Abstract
Deep-sea hydrothermal plumes are considered natural laboratories for understanding ecological and biogeochemical interactions. Previous studies focused on interactions between microorganisms and inorganic, reduced hydrothermal inputs including sulfur, hydrogen, iron, and manganese. However, little is known about transformations of organic compounds, especially methylated, sulfur-containing compounds, and petroleum hydrocarbons. Here, we reconstructed nine gammaproteobacterial metagenome-assembled genomes, affiliated with Methylococcales, Methylophaga, and Cycloclasticus, from three hydrothermal ecosystems. We present evidence that these three groups have high transcriptional activities of genes encoding cycling of C1-compounds, petroleum hydrocarbons, and organic sulfur in hydrothermal plumes. This includes oxidation of methanethiol, the simplest thermochemically-derived organic sulfur, for energy metabolism in Methylococcales and Cycloclasticus. Together with active transcription of genes for thiosulfate and methane oxidation in Methylococcales, these results suggest an adaptive strategy of versatile and simultaneous use of multiple available electron donors. Meanwhile, the first near-complete MAG of hydrothermal Methylophaga aminisulfidivorans and its transcriptional profile point to active chemotaxis targeting small organic compounds. Petroleum hydrocarbon-degrading Cycloclasticus are abundant and active in plumes of oil spills as well as deep-sea vents, suggesting that they are indigenous and effectively respond to stimulus of hydrocarbons in the deep sea. These findings suggest that these three groups of Gammaproteobacteria transform organic carbon and sulfur compounds via versatile and opportunistic metabolism and modulate biogeochemistry in plumes of hydrothermal systems as well as oil spills, thus contributing broad ecological impact to the deep ocean globally.
Subject terms: Biogeochemistry, Water microbiology
Introduction
Deep-sea hydrothermal vents are unique ecosystems, widely distributed at mid-ocean ridges and other types of spreading centers throughout the global oceans [1]. Electron donors enriched in deep-sea hydrothermal plumes include some organic compounds, such as methanethiol (MT), methanol, formate, acetate, methylamine, alkanes, and aromatics, and these organic compounds can support the growth of microbes in hydrothermal plumes [2–5]. These organic molecules, particularly MT, may have been abundant on the early Hadean-Archaean Earth, and could have played a role in the emergence of early hyperthermophilic microbial life [2]. Deep-sea hydrothermal plumes have been considered natural laboratories for understanding the ecology, physiology, and function of microbial groups that mediate much of the transfer of elements and energy from the subsurface to deep-sea and from the geosphere to the biosphere [6].
Microorganisms have expansive diversity, metabolic versatility, and functional redundancy in the hydrothermal sediments, and they are responsible for the important organic carbon turnover and nitrogen and sulfur cycling processes [7–10]. Furthermore, metagenomic and metatranscriptomic analyses have provided insights into the roles of dominant microbial populations involving the oxidation of ammonia [11], hydrogen [12, 13], and sulfur [12, 14] in deep-sea hydrothermal vent plumes. Compared to this relatively well documented biotic cycling of inorganic substrates and role of chemoautotrophic microorganism in mediating plume biogeochemistry, the transformation of organic carbon, nitrogen, and sulfur, especially methylated compounds and petroleum hydrocarbons, have received less attention in deep-sea hydrothermal plumes. The corresponding microbial metabolisms have been documented in a limited number of studies, including organic matter utilizing bacteria and archaea [15, 16] and the presence of various hydrocarbon monooxygenase genes in deep-sea hydrocarbon plumes [17].
While largely unknown in the hydrothermal plume setting, microbes that use methyl-, sulfur-, and petroleum organic compounds have recently been identified in other marine environments. Previous researches on Gammaproteobacteria within hydrothermal plumes have unraveled the important roles on hydrogen and sulfur oxidation (SUP05 group) [13] and methane oxidation (Methylothermaceae) [18]. Nevertheless, several other gammaproteobacterial groups with functions beyond these lithotrophic and methanotrophic activities are not as well studied. Members belonging to genus Methylophaga are a unique group of aerobic, halophilic, non-methane-utilizing methylotrophs that have been isolated from various marine environments and brackish waters [19]. They were found to be dominant in waters influenced by the Deepwater Horizon oil spill [20] and identified as a major organism involved in the methanol breakdown process [21]. Other gammaproteobacterial methano-/methylotrophs and petroleum hydrocarbon degraders, such as Methylococcales and Cycloclasticus, have also been found to be prevalent in marine methane/oil seeps [22, 23]. Cycloclasticus has been discovered from sponge symbionts and oil spill plumes as an obligate oil-degrading microbial genus [20, 23], however, its metabolic potential and functional activities remain elusive in hydrothermal environments. We hypothesized these microorganisms can also live and thrive in hydrothermal plume ecosystems, due to the similar hydrocarbon-rich settings in oil spill and hydrothermal plumes [6, 17].
Unlike other well-studied organisms with genomic characterization, such as Alcanivorax [24, 25] on alkane degradation, Marinobacter [26, 27] and Acinetobacter [28, 29] on hydrocarbon degradation, and SAR324 [30] on alkane and sulfur degradation, these three groups of Gammaproteobacteria have been seldom studied before, except for limited reports of hydrocarbon degrading Cycloclasticus genomes [20, 23]. The distribution pattern of these microorganisms in the marine water column and how they are triggered by the presence of organic compounds are also intriguing, of great ecological significance and still remain unknown at the present time.
Here, we reconstructed metagenome-assembled genomes (MAGs) from the hydrothermal plume and background metagenomes of three hydrothermal fields, and mapped corresponding metatranscriptomic reads to enable genome-specific measurements and quantitative analyses of metabolic activities. Based on these results, we evaluated metabolism with regard to transformations of methyl-, sulfur-, and petroleum organic compounds by three groups of Gammaproteobacteria in deep-sea hydrothermal systems, and discussed their potential influence on plume biochemistry.
Materials and methods
Sample information and metagenome/metatranscriptome sequencing
Hydrothermal plume and surrounding background samples were acquired from the corresponding cruises: R/V New Horizon in Guaymas Basin, Gulf of California (Sampling dates: July 2004), R/V Atlantis in Mid-Cayman Rise, Caribbean Sea (Sampling dates: January 2012) and two consecutive cruises of the R/V Thomas G Thompson in Eastern Lau Spreading Center (ELSC), Lau Basin, western Pacific Ocean (Sampling dates: May–July 2009). Guaymas Basin hydrothermal plume and background samples were collected by CTD and filtered shipboard onto 142 mm diameter 0.2 μm woven polyethersulfone (SUPOR®) filters, and then, preserved with RNAlater right after the collection [12]. Cayman hydrothermal plume and surrounding background samples were collected by Suspended Particulate Rosette (SUPR) filtration device mounted to the remotely operated vehicle (ROV) Jason II [31]. The samples were filtered in situ onto 142 mm diameter 0.2 μm woven polyethersulfone (SUPOR®) filters, and then preserved in situ [31]. The ELSC samples were collected by the SUPR filtration device of ROV Jason II and filtered in situ onto 47 mm diameter 0.8 μm polycarbonate filters, and then preserved shipboard with RNAlater (~6 h post filtration) [32, 33]. Details regarding geochemical analysis, and metagenomic/metatranscriptomic sequencing and QC-processing are described in the previous publications for Guaymas Basin samples [13, 34], Mid-Cayman Rise samples [16, 31, 32], and Lau Basin samples [35]. Detailed cruises and sampling information refer to Supplementary Dataset S1.
Assembling and metagenomic binning
QC-processed reads were assembled de novo by MEGAHIT v1.1.2 with settings as “--k-min 45 --k-max 95 --k-step 10” [36]. Hydrothermal plume and background (P&B) metagenomes from the same hydrothermal site were assembled together. The QC-processed reads were re-mapped to assemblies by Bowtie 2 v2.2.8 with default settings [37]. For each hydrothermal site, hydrothermal plume and background reads were mapped to corresponding assemblies separately; bam files of plume and background reads for individual assemblies were used for downstream binning. Separate mapping of hydrothermal plume and background metagenome reads to P&B de novo co-assemblies from each site resulted in two sets of scaffold coverages that were subsequently used in MAG binning. Subsequently, the assemblies were subjected to a MetaBAT v0.32.4 based binning with 12 combinations of parameters (including the parameters of sensitivity mode, proportion of shared membership in bootstrapping, and minimal contig length) [38]. DAS-Tool v1.0 was implemented to screen MetaBAT MAGs resulted from 12 sets of combination runnings (12-batch of resulted MAGs) for the best set of high quality and completeness MAGs [39]. Outlier scaffolds with abnormal coverage, tetranucleotide signals, and GC pattern within potential high contamination MAGs (value checked by CheckM v1.0.7) and erroneous SSU sequences within MAGs were screened out and decontaminated by RefineM v0.0.20 with default settings [40, 41]. After that, further MAG refinement for decontaminating certain MAGs was manually inspected based on VizBin [42]. MAGs are picked using a threshold of <10% contamination and >50% completeness (Given the incompleteness of MAGs, the absence of metabolic pathways within MAGs does not necessarily indicate their absence in the actual genome).
MAGs genomic property and annotation
The genome taxonomy was determined by RefineM v0.0.20 [40] and GTDB database (http://gtdb.ecogenomic.org). The genomic properties, including genome coverage, genome and 16S rRNA taxonomy, tRNAs, genome completeness, and scaffold parameters, were all obtained using the homemade bioinformatic pipeline which was deposited in GitHub (https://github.com/ChaoLab/GenomePropertyParsingPipeline). Relative genome coverage was calculated by normalizing each metagenome to 100M reads. For ORF annotation, GhostKOALA v2.0, KAAS v2.1, and eggNOG-mapper v4.5.1 were applied to thoroughly annotate ORFs to KOs with default settings [43–45]. GhostKOALA and KAAS annotations provide results in KO identifier style. For eggNOG-mapper annotation, we use the following method to translate its annotations to KO identifiers: (a) we use its first KO hit as the final result; (b) if it only has a COG hit, we translate the COG identifier to KO identifier by ‘ko2cog.xl’ provided by KEGG database and take this as the final result. When combining the results of these software, we use the annotation (the KO identifier) from the first software as the final annotation; if there is no annotation from the first software, then we will move to the next software accordingly. Annotation using NCBI nr database (Mar 6, 2017 updated) was done by DIAMOND BLASTP v0.9.28.129 [46] with default settings. Only the top ten hits were retained for each BALSTP run. Within the top ten BLASTP hits of each annotation, we used the first ranked hit as the final annotation. When the first ranked hit is “hypothetical protein”, we will move downward and use the next one, until we find an annotation result which is not “hypothetical protein”. The pipeline for the above annotation modification was deposited in GitHub (https://github.com/ChaoLab/AnnotationModify).
Phylogenetic construction and genome alignment
To define the genome phylogeny, 43 aligned and masked phylogenetically informative marker proteins of MAGs from this study and their closely related reference genomes were retrieved by CheckM. Concatenated markers were applied to construct the phylogenomic tree (genomes that contained <25% informative sites in the concatenated marker alignment were not included in tree construction). The phylogenomic tree was constructed by IQ-TREE v1.6.3 [47] with its self-implemented ModelFinder procedure (best-fit model automatically chosen case by case) and the settings were as follows: “-m MFP -bb 1000 -mset WAG,LG,JTT,Dayhoff -mrate E,I,G,I+G -mfreq FU -wbtl”. To identify synteny genomic regions among genomes, Mauve v2.4.0 was applied to align genomes with progressiveMauve aligning algorithm [48] and 70% cutoff of sequence similarity (nucleotide) within the shared region was used. The gene operons were identified among genomes according to the accurate reports on the reference genomes and their gene arrangement patterns. We built phylogenetic trees for functionally important proteins (e.g., MxaF, XoxF, MtoX, etc.) that had complex phylogenetic subclades. In each phylogenetic tree, proteins that had biochemical activity evidence from references were labeled accordingly. The functions and phylogenetic affiliation information of the rest proteins were deduced from the phylogenetic tree accordingly.
Transcriptomic level analysis and metagenome mapping
The non-rRNA containing metatranscriptomic datasets were used to map the MAG gene sequences respectively using Bowtie 2 v2.2.8 with the “--very-sensitive” option (the same as running with options “-D 20 -R 3 -N 0 -L 20 -i S,1,0.50”) to achieve sensitive and accurate mapping performance. The RPKM (mapped read number per million total reads per kilobase gene length) was calculated by counting both forward and reserve reads (for single-end cDNA library, one end read was used) mapped on each gene sequence. To compare the plume and background gene expression levels, we also normalized the RPKM value by the genome coverage of each genome in the corresponding metagenomes (All the normalized RPKM values were set to 10×, to make the values easy to read and compare. If a given genome was only reconstructed from either plume or background metagenome, we use it as the mapping target for both the plume and background metagenomes) to exclude the influence of genome abundance on the gene transcriptomic level comparison. If it is not additionally stated, nRPKM (normalized RPKM) in the following text stands for genome coverage-normalized RPKM.
A collection of 371 marine water column metagenomes around the world were used as the database to evaluate the relative abundance of MAGs and reference genomes in open oceans around the world (Supplementary Dataset S2). The metagenomic mapping result of each MAG was represented by RPKG (reads per kb per genome equivalent), which serves as a good standard to estimate genome abundance within metagenomes.
Results
Sampling site
Deep-sea hydrothermal plume samples were collected from Guaymas Basin, Mid-Cayman Rise, and the ELSC, respectively. Guaymas Basin in the Gulf of California is located at 2000 m depth and characterized by active seafloor spreading and rapid deposition of organic-rich, diatomaceous sediments. Active hydrothermal venting is predominantly found in the southern portion of the basin where hydrothermal vents, mounds, and chimneys appear on the seafloor [49]. The Mid-Cayman Rise is the Earth’s deepest and slowest spreading mid-ocean ridge. In this study, two hydrothermal vent sites associated with the Mid-Cayman Rise were investigated: Piccard (Cayman Deep), the deepest known hydrothermal vent site, which is basalt-hosted and located at a depth of 4960 m; and Von Damm (Cayman Shallow), an ultramafic-influenced hydrothermal vent adjacent to Piccard (~20 km) at a depth of 2350 m, which is near the summit of an oceanic core complex [16, 50]. The ELSC is located in Lau Basin, a back-arc basin in the Western Pacific Ocean [16]. In this study, six hydrothermal vent sites along the ELSC were investigated. These vents exhibit a range of geochemical properties with vents at the southern end exhibiting higher concentrations of H2S, Fe and Mn, and lower pH [51].
Genome reconstruction and phylogeny
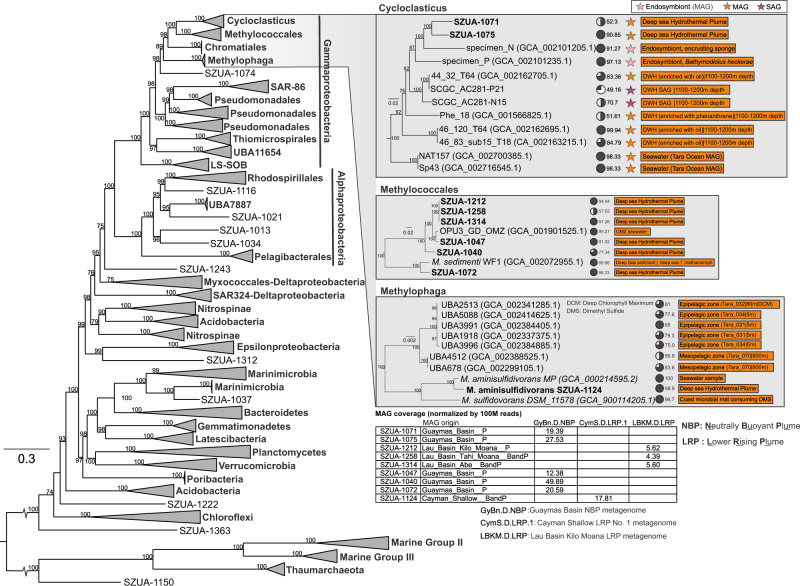
Among metagenomes from these three hydrothermal sites, we reconstructed 206 draft MAGs from the plume and background samples of >50% genome completeness and <10% genome redundancy (Supplementary Dataset S3). This included 22 archaeal MAGs affiliated to Thaumarchaeota and Marine Group II & III, and 184 MAGs affiliated to various bacterial groups, mainly the Alpha-, Gamma-, Delta- and Epsilonproteobacteria (84 MAGs) (Fig. 1). The three major gammaproteobacterial lineages, including Cycloclasticus, Methylophaga, and Methylococcales, were reconstructed with high genome completeness and low genome redundancy (Supplementary Dataset S3). The phylogenomic tree built by concatenated 43-marker protein alignments represents the overall placement of MAGs (Fig. 1). In the Cycloclasticus subtree (Fig. 1 and Supplementary Fig. S1), the closest genomes are from uncultured Deepwater Horizon oil spill (DWH) impacted pelagic samples or mussel/sponge endosymbionts, which are separated from the “isolate” strains (cultured isolates but not unculturable stains or symbiotic strains). In the Methylococcales subtree (Fig. 1 and Supplementary Fig. S2), both the phylogenomic tree and ANI cluster dendrogram support the close genomic relationship between MAGs from this study and deep-sea water column/sediment strains. For Methylophaga (Fig. 1 and Supplementary Fig. S3), both methods show that Methylophaga aminisulfidivorans SZUA-1124 clusters with the seawater isolate from the same species and shares a further relationship with TARA metagenome-binned epi-/mesopelagic MAGs. The normalized MAG coverage indicates these gammaproteobacterial MAGs (Hydro-γ-MAGs hereafter) are present with considerable abundance in hydrothermal plumes in distinct geological settings within three different ocean basins (Fig. 1).
Fig. 1. Phylogenetic tree of the hydrothermal plume and background MAGs based on concatenated marker proteins.
The piecharts in the tree stand for genome completeness. The MAG coverage shown in the table was normalized by per 100 million reads in each corresponding metagenome. MAG origin stands for the metagenomes that were used to assemble and bin the MAGs; ‘P’ stands for only plume metagenome, and ‘BandP’ stands for both plume and background metagenomes together. Only the metagenomes with the largest MAG coverage values were listed in the table within the figure. MAGs from this study are labeled bold, others are from online available assemblies. Abbreviations for metagenomes refer to Supplementary Dataset S1.
Gene operons of methyl-metabolism
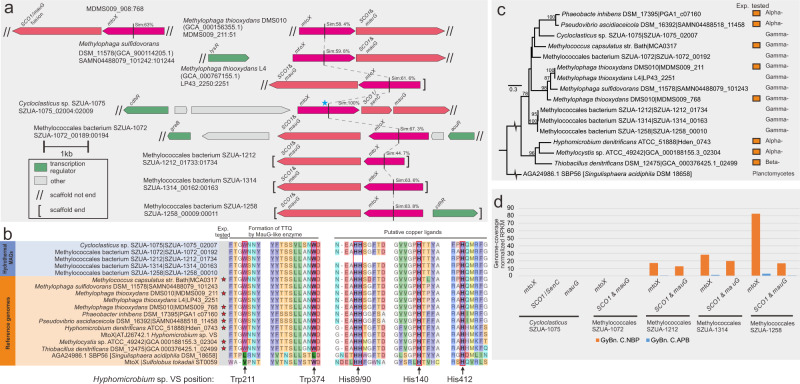
We identified gene operons for methane oxidation, methylamine utilization (details in Supplementary Information), and MT oxidation. Methanethiol (MT) is an intermediate in the breakdown of abundant organosulfur compounds [52] and can itself be degraded by MT oxidase (MTO), encoded in the operon containing mtoX, SCO1/senC (encoding a copper chaperone), and mauG (sometimes it is a SCO1/mauG fusion, encoding a diheme cytochrome c peroxidase for generating MTO co-factor, TTQ) [52, 53]. Cycloclasticus sp. SZUA-1075 and four of the Methylococcales bacteria contain transcriptionally active mto operons with conserved key residues for the formation of TTQ and copper ligands (Fig. 2a, b). The phylogenetic tree places these mtoX genes from 5 Hydro-γ-MAGs to affiliate with other Alpha- and Betaproteobacteria, of which the active functions of MTO have been tested with genomic and enzymatic evidence (Fig. 2c) [52]. Furthermore, Guaymas Basin metatranscriptome results suggest that mto genes from three Methylococcales bacteria (SZUA-1212, 1314, and 1258) have very high transcriptional expression, and their transcription levels are considerably higher in the plume than in the background (Fig. 2d).
Fig. 2. Genomic, phylogenetic, and transcriptomic analyses of mto genes.
a Schematic diagram of aligned mto gene operon of Cycloclasticus sp. and Methylococcales from this study and reference strains. The star labeled mto gene is used as the standard for calculating sequence similarity. b Conserved amino acids for key functions of the MtoX protein. Trp211 and Trp 374 are conserved key residues for the formation of tryptophan tryptophylquinone prosthetic group (TQQ) by MauG-like enzyme, and His89/90, His140 and His412 are conserved key residues of copper ligands. The strains that were experimentally tested with methanethiol oxidizing function (Exp. tested) and successful PCR amplification of mtoX gene are labeled with stars. c Phylogenetic tree of MtoX rooted with a distant SBP56 protein constructed by IQ-TREE. The strains that were experimentally tested (Exp. tested) with methanethiol oxidizing function were labeled with yellow square. d The transcriptional expression of mto genes in the identified genomes. The X-axis stands for the genome-coverage normalized RPKM. Metatranscriptome abbreviations: GyBn Guaymas Basin, C cDNA, NBP neutrally buoyant plume, APB above plume background.
Methyl-compound and C1 metabolism
The Piccard and Von Damm (Cayman Deep/Shallow) metatranscriptome mapping results suggest the high expression of methanol dehydrogenase mxaF gene and Clade 1, 2, and 3 pyrroloquinoline quinone-dependent methanol dehydrogenase xoxF genes of M. aminisulfidivorans SZUA-1124 (Supplementary Fig. S4,a). Interestingly, only the specific Clade 1 xoxF copy (SZUA-1124_00001) has a high expression level, while the other copy shows little transcriptional activity. Meanwhile, the metatranscriptomic data suggest that the xoxF gene had higher abundance of transcripts than mxaF based on the normalized gene expression calculation by genome abundance. M. aminisulfidivorans SZUA-1124 was originally reconstructed from the Cayman Shallow metagenome (Supplementary Dataset S1); the corresponding metatranscriptome mapping results suggest the active expression of genes in methylamine utilization pathways. These results suggest that methylamine utilization occurs mainly in the hydrothermal plume, while the methanol oxidation occurs in both plume and background (Supplementary Fig. S4a). M. aminisulfidivorans SZUA-1124 and three Guaymas Basin Methylococcales bacteria all have high transcriptional activities on formate dehydrogenation and formaldehyde assimilation by the ribulose monophosphate (RuMP) cycle and the Wood–Ljungdahl (WL) pathway (Supplementary Fig. S4b). Nevertheless, for other C1 metabolisms, e.g., formaldehyde dehydrogenation and formamide degradation, no transcripts could be detected, although some Hydro-γ-MAGs possess the encoding genes.
PAH degradation, methane and alkane incorporation
The Cycloclasticus sp. SZUA-1075 MAG (completeness > 90%) has the most complete set of genes for degradation of polycyclic-aromatic hydrocarbons (PAHs), including degradation of various PAHs by dioxygenation and monooxygenation of benzene, toluene and cyclohexane and polycyclic-aromatic ring removal (Supplementary Fig. S5a). Nearly all of them have transcriptional activities in Guaymas Basin plume samples; monooxygenation of benzene to catechol was also active in background seawater (Supplementary Fig. S5a). Other minor dioxygenase encoding genes are also actively expressed by Guaymas Basin Methylococcales bacteria in the plume (Supplementary Fig. S5a).
The phmoA (gene for particulate hydrocarbon monooxygenase subunit A) phylogenetic tree places one identified phmoA gene (SZUA-1075_01584) from Cycloclasticus sp. SZUA-1075 adjacent to those of mussel/sponge endosymbiotic and DWH Cycloclasticus within Group Z, which mediate short-chain alkane oxidation based on genomic and proteomic evidence [23], and the other phmoA gene (SZUA-1075_00092) adjacent to only those of DWH Cycloclasticus but not mussel/sponge endosymbionts within Group X (Supplementary Fig. S6) [17]. The latter gene could be involved in the oxidation of alkanes of various length [23]. The phylogenetic trees of pqq-adh and aor (genes encoding enzymes for alkane oxidation to carboxylic acids) indicate gene copies of SZUA-1075 always form highly supported monophyletic clades with those of mussel/sponge endosymbionts and DWH strains (Supplementary Figs. S7 and S8). These ‘DWH and endosymbiont’ clade gene copies also have higher transcript abundance than those genes from ‘isolate’ clade (Supplementary Fig. S8 and S5b). It is suggested that ‘DWH and endosymbiont’ clade pqq-adh and aor genes are involved with the exact functions of alkane oxidation while the ‘isolate’ clade genes acquire divergent functions [23]. Together with the existence of methylcitrate and beta-oxidation pathways, these results suggest that the alkane oxidation pathway is active in SZUA-1075 [23]. Guaymas Basin metatranscriptome indicates that SZUA-1075 has transcriptional activities on incorporating both short (<C3) and long (>C6) alkanes; however, the former has relatively higher expression (Supplementary Fig. S5b). Meanwhile, the Guaymas Basin metatranscriptome also supports highly expressed Methylococcales pmoA in both plume and background as observed previously [13, 17] (Supplementary Fig. S5b).
Active, concurrent thiosulfate and methane oxidation
Genomic and transcriptomic evidence for thiosulfate oxidation was found in Cycloclasticus and Methylococcales genomes, but not in the M. aminisulfidivorans SZUA-1124 genome (Supplementary Fig. S5c). The concurrent expression of pmoA/phmoA and soxABX within Cycloclasticus and Methylococcales genomes also indicates they could depend on abundant energy sources from surrounding environments. As predicted by Cycloclasticus sp. SZUA-1075 and Methylococcales genomic contents, elemental sulfur could be formed by both flavocytochrome c/sulfide dehydrogenase (fccAB encoded) and sulfide quinone oxidoreductase (sqr encoded), while the metatranscriptome results suggest the former gene has higher transcriptional levels (Supplementary Fig. S5c). Cycloclasticus spp. from this study also contain genomic capacities and transcriptional activities on transforming important organosulfur compounds [54], including MT, dimethyl sulfoxide (DMSO), and methanesulfonate (MSA) (Supplementary Fig. S5c).
Metabolism of Methylophaga aminisulfidivorans SZUA-1124
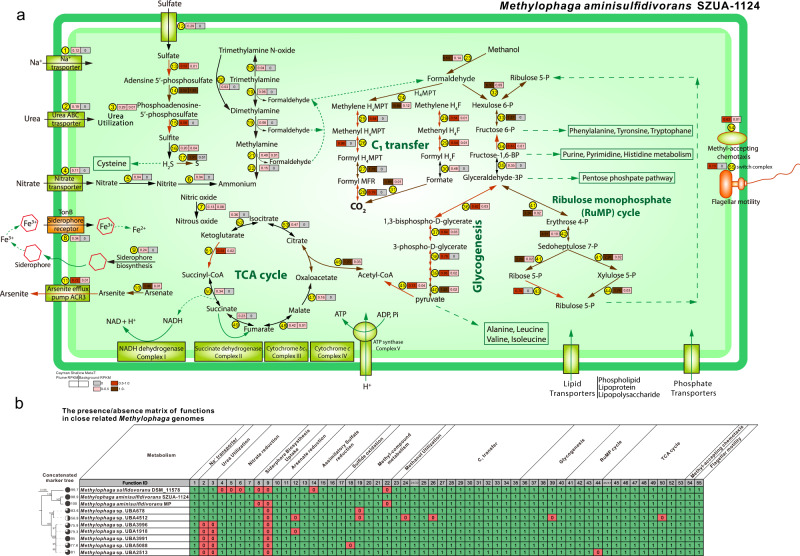
The Methylophaga aminisulfidivorans SZUA-1124 MAG from this study is currently the only reported high-quality (with 98.9% genome completeness, 1.09% genome redundancy, and 30 contigs) Methylophaga genome reconstructed from hydrothermal environments (Fig. 3). Beyond C1 and methyl-compound metabolisms described above, it could actively transport and utilize urea and reduce nitrate (Fig. 3). The assimilatory sulfate reduction and sulfide oxidation are of high activity (Fig. 3), meanwhile, it could actively reduce arsenate and export them out of the cell. It could actively synthesize siderophore to incorporate iron (III) into the cell, which is absent in other closely related Methylophaga genomes (Fig. 3, Dataset S4 and S5) [55].
Fig. 3. The metabolism of Methylophaga aminisulidivorans SZUA-1124.
a The reconstructed metabolic pathway of Methylophaga aminisulidivorans SZUA-1124. The expression levels of individual protein-encoding gene/genes were calculated and labeled accordingly in RPKM. The left square represents the average RPKM value from all Cayman Shallow plume metatranscriptomes, and the right square represents that from all Cayman Shallow background metatranscriptomes. The square and arrow colors correspond to different RPKM value ranges. The protein details of labeled IDs refer to Supplementary Dataset S8. b The presence/absence matrix of functions in closely related Methylophaga genomes. The function IDs are the same as those in Panel (a). The detailed annotations of these close related Methylophaga genomes refer to Supplementary Dataset S8.
No peptide/protein and sugar hydrocarbon transporter encoding genes could be identified, and no complete and effective alkane/PAH-degrading pathways could be reconstructed, suggesting that SZUA-1124 could only depend on small carbons (C1 and methyl-compounds) as energy sources. The active expression of glycogenesis and RuMP cycle genes suggests that SZUA-1124 has a well-established anabolic system for effectively synthesizing sugar hydrocarbons and amino acids for cell life functions (Fig. 3). Similar to that in Oceanospirillales single-cell amplified genome and hydrocarbon plume metagenome and metatranscriptome [56], the active expression of methyl-accepting chemotaxis and flagellar motor switch protein-encoding genes in SZUA-1124 suggests that it could rapidly respond when its targeted substrates (methyl-compounds) become available [57].
Distribution in the ocean
To evaluate the prevalence of Hydro-γ-MAGs in the global ocean and their distribution patterns, we used their genomes as the query to recruit reads from 371 NCBI SRA (sequence read archive) metagenome archives, which provide wide coverage on global ocean waters of different depths. The RPKG (reads per kb per genome equivalent) serves as a good standard to estimate genome abundance within marine metagenomes, and RPKG > 1 stands for a clear sign that a genome is well-represented [58]. There are two general patterns (Supplementary Dataset S5): Cycloclasticus sp. SZUA-1075, Methylococcales bacterium SZUA-1212, and Methylophaga aminisulfidivorans SZUA-1072 only have significant distribution in hydrothermal environments, while the other Hydro-γ-MAGs have detectable distribution in seawater at various depths globally (Supplementary Fig. S9). In the latter group, Methylococcales bacterium SZUA-1258 has an even distribution throughout all depths [expect for high altitude Baltic Sea [59] and North Sea (PRJNA182070) metagenomes], while, the other MAGs are predominantly represented in mesopelagic and bathypelagic waters (Supplementary Fig. S9).
Discussion
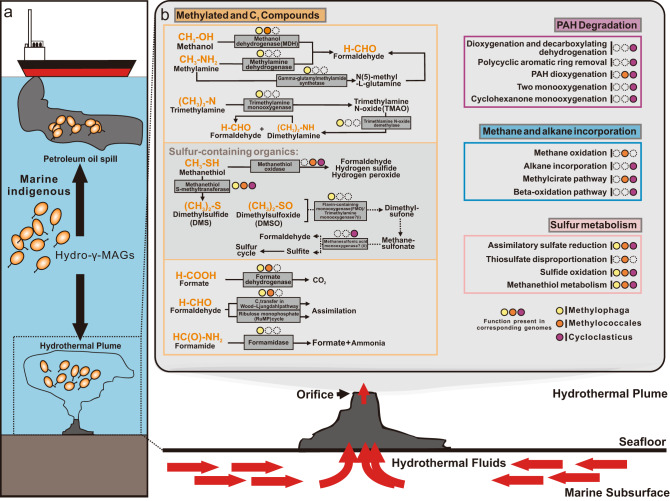
We reconstructed detailed pathways for the metabolism of five groups of substrates (methylated and C1 compounds, sulfur-containing organics, PAH, methane and alkane, and sulfur cycling-related compounds) that had been understudied heretofore in deep-sea hydrothermal plumes (Fig. 4). Placement of these pathways into their genomic context allowed us to make predictions about which taxa are able to use these different substrates. For the metabolism of methylated and C1 compounds, two groups of methylotrophic microorganisms, Methylococcales and Methylophaga could utilize methanol and formate, and assimilate formaldehyde [5], while methylamine pathways are exclusively found in Methylophaga genome. Capacities for PAH and alkane degradation were primarily in the Cycloclasticus genome, while methane oxidation could only be found in Methylococcales genome.
Fig. 4. Schematic figure indicating ecology and function of Hydro-γ-MAGs.
a Schematic diagram showing how indigenous Hydro-γ-MAGs are distributed in the marine water column and may utilize the chemicals in petroleum oil spills and hydrothermal plumes. b Summary diagram representing the metabolic functions that are mediated by Hydro-γ-MAGs in the hydrothermal environments. Remarks: (i) The flavin-containing monooxygenase [annotated as trimethylamine (TMA) monooxygenase here] was also reported to have broad substrate specificity and could oxidize DMS and DMSO; [71] (ii) The original annotations here are toluene monooxygenase (TmoCF), which have high sequence similarity to methanesulfonic acid monooxygenase (MsmCD) [72]. The function assigned here (oxidizing methanesulfonate) needs further validation.
Meanwhile, metatranscriptome comparison indicates functional variation between plume and background, and among different gene orthologs. For instance, in the Methylophaga aminisulfidivorans SZUA-1072 genome (Supplementary Fig. S4 and details in Supplementary Information), the methanol dehydrogenase gene (xoxF) Clade 1 copy (SZUA-1124_00001) has the highest expression level among all xoxF genes, while the other copy (SZUA-1124_02641) has nearly no expression. A possible evolutionary explanation is that SZUA-1124_00001 is the copy with original methanol oxidizing function and other paralogs have site substitutions, which make them either unexpressed/functionless or subfunctionalized [60]. Meanwhile, the higher expression level of xoxF (SZUA-1124_00001) relative to mxaF (Supplementary Fig. S4) indicates that xoxF is the more important functional gene for methanol oxidation. XoxF-MDHs bind rare-earth elements [such as cerium (III) and lanthanum (III)] as a co-factor that assists PQQ (Pyrroloquinoline Quinone, the prosthetic group for catalyzing) in catalysis. It is speculated that the presence of rare-earth elements at the active sites could confer a superior catalyzing efficiency to their Ca2+-binding MxaF counterparts [61].
Thermogenic interactions between upwelling vent fluids and dissolved organic matter (DOM) (likely subsurface microbial biomass) could be responsible for the production of MT and other organosulfur compounds [2]. The low temperature mixed fluids in Guaymas Basin and Cayman are enriched in MTs (up to 103–104 nM) (Supplementary Dataset S4) [2], consistent with the high expression level of the MT oxidation operon in the hydrothermal plume. The potential oxidation products of MT [52], e.g., formaldehyde and hydrogen sulfide, could be further utilized by plume microbes. Meanwhile, Hydro-γ-MAGs could actively catalyze the methylation of MT to dimethyl sulfide (DMS), further contributing to the microbial removal of MT [52, 62]. This study, to our knowledge, is the first functional evidence that hydrothermal plume microorganisms could utilize the simplest thermochemically-derived sulfur organics for energy yields.
In further support of the above statement that these Gammaproteobacteria are able of transforming important organosulfur compounds, spectroscopic data indicate that the particulate sulfur pool in the Von Damm buoyant plume is dynamic and diverse along plume rising path (Supplementary Dataset S6). The particulate sulfur pool contained a wide variety of organic and inorganic sulfur-bearing functional groups including diverse reduced, intermediate, and oxidized sulfur species (Supplementary Dataset S6). The organic sulfur compounds could be generated by the degradation of pre-existing sedimentary organics in the subsurface, and along the venting path, they were entrained to the plume environments and present in the sulfur pool [63]. Organic sulfur compounds may also form in the plume through the reaction of hydrogen sulfide and polysulfides with organic matter, a process referred to as sulfurization [63]. As these sulfurization products [thiol and organic monosulfide (-R-S-H) and thiophene (aromatic-R-S-R′, R could also be “H”)] are persistent in the buoyant plume, it is more likely that the latter process significantly contributes to the presence of organic sulfur compounds [63]. Components of the sulfur pool that contain the MT bond, are likely biogeochemically cycled by MT-utilizing microorganisms (either via methyl-transferring or MT bond oxidizing pathways), thereby contributing to element and energy transformation activities of plume microbiome. This is also consistent with the functional capacity of Methylophage from Cayman plume on utilizing MT (Fig. 4).
Hydro-γ-MAGs could perform Sox-dependent thiosulfate oxidation with sulfate as the product (Supplementary Fig. S5c); while, other bacteria could actively oxidize methane for energy yields. In addition, they could actively oxidize sulfide to element sulfur as the energy source, consistent with the chemical datasets showing high H2S concentrations (100–101 mM) in Guaymas Basin and Cayman plume samples [2, 64]. This suggests Hydro-γ-MAGs are well adapted to utilize abundant electron donors in hydrothermal environments.
The genome of Methylophaga aminisulfidivorans indicates the complete degradation pathway for methylamines and also suggests the utilization of various hydrothermal vent C1-compounds, including formate, formaldehyde, urea, and methanol (Supplementary Dataset S7). Moreover, the arsenic detoxification pathway, which reduces As (V) to As (III) and extrudes As (III) by arsenate reductase (ArsC) and arsenite transporter (ArsB), is present (Supplementary Dataset S7). Hydrothermal vent fluids are a significant source of arsenic to the deep-sea [65]; collectively, this indicates Methylophaga aminisulfidivorans is well adapted to the chemical background in close proximity to hydrothermal plume, being functionally capable to utilize these organic compounds.
These Hydro-γ-MAGs are major functional players involved in the use of C1-compounds, petroleum hydrocarbons, PAHs, and sulfur cycling (Supplementary Fig. S10) in specific hydrothermal plume environments. The significantly enriched distribution and elevated expression level revealed by this study suggest that they are specifically triggered by these hydrothermally sourced substrates in the plume environments (Supplementary Fig. S10). Hydro-γ-MAGs are prevalent in global marine water columns and have methyl-accepting chemotaxis mechanisms for searching preferable eco-niches with substantial substrates (for Cycloclasticus and Methylophaga) (Supplementary Fig. S10 and Supplementary Dataset S5). Though they are abundant and active in hydrothermal plume environments revealed from this study, these properties enable them to be widely adapted and significantly triggered in environments with similar substrates throughout the marine water column. For instance, as stimulated by similar petroleum hydrocarbons, e.g., alkanes, aromatics, methane, and other C1-compounds, these Hydro-γ-MAGs phylotypes (especially for Cycloclasticus) also play a significant functional role in petroleum oil spill environments.
As indicated by previous research, the heterotrophic degradation of marine DOM stimulates the activity of methylotrophs (e.g., Methylophaga) [66], potentially through degradation of DOM methyl sugars to the substrates of methylotrophs such as methanol or formaldehyde [66]. This successive and synergistic incorporation of DOM could in part explain the ubiquitous distribution of marine methylotrophs in the open ocean [66]. The linkage of heterotrophic DOM degradation and methyl-compound incorporation indicates the metabolic handoff of two groups of microorganisms [67]. The wide accessibility of DOM in the ocean suggests that these methylotrophs can be widely distributed in the marine water column and have a broader ecological impact [68]. The results from our study indicate that such marine methylotrophs could play a metabolic opportunistic role in hydrothermal plumes through their chemotactic properties, thus further improving our understanding of their significance in the ocean carbon cycle. Meanwhile, it will be also interesting to study the contribution of DOM-driven syntrophic activities in plume biogeochemical cycles.
Conclusion
This study reveals the metabolic potential and functional activity of Hydro-γ-MAGs in using methyl-, sulfur- and petroleum organic compound in deep ocean hydrothermal plumes, highlighting the essential importance of these compounds in the ecology and biogeochemistry of plumes and the connections between habitats of hydrothermal plumes, the open ocean, and oil spills. Based on these insights we propose a conceptual model, in which Hydro-γ-MAGs are indigenous microorganisms widely distributed in the global marine water column but their abundance and activity are maintained at a relatively low level due to limited availability of their carbon sources and electron donors for energy metabolism in the pelagic background [69]. As a response to stimulus (hydrothermal plume or petroleum oil spill), they find new habitats with a substantial supply of hydrocarbons and vigorously incorporate the substrates and replicate themselves to become the dominant taxa [20, 56, 70], thus playing significant roles in the biogeochemistry of deep-sea hydrothermal plumes.
Supplementary information
Acknowledgements
Thanks to the suggestions and critical reviews from anonymous reviewers. Thanks to the proofreading efforts and comments from Patricia Tran from University of Wisconsin – Madison. This study was funded by National Natural Science Foundation of China (No. 91851105, 31970105), the Science and Technology Innovation Committee of Shenzhen (No. JCYJ20170818091727570), the Key Project of Department of Education of Guangdong Province (No. 2017KZDXM071), Gordon and Betty Moore Foundation (GBMF2609 to GJD), and the National Science Foundation (OCE 1029242). We are grateful to the captains, crew, and chief scientists (Fred Prahl, Brian Popp, Anna-Louise Reysenbach, and Chris German) of the cruises on which samples were collected.
Data availability
The Sequence Read Archive IDs for metagenomic and metatranscriptomic are summarized in Supplementary Dataset S3. The MAGs that were resolved from this study are deposited under NCBI BioProject PRJNA488180. The further analyses on gene operons of methyl-metabolism, transcriptomic ratio of Hydro-γ-MAGs, and Cycloclasticus expression in DWH are present in Supplementary Information and Figs. S11–S18.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00745-5) contains supplementary material, which is available to authorized users.
References
- 1.Dick GJ. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol. 2019;17:271–83. doi: 10.1038/s41579-019-0160-2. [DOI] [PubMed] [Google Scholar]
- 2.Reeves EP, McDermott JM, Seewald JS. The origin of methanethiol in midocean ridge hydrothermal fluids. Proc Natl Acad Sci USA. 2014;111:5474. doi: 10.1073/pnas.1400643111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang SQ, Butterfield DA, Schulte M, Kelley DS, Lilley MD. Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim Cosmochim Acta. 2010;74:941–52. [Google Scholar]
- 4.Konn C, Charlou JL, Donval JP, Holm NG, Dehairs F, Bouillon S. Hydrocarbons and oxidized organic compounds in hydrothermal fluids from Rainbow and Lost City ultramafic-hosted vents. Chem Geol. 2009;258:299–314. [Google Scholar]
- 5.Proskurowski G, Lilley MD, Seewald JS, Fruh-Green GL, Olson EJ, Lupton JE, et al. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science. 2008;319:604–7. doi: 10.1126/science.1151194. [DOI] [PubMed] [Google Scholar]
- 6.Dick G, Anantharaman K, Baker B, Li M, Reed D, Sheik C. The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Microbiol. 2013;4:124. [DOI] [PMC free article] [PubMed]
- 7.Baker BJ, Lazar CS, Teske AP, Dick GJ. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome. 2015;3:14. [DOI] [PMC free article] [PubMed]
- 8.Dombrowski N, Seitz KW, Teske AP, Baker BJ. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:106. doi: 10.1186/s40168-017-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 2018;9:4999. doi: 10.1038/s41467-018-07418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Liu Y, Xu W, Pan J, Luo Z-H, Li M. Genome and community-level interaction insights on wide carbon utilizing and element cycling function of Hydrothermarchaeota from hydrothermal sediment. mSystems. 2020;5:e00795–00719. doi: 10.1128/mSystems.00795-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker BJ, Lesniewski RA, Dick GJ. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 2012;6:2269. doi: 10.1038/ismej.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anantharaman K, Breier JA, Sheik CS, Dick GJ. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci USA. 2013;110:330. doi: 10.1073/pnas.1215340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ. The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J. 2012;6:2257. doi: 10.1038/ismej.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantharaman K, Duhaime MB, Breier JA, Wendt K, Toner BM, Dick GJ. Sulfur oxidation genes in diverse deep-sea viruses. Science. 2014;344:757–60. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Jain S, Dick GJ. Genomic and transcriptomic resolution of organic matter utilization among deep-sea bacteria in guaymas basin hydrothermal plumes. Front Microbiol. 2016;7:1125. [DOI] [PMC free article] [PubMed]
- 16.Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ. Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun. 2015;6:8933. doi: 10.1038/ncomms9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Jain S, Baker BJ, Taylor C, Dick GJ. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ Microbiol. 2014;16:60–71. doi: 10.1111/1462-2920.12182. [DOI] [PubMed] [Google Scholar]
- 18.Skennerton CT, Ward LM, Michel A, Metcalfe K, Valiente C, Mullin S, et al. Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (Family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Front Microbiol. 2015;6:1425. [DOI] [PMC free article] [PubMed]
- 19.Villeneuve C, Martineau C, Mauffrey F, Villemur R. Methylophaga nitratireducenticrescens sp. nov. and Methylophaga frappieri sp. nov., isolated from the biofilm of the methanol-fed denitrification system treating the seawater at the Montreal Biodome. Int J Syst Evol Microbiol. 2013;63:2216–22. doi: 10.1099/ijs.0.044545-0. [DOI] [PubMed] [Google Scholar]
- 20.Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7:2315. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sargeant S, Murrell J, Nightingale P, Dixon J. Seasonal variability in microbial methanol utilisation in coastal waters of the western English Channel. Mar Ecol Prog Ser. 2016;550:53–64. [Google Scholar]
- 22.Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. Global dispersion and local diversification of the methane seep microbiome. Proc Natl Acad Sci USA. 2015;112:4015–20. doi: 10.1073/pnas.1421865112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, et al. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol. 2017;2:17093. doi: 10.1038/nmicrobiol.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneiker S, Martins dos Santos VA, Bartels D, Bekel T, Brecht M, Buhrmester J, et al. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol. 2006;24:997–1004. doi: 10.1038/nbt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai Q, Li W, Shao Z. Complete genome sequence of Alcanivorax dieselolei type strain B5. J Bacteriol. 2012;194:6674. doi: 10.1128/JB.01813-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rani S, Jeon WJ, Koh HW, Kim YE, Kang MS, Park SJ. Genomic potential of Marinobacter salinus Hb8(T) as sulfur oxidizing and aromatic hydrocarbon degrading bacterium. Mar Genom. 2017;34:19–21. [Google Scholar]
- 27.Singer E, Webb EA, Nelson WC, Heidelberg JF, Ivanova N, Pati A, et al. Genomic potential of marinobacter aquaeolei, a biogeochemical “Opportunitroph”. Appl Environ Microbiol. 2011;77:2763–71. doi: 10.1128/AEM.01866-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fondi M, Rizzi E, Emiliani G, Orlandini V, Berna L, Papaleo MC, et al. The genome sequence of the hydrocarbon-degrading Acinetobacter venetianus VE-C3. Res Microbiol. 2013;164:439–49. doi: 10.1016/j.resmic.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Jung J, Baek JH, Park W. Complete genome sequence of the diesel-degrading Acinetobacter sp. strain DR1. J Bacteriol. 2010;192:4794–5. doi: 10.1128/JB.00722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheik CS, Jain S, Dick GJ. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol. 2013;16:304–17. doi: 10.1111/1462-2920.12165. [DOI] [PubMed] [Google Scholar]
- 31.Breier JA, Sheik CS, Gomez-Ibanez D, Sayre-McCord RT, Sanger R, Rauch C, et al. A large volume particulate and water multi-sampler with in situ preservation for microbial and biogeochemical studies. Deep Sea Res Part I: Oceanographic Res Pap. 2014;94:195–206. [Google Scholar]
- 32.Breier JA, Rauch CG, McCartney K, Toner BM, Fakra SC, White SN, et al. A suspended-particle rosette multi-sampler for discrete biogeochemical sampling in low-particle-density waters. Deep Sea Res Part I. 2009;56:1579–89. [Google Scholar]
- 33.Breier J, Toner B, Fakra S, Marcus M, White S, Thurnherr A, et al. Sulfur, sulfides, oxides and organic matter aggregated in submarine hydrothermal plumes at 9°50′N East Pacific Rise. Geochim Cosmochim Acta. 2012;88:216–36. [Google Scholar]
- 34.Dick GJ, Tebo BM. Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ Microbiol. 2010;12:1334–47. doi: 10.1111/j.1462-2920.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- 35.Sheik CS, Anantharaman K, Breier JA, Sylvan JB, Edwards KJ, Dick GJ. Spatially resolved sampling reveals dynamic microbial communities in rising hydrothermal plumes across a back-arc basin. ISME J. 2014;9:1434. doi: 10.1038/ismej.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sieber CM, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 41.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laczny CC, Sternal T, Plugaru V, Gawron P, Atashpendar A, Margossian HH, et al. VizBin-an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome, 2015;3:1. [DOI] [PMC free article] [PubMed]
- 43.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2014;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teske A, de Beer D, McKay LJ, Tivey MK, Biddle JF, Hoer D, et al. The guaymas basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Front Microbiol. 2016;7:75. [DOI] [PMC free article] [PubMed]
- 50.Reveillaud J, Reddington E, McDermott J, Algar C, Meyer JL, Sylva S, et al. Subseafloor microbial communities in hydrogen-rich vent fluids from hydrothermal systems along the Mid-Cayman Rise. Environ Microbiol. 2016;18:1970–87. doi: 10.1111/1462-2920.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anantharaman K, Breier JA, Dick GJ. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2015;10:225. doi: 10.1038/ismej.2015.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyice O, Myronova N, Pol A, Carrion O, Todd JD, Smith TJ, et al. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2018;12:145–60. doi: 10.1038/ismej.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Graichen ME, Liu A, Pearson AR, Wilmot CM, Davidson VL. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42:7318–25. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 54.Gondwe M, Krol M, Klaassen W, Gieskes W, de Baar H. Comparison of modeled versus measured MSA:nss SO4 = ratios: a global analysis. Global Biogeochem Cycles 2004;18:2.
- 55.Li M, Toner BM, Baker BJ, Breier JA, Sheik CS, Dick GJ. Microbial iron uptake as a mechanism for dispersing iron from deep-sea hydrothermal vents. Nat Commun. 2014;5:3192. doi: 10.1038/ncomms4192. [DOI] [PubMed] [Google Scholar]
- 56.Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6:1715–27. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 58.Haro-Moreno JM, Rodriguez-Valera F, Lopez-Garcia P, Moreira D, Martin-Cuadrado A-B. New insights into marine group III Euryarchaeota, from dark to light. ISME J. 2017;11:1102–17. [DOI] [PMC free article] [PubMed]
- 59.Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B, et al. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 2014;8:1892. doi: 10.1038/ismej.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–8. [Google Scholar]
- 61.Keltjens JT, Pol A, Reimann J, Op den Camp HJM. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 62.Carrión O, Curson ARJ, Kumaresan D, Fu Y, Lang AS, Mercadé E, et al. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun. 2015;6:6579. doi: 10.1038/ncomms7579. [DOI] [PubMed] [Google Scholar]
- 63.Cron BR, Sheik CS, Kafantaris FCA, Druschel GK, Seewald JS, German CR, et al. Dynamic biogeochemistry of the particulate sulfur pool in a buoyant deep-sea hydrothermal plume. ACS Earth Space Chem. 2020;4:168–82. [Google Scholar]
- 64.Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545. doi: 10.1038/ismej.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X-G, Lyu S-S, Garbe-Schönberg D, Lebrato M, Li X, Zhang H-Y, et al. Heavy metals from Kueishantao shallow-sea hydrothermal vents, offshore northeast Taiwan. J Mar Syst. 2016;180:211–9.
- 66.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–7. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansell DA, Carlson CA, Repeta DJ, Schlitzer R. Dissolved organic matter in the ocean a controversy stimulates new insights. Oceanography. 2009;22:202–11. [Google Scholar]
- 69.Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2004;437:349–55. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 70.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–8. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA. 2011;108:17791–6. [DOI] [PMC free article] [PubMed]
- 72.Leahy JG, Batchelor PJ, Morcomb SM. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev. 2003;27:449–79. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Sequence Read Archive IDs for metagenomic and metatranscriptomic are summarized in Supplementary Dataset S3. The MAGs that were resolved from this study are deposited under NCBI BioProject PRJNA488180. The further analyses on gene operons of methyl-metabolism, transcriptomic ratio of Hydro-γ-MAGs, and Cycloclasticus expression in DWH are present in Supplementary Information and Figs. S11–S18.