalomycin F (Supplementary Fig. 9) protect C. reinhardtii.
Fig. 3. Co-cultivation of C. reinhardtii and A. nidulans protects the alga from azalomycin F.
a Microscopy pictures of C. reinhardtii co-cultivated with or without A. nidulans and treated with or without azalomycin F. Size bars: 50 µm. b Autofluorescence of C. reinhardtii treated with azalomycin F (crosshatched bars) in monoculture (Control) and co-culture with A. nidulans. Loss of red autofluorescence indicates cell death. Scale bar 50 µm. b, c Autofluorescence of C. reinhardtii. b C. reinhardtii was either immediately treated with azalomycin F (−5 h) or allowed to associate with A. nidulans 5 h prior to azalomycin F addition (+5 h). c Correlation of protection of C. reinhardtii with an increased number of spores inoculated into the medium 18 h prior to addition of C. reinhardtii. Azalomycin F was added 5 h after coincubation. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns not significant; data obtained from at least three biological replicates, error bars represent SDs.
Protection of C. reinhardtii by A. nidulans against azalomycin F can be neutralized by fungal polar lipids
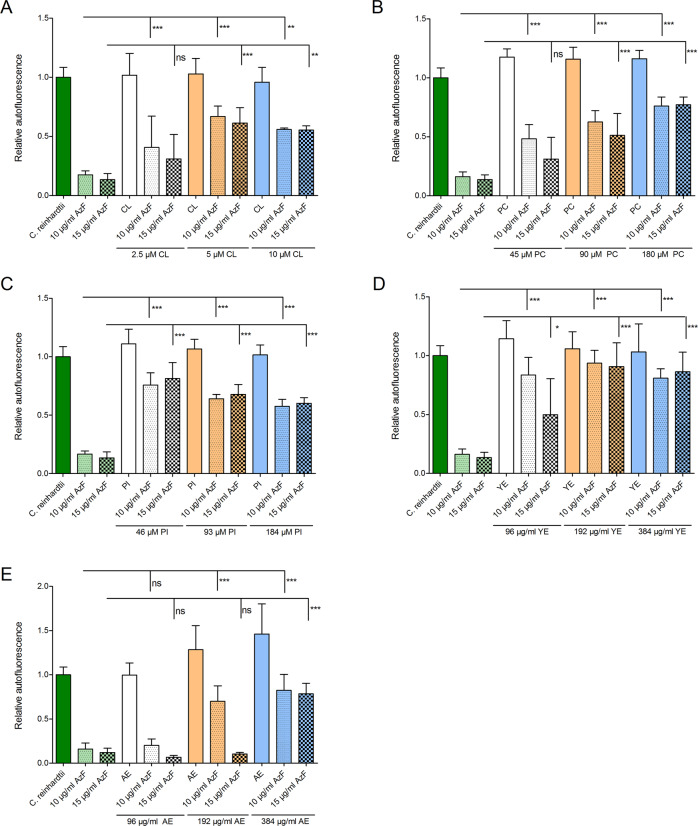
To elucidate the mechanisms underlying the protection of C. reinhardtii, against azalomycin F by A. nidulans, we sought to identify the target of azalomycin F. Recently, the binding of azalomycin F5a to membrane components of methicillin-resistant Staphylococcus aureus was proposed as a determinant of its antibacterial activity [26, 36]. Consequently, we reasoned that the addition of lipids to the culture medium could neutralize the algicidal activity of azalomycin F. As shown in Fig. 4a–d, cardiolipin (CL), 16:0-i15:0 phosphatidylcholin (PC), phosphatidylinositol (PI), and a commercially available extract of polar lipids of S. cerevisiae (YE) all significantly lowered the detrimental effect of azalomycin F on C. reinhardtii. Since 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG) and the sphingolipid d18:1/18:0 C18 glucosyl(β)ceramide (GC) did not affect the activity of azalomycin F (Supplementary Fig. 12), these findings indicate that there is binding specificity toward distinct polar lipids.
Fig. 4. Influence of lipids on the algicidal activity of azalomycin F measured by C. reinhardtii autofluorescence.
Algal cells were treated with 0, 10, and 15 µg/mL azalomycin F and various concentrations of polar lipids. a CL, cardiolipin; b PC, phosphatidylcholin; c PI, phosphatidylinositol; d YE, extract of polar lipids of S. cerevisiae; e AE, extract of polar lipids of A. nidulans. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns not significant; data were obtained from at least three biological replicates, error bars represent SDs.
Based on these findings, we hypothesized that a polar lipid extract of A. nidulans should also neutralize the effect of azalomycin F on C. reinhardtii. Indeed, addition of A. nidulans polar lipids to the culture of C. reinhardtii prevented the algae from decolorization in a dose-dependent manner (Fig. 4e). Addition of 384 µg/mL of the polar lipid extract of A. nidulans (AE) to C. reinhardtii treated with azalomycin F restored the autofluorescence to the level of untreated controls. Thus, binding of azalomycin F to membrane polar lipids is required for its antibacterial activity [26, 36] as well as for its antifungal [37] and algicidal activity, as shown here.
Aspergillus nidulans attracts and fosters the growth of Chlamydomonas reinhardtii
We were able to show that an algal–fungal association protects C. reinhardtii from azalomycin F. Figure 5a shows such an association of A. nidulans and C. reinhardtii. To address the question whether this assembly is actively or passively established, a chemotaxis assay was performed. Figure 5b shows that significantly (P ≤ 0.01) more C. reinhardtii cells swam into a glass capillary loaded with supernatant of an A. nidulans culture than into a capillary filled with fresh medium (Fig. 5b). PBS served as a negative control showing unspecific swimming without an obvious chemotactic cue. Also supernatants of the other fungi tested, i.e., S. cerevisiae and S. macrospora, attracted the C. reinhardtii cells, indicating that C. reinhardtii actively swims toward the fungi.
Fig. 5. Association of A. nidulans with C. reinhardtii.
a Microscopy picture showing C. reinhardtii cells (green) in mycelium of A. nidulans. Scale bar: 50 µm. b Numbers of C. reinhardtii cells found in capillaries filled with culture supernatant of A. nidulans, medium or PBS. ** represents P ≤ 0.01, ***P ≤ 0.001, calculated from at least three biological replicates; error bars indicate SDs. c Optical density of C. reinhardtii cells reached after co-cultivation with A. nidulans in co-cultivation chambers, separated by a PVDF membrane pore size 0.1 µm. d Dry masses of A. nidulans grown in mono- and co-culture.
In order to measure any effects on algal or fungal growth, co-cultivation chambers [25] were applied to separate the organisms by a PVDF membrane with a pore size of 0.1 µm. When co-cultured with A. nidulans, C. reinhardtii exhibited a more than two-fold increased growth rate (µmax = 2.5) and higher overall cell density compared with the monoculture (Fig. 5c). In contrast, A. nidulans produced a lower biomass in co-culture, compared with the monoculture at the end of the experiment (Fig. 5d). In the light of our data showing protection of the alga by fungal polar lipids, it was conceivable that more lipids were formed as an adaptive response and at the same time, energy would be redirected to lipid metabolism of the fungus explaining lower growth of A. nidulans in co-culture with C. reinhardtii. Therefore, we measured the expression of polar lipid biosynthesis genes of A. nidulans in presence of C. reinhardtii by qRT-PCR. As shown in Supplementary Fig. 13, overexpression of polar lipid biosynthesis genes was not observed in presence of C. reinhardtii excluding both possibilities. Taken together, our findings suggest a chemotactically driven attraction of the alga to the fungus, which ultimately provides benefit to the alga by protecting against toxins.
Discussion
Although algal–fungal co-cultures have been successfully used for biotechnological processes [38], as well as establishing algal–fungal [39] and tripartite algal–fungal–bacterial interaction systems [40], the responses of microbial consortia to harmful invaders remained largely understudied. An important step toward the goal of understanding important ecological interactions and their evolutionary selection is the study of increasingly complex biosystems in the laboratory. Among the simplest naturally occurring examples of such interactions are lichens that represent an early evolutionary emergence of organismal co-existence [3, 41]. Lichens typically consist of a photobiont, such as a green alga or a cyanobacterium and a fungal partner, the mycobiont [42]. Recent studies also reported the association of bacterial consortia with lichens [9, 10]. How such algal–fungal associations may cooperate to facilitate responses to, for instance, invasion by toxin-producing microbes is largely unknown. Here, we discovered a lichen-like association of a fungus and a green alga that helps to protect the alga against toxic bacterial compounds (Fig. 6).
Fig. 6. Graphical model of the tripartite interaction of S. iranensis, A. nidulans, and C. reinhardtii.
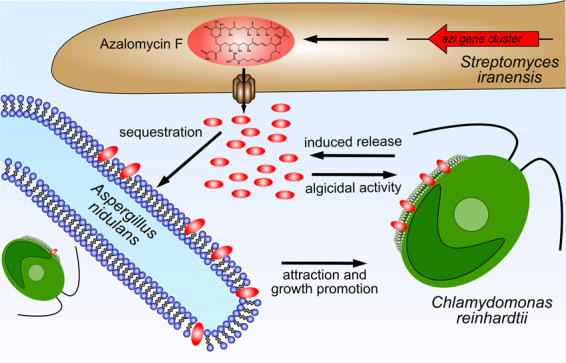
S. iranensis produces algicidal azalomycin F and releases it in presence of C. reinhardtii. As a counter measure, C. reinhardtii is attracted by A. nidulans and takes shelter in its mycelium. A. nidulans provides a high surface to azalomycin F and sequesters the compound. This way less azalomycin F is accessible for C. reinhardtii and the alga is less affected by the compound.
Bacterial azalomycin F exhibits anti-Gram-positive, antifungal, and algicidal activity
We have demonstrated that S. iranensis kills the green alga C. reinhardtii when both organisms were grown in co-culture. The algicidal compound was identified as the polyketide azalomycin F that also has antifungal and antibacterial activity against Gram positives [29, 31]. The reported algicidal activity found here points to a potential role of toxins in shaping algal–fungal associations.
For only a few natural products their ecological meaning has been elucidated. For example, terpenoids serve various functions ranging from chemical communication to defense against predators in plants [43]. The anaerobic bacterium Clostridium puniceum uses the polyphenolic metabolite clostrubin to be able to grow in an oxygen-containing environment and to defend its habitat against competitors [44]. Bacterial cyclic lipopeptides are known to play many biological roles, from promoting motility in the case of surfactin [45], to immobilize algal cells for predation [46]. A recent example by us showed that S. rapamycinicus induced the production of the novel compound fumigermin in A. fumigatus. The compound inhibits the germination of the inducing S. rapamycinicus [47]. Here, we identified an ecological function of a tripartite interaction between an alga and a fungus that protects the alga against a toxic bacterial natural product.
Streptomycetes are known to synthesize algicidal compounds such as NIG355, a nigericin derivative that inhibits the growth of a dinoflagellate [48] and the harmful algal bloom-forming haptophyte Phaeocystis globosa [49]. The observed light-dependency of the algicidal activity of azalomycin F remains obscure. The toxicity is independent of photosynthesis as photosynthesis-defective mutants of C. reinhardtii were effectively killed by azalomycin F. In addition, the significance of this observation for an ecological setting is unclear, as C. reinhardtii will almost always be illuminated to some extent in its natural habitat. It should be noted, however, that C. reinhardtii has at least 18 photoreceptors that absorb light in the UV as well as blue and red visible spectrum [50], and the biophysical properties and biological functions of only a few of these are known. It is conceivable that by disturbing the plasma membrane azalomycin F affects the positioning or presence of photoreceptors with a potentially lethal outcome (Fig. 6). In addition, we tested mutants that are deficient in assembly of the photosynthetic complex [33, 34]. Although the mutants still produce chlorophyll they are deficient in photosynthesis. It is thus conceivable that the presence of chlorophyll is already sufficient to render the cells susceptible to azalomycin F in a light-dependent manner.
Azalomycin F is specifically released in the presence of C. reinhardtii
Azalomycin F is produced by S. iranensis in monoculture but the majority of the compound is associated with the bacterial cells and not in the supernatant. Only co-cultivation of S. iranensis with C. reinhardtii triggered the release of azalomycin F, which, in turn, led to the killing of the alga. It is yet unclear whether azalomycin F resides inside the cell and is exported, or, whether it is attached to the bacterial membrane and simply detached from it upon a certain signal.
Typically, biosynthesis of an antibiotic is directly coupled to its export to avoid self-intoxication. For example, the exporters of the gyrase inhibitors simocyclinone [51] or of actinorhodin [52] are co-expressed with the corresponding biosynthesis genes. The export machinery of azalomycin F is as yet unknown as no putative exporter is encoded in the biosynthetic gene cluster [21]. That a mechanism of detection of the alga and subsequent active release, secretion or detachement of azalomycin F must be at work derives from our observation that C. reinhardtii is killed in conditions that prevent cellular contact.
Azalomycin F activity is neutralized by the presence of fungal membranes
We observed that C. reinhardtii is protected against bacterial azalomycin F by taking shelter in the mycelium of A. nidulans. Furthermore, we provide evidence that lipid binding is a means of neutralizing azalomycin F algicidal activity [26, 36]. We showed that azalomycin F has specificity for some polar lipids and that the polar membrane lipid phosphatidylinositol, which is important for signaling in eukaryotic cells [53–55], is also bound by azalomycin F. Ergosterol, an important cell membrane component of both A. nidulans and C. reinhardtii [56, 57], is not bound by azalomycin F, which further underlines its specificity to polar lipids (Supplementary Fig. 12). This differs markedly from other antifungal macrolides such as amphotericin B, which is considered to primarily bind ergosterol [58]. We thus propose that polar membrane lipids are the target of azalomycin F as well as the key to the protective mechanism by competitive binding (Fig. 6). A similar strategy has been applied to neutralize bacterial toxins as an antibiotic adjunctive therapy in severe pneumococcal pneumonia. Liposomal nanoparticles acted as traps for a broad panel of bacterial toxins that are known to be inserted in cellular membranes [59, 60]. Thus, this medical concept, which is promising to treat infectious diseases, appears to have existed in nature for a long time.
It is interesting to note that S. macrospora could not protect C. reinhardtii from azalomycin F despite attracting the alga. This coincides with susceptibility of S. macrospora against azalomycin F that is not observed for S. cerevisiae and A. nidulans. It is tempting to speculate that only fungi resistant against azalomycin F have the capability to protect C. reinhardtii. The reason for the susceptibility of S. macrospora against azalomycin F awaits further clarification.
Fungal hyphae can provide various benefits to partner organisms, as exemplified by the symbiosis of fungi and terrestrial plants in the Mycorrhiza. Here, the fungus gains fixed carbon products from the phytobiont, in exchange provides nitrogen, phosphate, and increased water supply to the land plant [61]. The fungal hyphae can be used as “fungal highways” by predatory bacteria to bridge unfavorable environments and find new, otherwise inaccessible prey [62]. Fungal mycelia have been also suggested to form microhabitats for bacteria. For example, Morchella crassipes fosters growth of associated Pseudomonas putida bacteria that in turn were also used as a carbon source by the fungus [63]. Another example is the bacterial community colonizing the truffle-fruiting body, which produces thiophene volatiles. These compounds might form the characteristic aroma of the truffle and are discussed to attract endozoochoric spore dispersers [64]. Similarly, we found that A. nidulans enhanced the growth of C. reinhardtii.
Algal–fungal association as a novel way of coping with biotic stress
Lutzoni et al. proposed that the common ancestor of the modern Eurotiales, including the genus Aspergillus, once was lichen forming but the recent fungi have lost this ability during the course of evolution [65]. However, our data indicate that there is residual capability of forming symbiosis in the filamentous fungi investigated here because, the motile single-celled green alga C. reinhardtii readily migrated into A. nidulans and S. macrospora hyphal shells in liquid and formed macroscopic structures, in which the algal cells gathered around fungal hyphae. Likewise, it was reported that auxotrophic mutants of A. nidulans unable to assimilate nitrite formed an obligate mutualism with C. reinhardtii. The algal partner was able to provide nitrogen to the fungus [19]. A. nidulans and C. reinhardtii may well have retained the capabilities of their lichen-forming ancestors to form a kind of algal–fungal mutualism. For C. reinhardtii, this is not surprising because species of the genus Chlamydomonas have been previously identified as mycetobiont algae in lichen-like symbiotic associations of wood-decaying fungi [66].
With reference to the co-evolution of fungi, algae, and terrestrial plants, Delaux et al. [67] postulated that plant genes relevant for symbiosis were already present in an algal ancestor. It is thus conceivable that a fungal ancestor of mycorrhiza and an ancient lichen-forming Aspergillus species likewise might have already possessed the genes promoting symbiosis. Indeed, it has been suggested that the fungal spread on land and the radiation of terrestrial plants are linked to their preexisting ability to establish symbioses [68]. A. nidulans and C. reinhardtii may thus have retained some genetic traits allowing for algal–fungal mutualism that provides a selective advantage over noncooperators, although both microorganisms no longer contribute to the formation of stable lichens and live solitarily.
Protection against environmental toxins may contribute to the evolution of lichen-like associations
C. reinhardtii cells located within the A. nidulans mycelium exhibited increased survival rates when treated with azalomycin F compared with C. reinhardtii that was prevented from entering the fungal mycelium. Presently, abiotic factors, such as water availability and radiation, are primarily considered as triggers for tight interactions [69, 70]. It stands to reason that at some point during algal–fungal co-evolution, attacks by invasive and harmful microorganisms will have occurred. Indeed, metagenomic data have indicated the presence of virulence-associated functions within the lichen microbial consortia, such as antibiotic biosynthesis genes and secretion systems. Furthermore, streptomycetes that produce antibiotic [11, 13, 14] and antifungal metabolites [12] were successfully isolated from lichens. Our data suggest that the ability to form algal–fungal associations is a viable strategy for coping with environmental toxins and biotic challenges by providing a selection advantage based on establishing symbiosis.
Supplementary information
Acknowledgements
We thank Christina Täumer, Karin Burmeister, and Karin Martin for valuable technical assistance and bacterial strain maintenance. Dr. Hendrik Huthoff is acknowledged for critical reading of the paper. We thank Prof. Stefanie Pöggeler for providing Sordaria macrospora and sharing valuable expertise. Financial support by the Deutsche Forschungsgemeinschaft (DFG)-funded excellence graduate school Jena School for Microbial Communication (JSMC) and the DFG-funded Collaborative Research Center 1127 ChemBioSys (projects A02, B01, and B02) is gratefully acknowledged. MGA is grateful for financial support from the European Research Council for a Marie Skłodowska-Curie Individual Fellowship (IF-EF) Project reference 700036. Open access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0731-2) contains supplementary material, which is available to authorized users.
References
- 1.Taylor TN, Remy W, Hass H. Parasitism in a 400-million-year-old green alga. Nature. 1992;357:493–4. [Google Scholar]
- 2.Taylor TN, Hass H, Remy W, Kerp H. The oldest fossil lichen. Nature. 1995;378:244. [Google Scholar]
- 3.Honegger R, Edwards D, Axe L. The earliest records of internally stratified cyanobacterial and algal lichens from the lower devonian of the welsh borderland. N Phytol. 2013;197:264–75. doi: 10.1111/nph.12009. [DOI] [PubMed] [Google Scholar]
- 4.Selosse MA, Le Tacon F. The land flora: a phototroph-fungus partnership? Trends Ecol Evol. 1998;13:15–20. doi: 10.1016/s0169-5347(97)01230-5. [DOI] [PubMed] [Google Scholar]
- 5.Schwendener S. Die Algentypen der Flechtengonidien. Universitätsbuchdruckerei von C Schultze, Basel. 1869.
- 6.Ahmadjian V, Jacobs JB. Relationship between fungus and alga in the lichen Cladonia cristatella Tuck. Nature. 1981;289:169–72. [Google Scholar]
- 7.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 8.Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, et al. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol. 2015;6:299. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grube M, Cernava T, Soh J, Fuchs S, Aschenbrenner I, Lassek C, et al. Exploring functional contexts of symbiotic sustain within lichen-associated bacteria by comparative omics. ISME J. 2015;9:412–24. doi: 10.1038/ismej.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grube M, Cardinale M, de Castro JV, Jr, Müller H, Berg G. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. ISME J. 2009;3:1105. doi: 10.1038/ismej.2009.63. [DOI] [PubMed] [Google Scholar]
- 11.Schneider O, Simic N, Aachmann FL, Rückert C, Kristiansen KA, Kalinowski J, et al. Genome mining of Streptomyces sp. YIM 130001 isolated from lichen affords new thiopeptide antibiotic. Front Microbiol. 2018;9:3139. doi: 10.3389/fmicb.2018.03139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Jiang Y, Lei H, Chen X, Ma Q, Han L, et al. Four new nanaomycins produced by Streptomyces hebeiensis derived from lichen. Chem Biodivers. 2017;14:e1700057. doi: 10.1002/cbdv.201700057. [DOI] [PubMed] [Google Scholar]
- 13.Parrot D, Antony-Babu S, Intertaglia L, Grube M, Tomasi S, Suzuki MT. Littoral lichens as a novel source of potentially bioactive Actinobacteria. Sci Rep. 2015;5:15839. doi: 10.1038/srep15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrot D, Legrave N, Delmail D, Grube M, Suzuki M, Tomasi S. Review—Lichen-associated bacteria as a hot spot of chemodiversity: Focus on uncialamycin, a promising compound for future medicinal applications. Planta Med. 2016;82:1143–52. doi: 10.1055/s-0042-105571. [DOI] [PubMed] [Google Scholar]
- 15.Netzker T, Flak M, Krespach MKC, Stroe MC, Weber J, Schroeckh V, et al. Microbial interactions trigger the production of antibiotics. Curr Opin Microbiol. 2018;45:117–23. doi: 10.1016/j.mib.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Fischer J, Müller SY, Netzker T, Jäger N, Gacek-Matthews A, Scherlach K, et al. Chromatin mapping identifies BasR, a key regulator of bacteria-triggered production of fungal secondary metabolites. eLife. 2018;7:e40969. doi: 10.7554/eLife.40969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–63. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stöcker-Worgötter E. Metabolic diversity of lichen-forming ascomycetous fungi: culturing, polyketide and shikimate metabolite production, and PKS genes. Nat Prod Rep. 2008;25:188–200. doi: 10.1039/b606983p. [DOI] [PubMed] [Google Scholar]
- 19.Hom EFY, Murray AW. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345:94–8. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Netzker T, Schroeckh V, Gregory MA, Flak M, Krespach MKC, Leadlay PF, et al. An efficient method to generate gene deletion mutants of the rapamycin-producing bacterium Streptomyces iranensis HM 35. Appl Environ Microbiol. 2016;82:3481–92. doi: 10.1128/AEM.00371-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Zhai G, Liu Y, Li Y, Shi Y, Hong K, et al. An iterative module in the azalomycin F polyketide synthase contains a switchable enoylreductase domain. Angew Chem Int Ed. 2017;56:5503–6. doi: 10.1002/anie.201701220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman D, Levine R. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–9. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sjoblad RD, Frederikse PH. Chemotactic responses of Chlamydomonas reinhardtii. Mol Cell Biol. 1981;1:1057–60. doi: 10.1128/mcb.1.12.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler RW, Weiss A, Kuegler S, Hermes C, Wichard T. Macroalgal-bacterial interactions: Role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta) Mol Ecol. 2018;27:1808–19. doi: 10.1111/mec.14472. [DOI] [PubMed] [Google Scholar]
- 25.Paul C, Mausz MA, Pohnert G. A co-culturing/metabolomics approach to investigate chemically mediated interactions of planktonic organisms reveals influence of bacteria on diatom metabolism. Metabolomics. 2013;9:349–59. [Google Scholar]
- 26.Xu L, Xu X, Yuan G, Wang Y, Qu Y, Liu E. Mechanism of azalomycin F5a against methicillin-resistant Staphylococcus aureus. BioMed Res Int. 2018;2018:6942452. doi: 10.1155/2018/6942452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pouneva I. Evaluation of algal viability and physiology state by fluorescent microscopic methods. Bulgarian J Plant Physiol. 1997;23:67–76. [Google Scholar]
- 28.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, et al. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45:W36–41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arai M. Azalomycin F, an antibiotic against fungi and Trichomonas. Arzneimittelforschung. 1968;18:1396–9. [PubMed] [Google Scholar]
- 30.Hong H, Sun Y, Zhou Y, Stephens E, Samborskyy M, Leadlay PF. Evidence for an iterative module in chain elongation on the azalomycin polyketide synthase. Beilstein J Org Chem. 2016;12:2164–72. doi: 10.3762/bjoc.12.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan GJ, Li PB, Yang J, Pang HZ, Pei Y. Anti-methicillin-resistant Staphylococcus aureus assay of azalomycin F5a and its derivatives. Chin J Nat Med. 2014;12:309–13. doi: 10.1016/S1875-5364(14)60061-3. [DOI] [PubMed] [Google Scholar]
- 32.Hong H, Fill T, Leadlay PF. A common origin for guanidinobutanoate starter units in antifungal natural products. Angew Chem Int Ed. 2013;52:13096–9. doi: 10.1002/anie.201308136. [DOI] [PubMed] [Google Scholar]
- 33.Bennoun P, Spierer-Herz M, Erickson J, Girard-Bascou J, Pierre Y, Delosme M, et al. Characterization of photosystem II mutants of Chlamydomonas reinhardii lacking the psbA gene. Plant Mol Biol. 1986;6:151–60. doi: 10.1007/BF00021484. [DOI] [PubMed] [Google Scholar]
- 34.Erickson JM, Rahire M, Malnoë P, Girard-Bascou J, Pierre Y, Bennoun P, et al. Lack of the D2 protein in a Chlamydomonas reinhardtiipsbD mutant affects photosystem II stability and D1 expression. EMBO J. 1986;5:1745–54. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masloff S, Pöggeler S, Kück U. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics. 1999;152:191–9. doi: 10.1093/genetics/152.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan G, Xu L, Xu X, Li P, Zhong Q, Xia H, et al. Azalomycin F5a, a polyhydroxy macrolide binding to the polar head of phospholipid and targeting to lipoteichoic acid to kill methicillin-resistant Staphylococcus aureus. Biomed Pharmacother. 2019;109:1940–50. doi: 10.1016/j.biopha.2018.11.067. [DOI] [PubMed] [Google Scholar]
- 37.Cheng J, Yang SH, Palaniyandi SA, Han JS, Yoon T-M, Kim T-J, et al. Azalomycin F complex is an antifungal substance produced by Streptomyces malaysiensis MJM1968 isolated from agricultural soil. J Korean Soc Appl Biol Chem. 2010;53:545–52. [Google Scholar]
- 38.Du ZY, Alvaro J, Hyden B, Zienkiewicz K, Benning N, Zienkiewicz A, et al. Enhancing oil production and harvest by combining the marine alga Nannochloropsis oceanica and the oleaginous fungus Mortierella elongata. Biotechnol Biofuels. 2018;11:174. doi: 10.1186/s13068-018-1172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du ZY, Zienkiewicz K, Vande Pol N, Ostrom NE, Benning C, Bonito GM. Algal-fungal symbiosis leads to photosynthetic mycelium. eLife. 2019;8:e47815. doi: 10.7554/eLife.47815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muggia L, Fernández-Brime S, Grube M, Wedin M. Schizoxylon as an experimental model for studying interkingdom symbiosis. FEMS Microbiol Ecol. 2016;92:fiw165. doi: 10.1093/femsec/fiw165. [DOI] [PubMed] [Google Scholar]
- 41.Grube M, Wedin M. Lichenized fungi and the evolution of symbiotic organization. Microbiol Spectr. 2016;4. [DOI] [PubMed]
- 42.Aschenbrenner IA, Cernava T, Berg G, Grube M. Understanding microbial multi-species symbioses. Front Microbiol. 2016;7:180. doi: 10.3389/fmicb.2016.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3:408–14. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 44.Shabuer G, Ishida K, Pidot SJ, Roth M, Dahse H-M, Hertweck C. Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory. Science. 2015;350:670–4. doi: 10.1126/science.aac9990. [DOI] [PubMed] [Google Scholar]
- 45.Kinsinger RF, Shirk MC, Fall R. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol. 2003;185:5627–31. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aiyar P, Schaeme D, García-Altares M, Carrasco Flores D, Dathe H, Hertweck C, et al. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat Commun. 2017;8:1756. doi: 10.1038/s41467-017-01547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stroe MC, Netzker T, Scherlach K, Krüger T, Hertweck C, Valiante V, et al. Targeted induction of a silent fungal gene cluster encoding the bacteria-specific germination inhibitor fumigermin. eLife. 2020;9:e52541. doi: 10.7554/eLife.52541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvey BM, Mironenko T, Sun Y, Hong H, Deng Z, Leadlay PF, et al. Insights into polyether biosynthesis from analysis of the nigericin biosynthetic gene cluster in Streptomyces sp. DSM4137. Cell Chem Biol. 2007;14:703–14. doi: 10.1016/j.chembiol.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 49.Zheng X, Zhang B, Zhang J, Huang L, Lin J, Li X, et al. A marine algicidal actinomycete and its active substance against the harmful algal bloom species Phaeocystis globosa. Appl Microbiol Biotechnol. 2013;97:9207–15. doi: 10.1007/s00253-012-4617-8. [DOI] [PubMed] [Google Scholar]
- 50.Greiner A, Kelterborn S, Evers H, Kreimer G, Sizova I, Hegemann P. Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell. 2017;29:2498–518. doi: 10.1105/tpc.17.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le TB, Fiedler HP, den Hengst CD, Ahn SK, Maxwell A, Buttner MJ. Coupling of the biosynthesis and export of the DNA gyrase inhibitor simocyclinone in Streptomyces antibioticus. Mol Microbiol. 2009;72:1462–74. doi: 10.1111/j.1365-2958.2009.06735.x. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Willems A, Au-Yeung C, Tahlan K, Nodwell JR. A two-step mechanism for the activation of actinorhodin export and resistance in Streptomyces coelicolor. MBio. 2012;3:e00191–12. doi: 10.1128/mBio.00191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wymann MP, Pirola L. Structure and function of phosphoinositide 3-kinases. Biochim Biophys Acta. 1998;1436:127–50. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- 54.Vanzela AP, Said S, Prade RA. Phosphatidylinositol phospholipase C mediates carbon sensing and vegetative nuclear duplication rates in Aspergillus nidulans. Can J Microbiol. 2011;57:611–6. doi: 10.1139/w11-034. [DOI] [PubMed] [Google Scholar]
- 55.Schink KO, Tan KW, Stenmark H. Phosphoinositides in control of membrane dynamics. Annu Rev Cell Dev Biol. 2016;32:143–71. doi: 10.1146/annurev-cellbio-111315-125349. [DOI] [PubMed] [Google Scholar]
- 56.Miller MB, Haubrich BA, Wang Q, Snell WJ, Nes WD. Evolutionarily conserved Δ25(27)-olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii. J Lipid Res. 2012;53:1636–45. doi: 10.1194/jlr.M027482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro BE, Gealt MA. Ergosterol and lanosterol from Aspergillus nidulans. Microbiology. 1982;128:1053–6. doi: 10.1099/00221287-128-5-1053. [DOI] [PubMed] [Google Scholar]
- 58.Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol. 2014;10:400–6. doi: 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laterre P-F, Colin G, Dequin P-F, Dugernier T, Boulain T, Azeredo da Silveira S, et al. CAL02, a novel antitoxin liposomal agent, in severe pneumococcal pneumonia: a first-in-human, double-blind, placebo-controlled, randomised trial. Lancet Infect Dis. 2019;19:620–30. doi: 10.1016/S1473-3099(18)30805-3. [DOI] [PubMed] [Google Scholar]
- 60.Pletz MW, Bauer M, Brakhage AA. One step closer to precision medicine for infectious diseases. Lancet Infect Dis. 2019;19:564–5. doi: 10.1016/S1473-3099(19)30070-2. [DOI] [PubMed] [Google Scholar]
- 61.Miransari M. Arbuscular mycorrhizal fungi and nitrogen uptake. Arch Microbiol. 2011;193:77–81. doi: 10.1007/s00203-010-0657-6. [DOI] [PubMed] [Google Scholar]
- 62.Otto S, Bruni EP, Harms H, Wick LY. Catch me if you can: dispersal and foraging of Bdellovibrio bacteriovorus 109J along mycelia. ISME J. 2017;11:386–93. doi: 10.1038/ismej.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pion M, Spangenberg JE, Simon A, Bindschedler S, Flury C, Chatelain A, et al. Bacterial farming by the fungus Morchella crassipes. Proc R Soc B Biol Sci. 2013;280:20132242. doi: 10.1098/rspb.2013.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Splivallo R, Deveau A, Valdez N, Kirchhoff N, Frey-Klett P, Karlovsky P. Bacteria associated with truffle-fruiting bodies contribute to truffle aroma. Environ Microbiol. 2015;17:2647–60. doi: 10.1111/1462-2920.12521. [DOI] [PubMed] [Google Scholar]
- 65.Lutzoni F, Pagel M, Reeb V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature. 2001;411:937–40. doi: 10.1038/35082053. [DOI] [PubMed] [Google Scholar]
- 66.Mukhin VA, Patova EN, Kiseleva IS, Neustroeva NV, Novakovskaya IV. Mycetobiont symbiotic algae of wood-decomposing fungi. Russ J Ecol. 2016;47:133–7. [Google Scholar]
- 67.Delaux P-M, Radhakrishnan GV, Jayaraman D, Cheema J, Malbreil M, Volkening JD, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA. 2015;112:13390–5. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lutzoni F, Nowak MD, Alfaro ME, Reeb V, Miadlikowska J, Krug M, et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun. 2018;9:5451. doi: 10.1038/s41467-018-07849-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kranner I, Cram WJ, Zorn M, Wornik S, Yoshimura I, Stabentheiner E, et al. Antioxidants and photoprotection in a lichen as compared with its isolated symbiotic partners. Proc Natl Acad Sci USA. 2005;102:3141–6. doi: 10.1073/pnas.0407716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larson DW. Lichen water relations under drying conditions. N Phytol. 1979;82:713–31. doi: 10.1111/j.1469-8137.1979.tb01666.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.