Abstract
Upon the discovery of numerous genes involved in the pathogenesis of neurodevelopmental disorders, several studies showed that a significant proportion of these genes converge on common pathways and protein networks. Here, we used a reversed approach, by screening the AnkyrinG protein-protein interaction network for genetic variation in a large cohort of 1009 cases with neurodevelopmental disorders. We identified a significant enrichment of de novo potentially disease-causing variants in this network, confirming that this protein network plays an important role in the emergence of several neurodevelopmental disorders.
Subject terms: Genetics research, Neurological disorders
Introduction
Neurodevelopmental disorders (NDDs) originate from a disturbed development of the central nervous system, with symptoms including deficits in personal, social, and cognitive functioning, which becomes apparent during early development [1]. According to the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-V), the NDD group comprises Attention-Deficit Hyperactivity Disorder (ADHD), Autism Spectrum Disorder (ASD), Intellectual Disability (ID), communication disorders, specific learning disorders, and motor disorders [1]. Based on common phenotypic features as well as overlapping genetic factors underlying the diseases, additional disorders are often classified as NDDs. These include epilepsy, but also psychiatric disorders such as schizophrenia and bipolar disorder. NDDs are clinically heterogeneous, varying widely in severity. Additionally complicating the clinical presentation and prediction of the course of disease, is the fact that NDDs are often comorbid. Of all patients with ID for example, ~22% suffers from epilepsy, 17% has ASD and almost 10% has ADHD, percentages that are significantly higher than the population prevalences [2]. Furthermore, there is an extreme heterogeneity in the genetic causes of NDDs, with many different genes involved in the pathogenesis of the same disorder, while each of the genes explains only a very small proportion of patients. Strikingly, upon the discovery of many novel disease genes and CNVs in massive screening studies, it became apparent that certain genetic aberrations can be associated with multiple disorders, including ID, ASD, schizophrenia, and epilepsy [3–11]. The identified causal genes were found to be enriched for components of specific pathways, amongst which chromatin remodelling and Wnt-signalling, but also for neuron-specific networks responsible for synaptic function and neuronal development [12–14]. These novel insights in the genetics of NDDs prompted a novel pathophysiology hypothesis, shifting from single variants as the cause of a distinct disorder, to disturbance of a cellular pathway or an entire network underlying a broader spectrum of disorders.
We hypothesised that the significant enrichment of independently discovered genes in specific networks implicates that the majority of genes, in addition to the established NDD genes, in a given network are likely NDD genes. As a proof of principle, we investigated the de novo genetic variation in all of the individual genes from a protein–protein interaction (PPI) network in an extensive cohort of NDD patients. We selected the AnkG PPI network as it can be delineated relatively easily and consists of a limited amount of genes. AnkG proteins bind to the spectrin cytoskeleton as well as to multiple membrane proteins, including cell adhesion molecules and voltage-gated sodium and potassium channels [15–18]. These membrane proteins are thereby restricted to a defined spatial position, which is essential for proper axon potential initiation and propagation along the axon [19]. ANK3 encodes the AnkG protein, of which the two largest isoforms are highly enriched at the axon initial segment and the nodes of Ranvier of myelinated neurons in the central nervous system [20]. The protein in its various isoforms is necessary for axon maintenance and dendritic spine morphology [21, 22]. Variants in ANK3 were implicated in several NDD phenotypes, including ID, ASD, and behavioural problems, whereas multiple SNPs in ANK3 have confirmed associations with bipolar disorder, schizophrenia, and post-traumatic stress disorder in several genome-wide association studies [23–29]. At the time we initiated our study, several proteins interacting with AnkG had already been associated with variable NDD phenotypes.
Results
Defining the AnkyrinG network
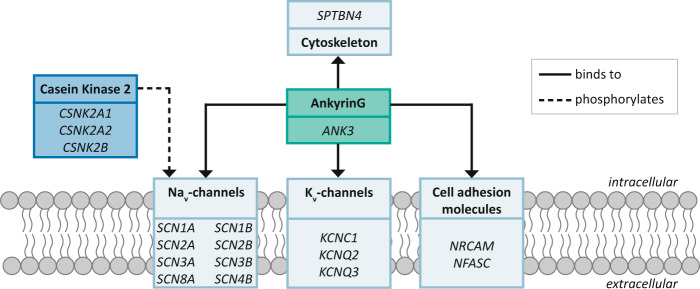
Although the interactions of AnkG with multiple proteins were reported, a comprehensive PPI network had not been described. We therefore performed an extensive literature search and identified a total of 17 proteins that either directly interact with AnkG, are part of a protein complex that interacts with AnkG or directly influence the binding affinity of particular proteins with AnkG (Fig. 1, supplementary table 1). Direct interactors include cell adhesion molecules, cytoskeleton-component β4-spectrin, and specific sodium and potassium channel components. The other proteins forming the sodium and potassium channels are indirect interactors of AnkG by means of their inclusion in the channel complexes. The genes encoding the components of Casein kinase 2 (CK2) were included in the study as CK2-mediated phosphorylation of sodium channels determines the binding affinity of the channel and AnkG [30].
Fig. 1. Schematic overview of the ANK3 protein-protein interaction network members.
Light blue boxes indicate genes encoding proteins or protein complexes that directly interact with AnkyrinG. Dark blue box indicates genes encoding the components of casein kinase 2 (CK2), which regulates the binding affinity of voltage-gated sodium channels for AnkyrinG through phosphorylation.
Coding de novo variants in the AnkG network are significantly enriched in patients with neurodevelopmental disorders
Using the molecular inversion probe (MIP) enrichment strategy [8], targeted sequencing of the 18 genes in the ANK3 PPI network (Fig. 1, Supplementary Table 1) was performed for 1009 patient-parent trios, enabling filtering for de novo variants. A total of 14 NextSeq 500 high output runs were used, with an average yield of 75.9 Gbp (±12.2 (SD)) and an average percentage of reads above Q30 of 93.2%. The total panel size was 66,662 bp, of which 992 bp were badly covered (regions of more than 10 subsequent bases with an average coverage below 5X in more than 20% of the samples). The average coverage over all samples was 794×. Upon data quality filtering, 911 complete trios remained with a total of 131 nonsynonymous de novo variants. Of these, we selected the 58 variants with a CADD (v1.3) Phred-scaled pathogenicity score larger than 20 for Sanger sequencing validation [31]. Upon this validation experiment, a total number of 14 confirmed de novo single nucleotide variants were identified in different ANK3 PPI network members. One variant affects a canonical splice site, nine are missense variants and four variants introduce a premature stopcodon (Table 1).
Table 1.
Overview of the potentially disease-causing de novo variants identified in the targeted screen of all ANK3 PPI network members.
| Gene | gDNA change (GRCh37 (hg19)) | cDNA change | Protein change | Variant Type | CADD_Phred score | Domain | Neurodevelopmental phenotype |
|---|---|---|---|---|---|---|---|
| CSNK2A1 | NC_000020.10:g.472926T>C | NM_177559.2:c.593A>G | p.(Lys198Arg) | missense | 30 | Activation loop (Lys198 responsible for recognition) | Developmental delay, hypotonia |
| CSNK2A1 | NC_000020.10:g.476394T>C | NM_177559.2:c.479A>G | p.(His160Arg) | missense | 24.7 | Active site | Epilepsy, developmental delay, autism spectrum disorder |
| KCNQ2 | NC_000020.10:g.62073826A>C | NM_172107.2:c.749T>G | p.(Val250Gly) | missense | 25 | Transmembrane helix 5 | Epileptic encephalopathy, intellectual disability |
| KCNQ2 | NC_000020.10:g.62044879C>T | NM_172107.2:c.1687G>A | p.(Asp563Asn) | missense | 26.7 | C-terminal cytoplasmic domain | Severe frontal epilepsy, autism spectrum disorder, severe intellectual disability, behavioural problems |
| SCN1A | NC_000002.11:g.166898844G>A | NM_001165963.1:c.2134C>T | p.(Arg712*) | nonsense | 38 | Cytoplasmic domain | Unexplained psychomotor problems |
| SCN1A | NC_000002.11:g.166852623C>T | NM_001165963.1:c.4481G>A | p.(Gly1494Glu) | missense | 29.3 | Cytoplasmic domain | Epilepsy (Dravet Syndrome), intellectual disability |
| SCN1B | NC_000019.9:g.35524482G>A | NM_001037.4:c.287G>A | p.(Arg96Gln) | missense | 26.3 | Extracellular domain | Developmental delay, mild intellectual disability, sleeping problems, severe psychiatric disorder (psychoses), craniosynostosis with scaphocephaly |
| SCN2A | NC_000002.11:g.166231477G>A | NM_001040142.1:c.4254 + 1G>A | p.? | splice site | 23.3 | — | Psychomotor retardation, epilepsy |
| SCN2A | NC_000002.11:g.166166923C>T | NM_021007.2:c.788 C>T | p.(Ala263Val) | missense | 29.0 | Transmembrane helix 5 repeat I | Intellectual disability, motor retardation, epilepsy/epileptiform EEG |
| SCN2A | NC_000002.11:g.166172167C>T | NM_021007.2:c.1570C>T | p.(Arg524*) | nonsense | 29.7 | Ankyrin-binding domain | Severe psychomotor retardation, autism spectrum disorder |
| SCN2A | NC_000002.11:g.166223837G>A | NM_021007.2:c.3631G>A | p.(Glu1211Lys) | missense | 34.0 | Transmembrane helix 1 repeat III | Unexplained psychomotor problems, infantile epileptic encephalopathy |
| SCN8A | NC_000012.11:g.52159701C>T | NM_014191.3:c.2791C>T | p.(Arg931*) | nonsense | 42 | Extracellular domain | Unexplained psychomotor problems, childhood absence epilepsy, behavioural problems, MRI abnormalities |
| SCN8A | NC_000012.11:g.52162973A>T | NM_014191.3:c.3226A>T | p.(Lys1076*) | nonsense | 43 | Ankyrin-binding domain | Developmental delay, intellectual disability, epilepsy |
| SCN8A | NC_000012.11:g.52162699C>G | NM_014191.3:c.2952C>G | p.(Asn984Lys) | missense | 26.5 | Cytoplasmic domain | Psychomotor retardation, therapy-resistent epilepsy |
The total mutation rate for all 18 genes was calculated by summing the gene-specific mutation rates of all the individual genes [32] and used to assess statistical significance of identifying 14 de novo (functional) variants in this gene set in a cohort of 911 individuals. A significant overrepresentation of de novo coding variants in the ANK3 PPI was observed in our cohort (p = 1.529e−10, Exact Poisson test). In addition, the enrichment for functional variants for each of the individual genes was determined, showing a significant enrichment of variants in four genes; CSNK2A1, KCNQ2, SCN2A, and SCN8A (Supplementary Table 2).
Discussion
We investigated whether the genes encoding proteins in the AnkG interactome are enriched for variants that are predicted to affect protein function in a cohort of neurodevelopmental disorder patients, mainly presenting with ID and/or ASD. When the gene panel was selected at the start of the study, protein interaction databases such as string-db, produced a limited and incomplete PPI for AnkG [33]. Therefore, we decided to perform an extensive literature study to identify the relevant members of this PPI. However, the amount of literature reports on protein interaction studies rises continuously, and the current string-db based AnkG PPI (Supplementary Fig. 1) is very similar to the AnkG interactome we identified through literature. Hence, we believe that extending a string-db generated interactome based on a detailed review of recent literature is a mandatory step to apply our protein-interaction based screening strategy.
We identified variants in six out of 18 genes in the AnkyrinG interactome in our cohort consisting of 911 trios with probands referred for ID and/or ASD. Four patients harbored a de novo genetic variation in SCN2A and three carried a variant in SCN8A (Table 1). The splice site and the three nonsense variants in these genes are generally accepted to affect protein function, whereas the three missense variants have CADD-Phred scores of 26.5, 29, and 34, respectively, suggesting at least a likely association of the variants with the phenotypes. In addition, we identified variants in CSNK2A1, KCNQ2, and SCN1A (two cases for each gene) as well as in SCN1B (Table 1). While the association of CSNK2A1 with NDDs was undocumented when we started this study, variants in CSNK2A1 are now known as the cause of Okur-Chung syndrome. This syndrome is characterized by developmental delay, ID with poor speech, behavioral problems, and additional features [34, 35]. Strikingly, we identified the exact same p.(Lys198Arg) de novo variant in CSNK2A1 in a patient with developmental delay and hypotonia in our cohort as described in a patient with developmental delay, ID, and hypotonia by Okur et al. [34]. This variant is located in the activation segment of the protein and affects a basic amino acid that is involved in the recognition of acidic amino acids surrounding the phosphorylation site in the substrate of the casein kinase 2 enzyme, of which CSNK2A1 is a subunit [36]. The exact same variant has now been identified in at least seven individuals (including the individual presented here) and may thus be considered as a hotspot mutation for Okur-Chung syndrome [34, 35, 37, 38]. In SCN1B, we identified a de novo p.(Arg96Gln) variant. Variants in SCN1B have so far only been associated with generalized epilepsy with febrile seizures plus and epileptic encephalopathy [39]. To our knowledge, only one de novo missense variant in SCN1B has been reported in cases with developmental delay or ID [40]. We identified a p.(Arg96Gln) missense variant, predicted to be likely disease-causing based on a CADD-Phred score of 26.3, in a patient with developmental delay, sleeping problems, and a severe psychiatric disorder. Our results thus strengthen the evidence for a broader NDD phenotype related to variants in SCN1B. Many of the epilepsy-related variants in SCN1B and the missense variant identified here are located in the extracellular immunoglobulin-like domain, which is required for its function as a modulator of the Nav α subunit [41–43]. Moreover, the Nav β1 subunit (encoded by SCN1B) contains an AnkG interaction site in the intracellular domain and has Ankyrin-recruiting activity [44]. This might implicate that variants in the β1 subunit are likely to result in NDD phenotypes, as disturbance of this particular subunit may result in a destabilization of the entire AnkG network. Finally, variants akin to the variants we identified in SCN1A and KCNQ2 have been reported as likely causative in patients with various NDDs in recent years [35, 45, 46].
To date, 12 out of the 18 ANK3 PPI genes have been implicated in neurodevelopmental disorders. Our study likely lacks the power to detect variants in the remaining six ANK3 PPI network members due to our cohort size and the low frequency of disease-associated variants in individual genes. Alternatively, these genes may be less vulnerable for variation and may only cause a phenotype when affected by biallelic or even bigenic variants [47]. In this study, we focused on de novo (heterozygous) variants in accordance with the de novo paradigm in neurodevelopmental disorders [48]. However, in follow-up studies it would be interesting to explore this dataset beyond autosomal dominant effects and investigate other inheritance patterns.
Second, we considered only those variants with a Phred-scaled CADD score of 20 and higher. Lowering this threshold might allow the identification of modifier or digenic effects [47]. However, the high false discovery rate (FDR) of our assay, requiring independent validation of detected variants, resulted in a prohibitive amount of variants for lower thresholds. Therefore, we highly recommend to take measures to reduce the FDR in MIPs based resequencing assays. First, applying double tiling and single-molecule MIP (smMIP) design offer advanced filtering options that largely alleviate the errors introduced by PCR and capture bias [49]. Second, we observed that cohort-based genotyping using GATK hampers the resolution of some of the quality metrics, including QualityByDepth (Supplementary Fig. 2). Nevertheless, as all variants reported here were independently verified, we can argue that the actual mutational burden on the network is even higher than reported here.
The identification of hundreds of genes that can cause NDDs when mutated, in particular for ID, ASD, and epilepsy, has allowed the identification of significantly enriched molecular pathways and protein networks involved in the pathogenesis of these disorders [8–10, 13, 35, 50]. We here provide evidence that the reverse may also be true, e.g, that interaction partners of genes disturbed in NDD patients, may cause similar phenotypes when mutated. We opted to investigate this hypothesis by selecting the ANK3 gene, which has been linked to several NDD phenotypes [19, 28, 29]. Based on literature, we selected a total of 17 proteins that directly interact with AnkG, are part of ion channel complexes of which at least one component directly interacts with AnkG or determine the binding affinity between AnkG and an interactor. We subsequently screened an NDD cohort of over 1000 ID trios for variants in the ANK3 PPI network and identified potential disease-causing de novo variants in 14 patients, including one splice site variant, nine missense and four nonsense variants. Based on the gene-specific mutation rates, this number of variants is significantly higher than may be expected to occur by chance in the AnkyrinG interactome. The studied PPI contains proteins that depend on the binding to each other and the integrity of the complex as a whole to fulfill their biological function. We expect similar variant enrichments for other protein complexes that meet these conditions, such as the BAF complex. This is an ATP-dependent chromatin remodeling complex of which multiple-member proteins have been linked to ID and/or ASD [51, 52]. The protein interaction approach demonstrated here may be less efficient starting from more promiscuous proteins such as FMRP. This protein product of the fragile X syndrome gene FMR1 interacts with many biochemical and cellular pathways [53, 54]. On the RNA level only at least 842 robust FMRP target mRNAs interactions have been identified [55]. Although these targets have been shown to be enriched amongst genes disrupted in ID and autism [56], an enrichment for neurodevelopmental disorder genes for FMRP interactors on all levels has not been reported.
Conclusion
Here, we studied the AnkG PPI network to determine the role of genetic variation in this network in the emergence of NDDs, possibly beyond known associated morbidities. Our results confirmed that multiple components of the AnkG PPI network can cause NDDs when mutated, by the discovery of a significant overrepresentation of potentially disease-causing variants in the network. We hypothesized that the significant enrichment of independently discovered genes in specific networks, implicates the remaining genes in these networks as likely candidate genes for (as yet undiagnosed) NDDs. As our results clearly support, this reversed or ‘guilty-by-association’ strategy may aid in compiling evidence for novel (very) rare genetic causes of NDDs.
Materials and methods
Samples
DNA samples were collected at the department of Medical Genetics of the University of Antwerp in Belgium (530 trios) and the Human Genetics department of the Radboudumc in Nijmegen, the Netherlands (479 trios). Included patients were diagnosed with intellectual disability and/or autism spectrum disorder, with or without other comorbidities. Clinical characteristics of the cohort are summarized in Supplementary Table 3. Previous genetic screenings included microarrays for CNV detection and in some cases Fragile-X syndrome testing or targeted single-gene sequencing based on phenotypic features, all with negative results. DNA of both parents had to be available to enable trio sequencing and subsequent de novo filtering. This study has been approved by the Committee for Medical Ethics UZA/UAntwerp.
Definition of the AnkG PPI network
To define the AnkG PPI network, a literature search using Pubmed was performed using the terms ‘ANK3’, ‘AnkG’, and ‘AnkyrinG’. In addition, more in depth and specific searches were performed based on results and references in the articles identified in the initial search. The target genes in the ANK3 PPI network were selected based on a search for interaction partners of AnkG and proteins that directly influence the interactions between AnkG and other proteins. Genes were included if functional evidence for interaction was presented in at least one article, or when interaction was strongly suspected based on literature. All relevant interaction partners published until August 2015 were included in the design of the targeted sequencing panel.
MIPs enrichment and sequencing
Enrichment of target sequences was done using the Molecular Inversion Probe (MIPs) technology, as described by O’Roak et al. [8]. In short, single tiled MIPs were designed using the MIPgen design pipeline [57]. Probes were synthesized by IDT (Leuven, Belgium) using standard desalting conditions. Upon balanced pooling, the probes were 5’ phosphorylated using T4 PNK (New England Biolabs, Ipswich, MA, USA). Targeted sequence capture was performed using Hemo Klentaq (New England Biolabs) and Ampligase DNA Ligase (Epicentre, Madison, WI, USA). Subsequent Exo-treatment was performed using EXO I and EXO III (New England Biolabs). PCR amplification was performed using iProof High Fidelity DNA Polymerase (Bio-Rad, Hercules CA, USA). Sequencing was done on an Illumina NextSeq 500 system (Illumina, San Diego, CA, USA), using NextSeq 500 v2 High Output kits (Illumina). Raw data have been deposited in the European Genome-Phenome Archive (EGA) under the study identifier “EGAS00001004326”.
Data analysis pipeline
Data processing was performed using an in-house developed pipeline (Radboudumc Nijmegen). Data files were demultiplexed and reads were aligned to the reference genome (version GRCh37/hg19) using BWA-MEM [58]. Subsequently, trimming of the ligation and extension arm sequences was performed based on genomic location referred to in the design file. Variant calling was done using GATK UnifiedGenotyper with default settings except for disabled downsampling [59]. A cohort approach performing multi-sample genotyping per sequencing run was applied [60]. Annotation of the variants was done using the Nijmegen research annotation pipeline.
De novo analysis
All variants called in patients were assessed for inheritance using the in-house developed trio analysis tool (Radboudumc Nijmegen). Variants in the patients were annotated as maternally or paternally inherited or de novo, producing a file containing all potentially de novo variants within the project. Variants in this file were filtered for a quality by depth >300, total coverage of the index call >50 and an alternative allele percentage between 20 and 80%. Trios for which at least one of the samples failed the quality parameters were excluded from analysis. Only non-synonymous variants in exonic regions and variants affecting canonical splice sites were considered based on in house annotation methods.
Sanger sequencing
Independent confirmation of variants detected in the MIPs screen was performed using Sanger sequencing. Primers were designed using Primer3Plus [61]. PCR was performed with the GoTaq G2 Polymerase kit (Promega Benelux, Leiden, the Netherlands) according to the guidelines of the manufacturer. PCR cleanup was performed using alkaline phosphatase (Roche, Basel, Switzerland) and exonuclease I (Bioké, Leiden, the Netherlands). Sanger sequencing reactions were done using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit with readout on an ABI3130XL sequencer (Applied Biosystems, Foster City, CA, USA). CLC DNA Workbench 5.0.2 (CLC bio, Aarhus, Denmark) was used for data analysis.
Statistical enrichment of de novo mutations
Gene-specific mutation rates (GSMR) of all the individual genes of the ANK3 PPI were retrieved from the dataset of Samocha et al. [32]. Total mutation rate for all 18 genes was calculated by summing the individual GSMRs. Statistical significance of identifying 14 de novo (functional) mutations in these 18 genes in a cohort of 911 individuals (1822 alleles) was assessed using the Exact Poisson test in R. In addition, enrichment for functional mutations in each of the individual genes was assessed with Exact Poisson tests. P-values were corrected with Bonferroni correction for the total number of 18 genes.
Supplementary information
Acknowledgements
This work was sponsored by a grant from the Fonds Wetenschappelijk Onderzoek - Vlaanderen (FWO) to RFK and GVDW and by a grant from the Netherlands Organization for Health Research and Development (912-12-109) to LELMV and BBAdV.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-0682-0) contains supplementary material, which is available to authorized users.
References
- 1.APA: American Psychiatric Association. Diagnostic and statistical manual of mental disorders DSM-V, Fifth ed., Text revision. Washington DC: American Psychiatric Association; 2013.
- 2.Oeseburg B, Dijkstra GJ, Groothoff JW, Reijneveld SA, Jansen DE. Prevalence of chronic health conditions in children with intellectual disability: a systematic literature review. Intellect Dev Dis. 2011;49:59–85. doi: 10.1352/1934-9556-49.2.59. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–8. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–7. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, Hippolyte L, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–8. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science (New York, NY) 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 11.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 12.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 15.Davis JQ, Lambert S, Bennett V. Molecular composition of the node of Ranvier: identification of ankyrin-binding cell adhesion molecules neurofascin (mucin+/third FNIII domain-) and NrCAM at nodal axon segments. J Cell Biol. 1996;135:1355–67. doi: 10.1083/jcb.135.5.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berghs S, Aggujaro D, Dirkx R, Jr, Maksimova E, Stabach P, Hermel JM, et al. betaIV spectrin, a new spectrin localized at axon initial segments and nodes of ranvier in the central and peripheral nervous system. J Cell Biol. 2000;151:985–1002. doi: 10.1083/jcb.151.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, et al. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins PM, Kim N, Jones SL, Tseng WC, Svitkina TM, Yin HH, et al. Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc Natl Acad Sci USA. 2015;112:957–64. doi: 10.1073/pnas.1416544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–9. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- 21.Sobotzik JM, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, et al. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci USA. 2009;106:17564–9. doi: 10.1073/pnas.0909267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schurmann B, et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 2014;84:399–415. doi: 10.1016/j.neuron.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan A, Yi Z, Wang Q, Sun J, Li Z, Du Y, et al. ANK3 as a risk gene for schizophrenia: New data in han Chinese and meta analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:997–1005. doi: 10.1002/ajmg.b.32112. [DOI] [PubMed] [Google Scholar]
- 25.Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry. 2009;14:487–91. doi: 10.1038/mp.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–53. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logue MW, Solovieff N, Leussis MP, Wolf EJ, Melista E, Baldwin C, et al. The ankyrin-3 gene is associated with posttraumatic stress disorder and externalizing comorbidity. Psychoneuroendocrinology. 2013;38:2249–57. doi: 10.1016/j.psyneuen.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P, et al. Mutations of ANK3 identified by exome sequencing are associated with autism ausceptibility. Hum Mutat. 2012;33:1635–8. doi: 10.1002/humu.22174. [DOI] [PubMed] [Google Scholar]
- 29.Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA, et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22:1960–70. doi: 10.1093/hmg/ddt043. [DOI] [PubMed] [Google Scholar]
- 30.Brechet A, Fache MP, Brachet A, Ferracci G, Baude A, Irondelle M, et al. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J Cell Biol. 2008;183:1101–14. doi: 10.1083/jcb.200805169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okur V, Cho MT, Henderson L, Retterer K, Schneider M, Sattler S, et al. De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum Genet. 2016;135:699–705. doi: 10.1007/s00439-016-1661-y. [DOI] [PubMed] [Google Scholar]
- 35.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarno S, Vaglio P, Meggio F, Issinger OG, Pinna LA. Protein kinase CK2 mutants defective in substrate recognition. Purification and kinetic analysis. J Biol Chem. 1996;271:10595–601. doi: 10.1074/jbc.271.18.10595. [DOI] [PubMed] [Google Scholar]
- 37.Chiu ATG, Pei SLC, Mak CCY, Leung GKC, Yu MHC, Lee SL, et al. Okur-Chung neurodevelopmental syndrome: Eight additional cases with implications on phenotype and genotype expansion. Clin Genet. 2018;93:880–90. doi: 10.1111/cge.13196. [DOI] [PubMed] [Google Scholar]
- 38.Akahira-Azuma M, Tsurusaki Y, Enomoto Y, Mitsui J, Kurosawa K. Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum Genome Var. 2018;5:18011. doi: 10.1038/hgv.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr, Phillips HA, et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–70. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 40.Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 41.Patino GA, Isom LL. Electrophysiology and beyond: multiple roles of Na+ channel beta subunits in development and disease. Neurosci Lett. 2010;486:53–9. doi: 10.1016/j.neulet.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffer IE, Harkin LA, Grinton BE, Dibbens LM, Turner SJ, Zielinski MA, et al. Temporal lobe epilepsy and GEFS+ phenotypes associated with SCN1B mutations. Brain: J Neurol. 2007;130:100–9. doi: 10.1093/brain/awl272. [DOI] [PubMed] [Google Scholar]
- 43.McCormick KA, Isom LL, Ragsdale D, Smith D, Scheuer T, Catterall WA. Molecular determinants of Na+ channel function in the extracellular domain of the beta1 subunit. J Biol Chem. 1998;273:3954–62. doi: 10.1074/jbc.273.7.3954. [DOI] [PubMed] [Google Scholar]
- 44.Malhotra JD, Koopmann MC, Kazen-Gillespie KA, Fettman N, Hortsch M, Isom LL. Structural requirements for interaction of sodium channel beta 1 subunits with ankyrin. J Biol Chem. 2002;277:26681–8. doi: 10.1074/jbc.M202354200. [DOI] [PubMed] [Google Scholar]
- 45.Fry AE, Rees E, Thompson R, Mantripragada K, Blake P, Jones G, et al. Pathogenic copy number variants and SCN1A mutations in patients with intellectual disability and childhood-onset epilepsy. BMC Med Genet. 2016;17:34. doi: 10.1186/s12881-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weckhuysen S, Mandelstam S, Suls A, Audenaert D, Deconinck T, Claes LRF, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71:15–25. doi: 10.1002/ana.22644. [DOI] [PubMed] [Google Scholar]
- 47.Papadimitriou S, Gazzo A, Versbraegen N, Nachtegael C, Aerts J, Moreau Y, et al. Predicting disease-causing variant combinations. Proc Natl Acad Sci USA. 2019;116:11878–87. doi: 10.1073/pnas.1815601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vissers LELM, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 49.Beyens M, Boeckx N, Van Camp G, Op de Beeck K, Vandeweyer G. pyAmpli: an amplicon-based variant filter pipeline for targeted resequencing data. BMC Bioinform. 2017;18:554. doi: 10.1186/s12859-017-1985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 51.Vandeweyer G, Helsmoortel C, Van Dijck A, Vulto-van Silfhout AT, Coe BP, Bernier R, et al. The Transcriptional Regulator ADNP Links the BAF (SWI/SNF) Complexes With Autism. Am J Med Genet C. 2014;166:315–26. doi: 10.1002/ajmg.c.31413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, Hibi-Ko Y, et al. Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature. Am J Med Genet. 2013;161A:1221–37. doi: 10.1002/ajmg.a.35933. [DOI] [PubMed] [Google Scholar]
- 53.Braat S, Kooy RF. Fragile X syndrome neurobiology translates into rational therapy. Drug Discov Today. 2014;19:510–9. doi: 10.1016/j.drudis.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB, Jr, Moine H, Kooy RF, et al. Fragile X syndrome. Nat Rev Dis Prim. 2017;3:17065. doi: 10.1038/nrdp.2017.65. [DOI] [PubMed] [Google Scholar]
- 55.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyle EA, O’Roak BJ, Martin BK, Kumar A, Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.