Abstract
β1–3-Linked galactosides such as Galβ1‒3GlcNAcβOR are common carbohydrate motifs found in human milk oligosaccharides (HMOSs), glycolipids, and glycoproteins. Efficient and scalable enzymatic syntheses of these structures have proven challenging due to the lack of access to a highly active β1‒3-galactosyltransferase (β3GalT) in large amounts. Previously reported E. coli β3GalT (EcWbgO) has been identified as a limiting factor for producing a β1–3-galactose-terminated human milk oligosaccharide lacto-N-tetraose (LNT) by fermentation. Here we report the identification of an EcWbgO homolog from C. violaceum (Cvβ3GalT) which showed a high efficiency in catalyzing the formation of LNT from lacto-N-triose (LNT II). With the highly active Cvβ3GalT, multigram-scale (>10 gram) synthesis of LNT from lactose was achieved using a sequential one-pot multienzyme (OPME) glycosylation process. The access to Cvβ3GalT enabled enzymatic synthesis of several fucosylated HMOSs with or without further sialylation including LNFP II, S-LNF II, LNDFH I, LNFP V, and DiFuc-LNT. Among these, LNFP V and DiFuc-LNT would not be accessible by enzymatic synthesis if an active β3GalT were not available.
Keywords: biocatalysis, enzymatic synthesis, glycosyltransferase, human milk oligosaccharide, LNT
Graphical Abstract
β1–3-Linked galactosides such as Galβ3GlcNAcβOR are common carbohydrate motifs found in human milk oligosaccharides (HMOSs), glycolipids, and glycoproteins.1–3 Galβ1‒3GlcNAc-containing blood group epitopes Lewis b and sialyl Lewis a are tumor associated carbohydrate antigens (TACAs), and their upregulation on cancer cells is associated with poor cancer prognosis.4 Galβ3GlcNAcβOR, which is called the Type 1 glycan structure, is also an important motif in HMOSs, a group of more than 100 oligosaccharides naturally presented in human milk at a concentration ranging from 5 to 15 g L−1.5–6 These compounds are non-digestible for human infants but have been found to contribute to the immediate and long-term benefits of breastfeeding, such as protecting breast-fed infants against infections and necrotizing enterocolitis,7–8 improving their intelligence, and reducing rates of childhood obesity, diabetes, leukemia, and suddent infant death syndrome.9 HMOSs display prebiotic properties which enrich beneficial bacteria such as B. infantis in the gut,10 serve as host-cell receptor decoys to disrupt pathogen adhesion,11–12 modulate intestinal surface glycan expression,13 affect cell growth and differentiation,14 and provide nutrients for the development of the brain and cognition of infants.1, 5
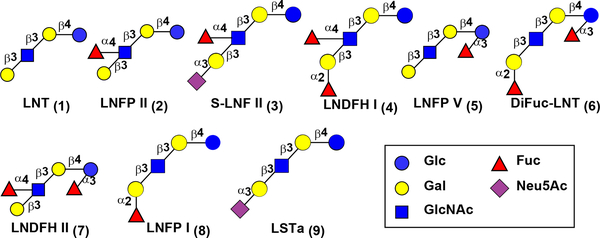
HMOSs consist of galactoside core structures with or without fucosylation and/or sialylation.1 Among the 20 HMOS core structures that have been identified, 11 contain at least one type 1 glycan-terminated branch.1 Lacto-N-tetraose (LNT, Galβ3GlcNAcβ3Lac) is the simplest type 1 glycan HMOS with a disaccharide unit Galβ3GlcNAc β1–3-linked to lactose (Lac, Galβ4Glc), the conserved reducing end disaccharide moiety in all HMOSs.15 LNT (1) and its fucosylated derivatives lacto-N-fucopentaose I (LNFP I, 8), LNFP II (2), lacto-N-difucosylhexaose I (LNDFH I, 4), and LNDFH II (7) (Figure 1) are among the most abundant HMOSs.12, 16 While the type 1 glycan structures predominate in human milk, they are less abundant and sometimes are completely absent in the milk of other mammals.16–17 Investigating the biological functions of individual type 1 glycan-containing HMOSs and their potential applications as prebiotics and antimicrobials requires access to sufficient amounts of structurally defined compounds.
Figure 1.
Structures of HMOSs that are synthesized in this work (1–6) and three LNT-containing HMOSs synthesized previously (7–9).1, 18–20
Chemical synthesis of these compounds21 involves multiple protection and deprotection processes. Though promising, efforts for enzymatic synthesis of β1–3-linked galactoside motifs have met limited success. B. infantis D-galactosyl-β1‒3-N-acetyl-D-hexosamine phosphorylase (BiGalHexNAcP) catalyzes the formation of β1–3-galactosyl linkages to short GlcNAc or GalNAc-terminated acceptor substrates efficiently22 but longer oligosaccharides such as lacto-N-triose (LNT II) were not suitable acceptor substrates.22–23 Synthesis of LNT from LNT II with three equivalents of ortho-nitrophenyl-β-galactoside (GalβoNP) by Bacillus circulans β1‒3-galactosidase (BcBgaC)-catalyzed transglycosylation resulted in only 20% yield.24 Its glycosynthase mutant E233G generated products with the desired Galβ3GlcNAc linkage in higher (59–86%) isolated yields using chemically synthesized α-galactopyranosyl fluoride as the donor substrate.25 Recently an endoglycosynthase strategy was explored using an oxazoline derivative of Galβ1‒3GlcNAc as the donor substrate for Bifidobacterium bifidum JCM 1254 β-D-hexosaminidase mutants to form LNT, achieving around 30% yield with two of the designed mutants.26
Efficient strategies for LNT production would be glycosyltransferase-mediated glycosylation using sequential one-pot multienzyme (OPME) reactions and fermentation,27 both with in situ production of UDP-sugar donors. E. coli β1‒3-galactosyltransferase (EcWbgO) was found to be active in catalyzing the formation of LNT from lacto-N-triose (LNT II).28 Metabolic engineering of E. coli to produce LNT by fermentation resulted in the production of 0.219 g L−1 LNT using 1% glucose as the carbon source,29 while the use of 1% galactose as the carbon source yielded 0.810 g L−1 LNT.30 In each case, EcWbgO was rate limiting, and its acceptor substrate LNT II (the intermediate for the formation of the desired LNT) was accumulated as the major product. The high KM value (3.4 mM) of EcWbgO toward UDP-Gal was believed to hinder its activity.28 Similarly, a strain of E. coli metabolically engineered to produce fucosylated LNT analogs accumulated LNT II intermediate and its α1‒3-fucoside, suggesting insufficient EcWbgO activity.31 Expression of EcWbgO as an N-terminal His6-tagged or a C-terminal His6-tagged fusion protein failed to produce detectable soluble proteins, but adding an N-terminal glutathione S-transferase (GST)-tag yielded 1.6 mg of purified fusion protein per liter culture.28 Cleavage of the GST tag was inefficient and the resulting EcWbgO was inactive. Recently, the expression of EcWbgO with a C-terminal 245-amino acid sequence of S. hyicus lipase pre-propeptide at a level of 54.5 mg per liter was described.32 It has been used for synthesizing linear long-chain LNT using UDP-Gal33–34 but has not been applied in multigram-scale of LNT with in situ generation of UDP-Gal by either OPME synthesis or fermentation. We aimed to identify a new β1‒3-galactosyltransferase with a high expression level and improved activity.
To search for an efficient β1‒3-galactosyltransferase for LNT synthesis, we expected that a suitable candidate could be obtained from homologs of EcWbgO. Clustal Omega multiple sequence alignment35 analysis of the BLAST36 results for EcWbgO homologs led to the selection of four candidates (Figure S1) including Chromobacterium violaceum (Cvβ3GalT), Pectobacterium parmentieri (Ppβ3GalT), Salmonella enterica (Sevβ3GalT), and Yersia intermedia (Yiβ3GalT). Synthetic genes with codon optimization for E. coli expression were cloned into pET-22b(+) vector for expressing C-His6-tagged fusion proteins. Of these four EcWbgO homologs, only Cvβ3GalT was expressed as a soluble protein in E. coli (Figure S2) and was therefore selected for additional characterization.
Cvβ3GalT was active in a broad pH range with optimal activities in the pH range of 6.5–9.5 (Figure S3). This broad pH tolerance is ideal for OPME reaction systems which involve numerous enzymes. Its activity was abolished in the presence of ethylenediaminetetraacetic acid (EDTA) or CaCl2 and enhanced by MgCl2 and MnCl2 (Figure S4). Similar to the conditions for EcWbgO,28 MnCl2 resulted in a higher enzymatic activity for Cvβ3GalT than MgCl2. Product was detected without the addition of a metal ion, and in the presence of LiCl, NaCl, or dithiothreitol (DTT), suggesting that Cvβ3GalT bond MgCl2 or MnCl2 tightly enough that the metal was not completely washed away during Ni2+-column purification.
Apparent kinetic constants for Cvβ3GalT (Table 1) were determined using LNT II-βProNHFmoc as the acceptor and UDP-Gal as the donor. The catalytic efficiency of Cvβ3GalT with LNT II-βProNHFmoc as the acceptor was about 23-fold greater than that of EcWbgO with LNT II as the acceptor,28 which was mainly contributed by a 116-fold higher turnover rate of Cvβ3GalT. Furthermore, the binding affinity of EcWbgO toward its acceptor substrate LNT II is 62-fold greater than toward its donor substrate UDP-Gal,28 which disfavors catalysis as glycosyltransferases follow an ordered sequential Bi-Bi mechanism in which the enzyme binds the nucleotide sugar before the acceptor for an effective catalytic process.37 The property of EcWbgO with a higher affinity toward its acceptor substrate than its donor substrate may explain why EcWbgO performed poorly during metabolic engineering efforts to produce LNT. In contrast, Cvβ3GalT displayed a preference for binding UDP-Gal (Km = 0.23 ± 0.06 mM) over LNT II-βProNHFmoc (Km = 0.74 ± 0.07 mM).
Table 1.
Kinetic Constants for Cvβ3GalT and EcWbgO.
| Enzyme | Substrate | kcat (min−1) | Km (mM) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|
| Cvβ3GalT | LNT II-βProNHFmoc | (2.9±0.1) × 102 | 0.74±0.07 | 3.9 × 102 |
| UDP-Gal | 79±4 | 0.23±0.06 | 3.4 × 102 | |
| GlcNAc | 2.55 | 0.32 | 7.89 | |
| EcWbgOa | LNT II | 2.50 | 5.5 × 10−2 | 16.7 |
| UDP-Gal | 13.90 | 3.4 | 4.09 | |
Reported previously28.
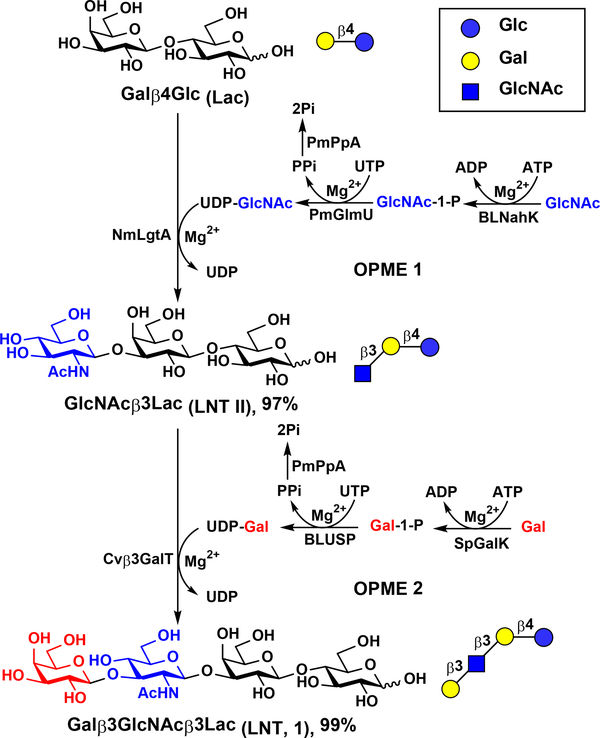
With Cvβ3GalT in hand as an excellent glycosyltransferase, preparative and large-scale syntheses of LNT from commercially available and inexpensive lactose were carried out using a sequential OPME glycosylation process.
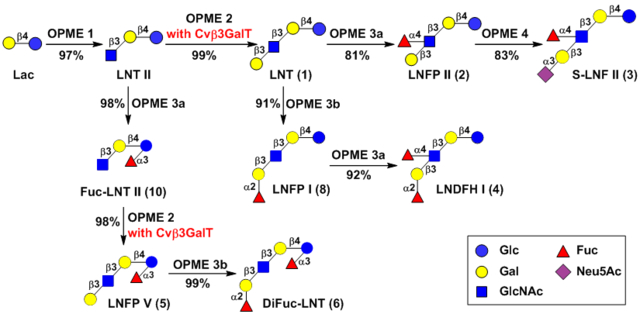
As shown in Scheme 1, trisaccharide GlcNAcβ3Galβ4Glc (LNT II) was initially synthesized in a 1.5-gram scale from lactose and N-acetylglucosamine (GlcNAc) using an OPME N-acetylglucosamine (GlcNAc)-activation and transfer system (OPME 1) containing Bifidobacterium longum strain ATCC55813 N-acetylhexosamine-1-kinase (BLNahK),38 Pasteurella multocida N-acetylglucosamine uridylyltransferase (PmGlmU),39 Pasteurella multocida inorganic pyrophosphatase (PmPpA),40 and Neisseria meningitidis β1–3-N-acetylglucosaminyltransferase (NmLgtA).41 An excellent 97% yield for purified LNT-II was achieved. LNT (Galβ3GlcNAcβ3Galβ4Glc) was then synthesized from LNT II and galactose in a 130-mg preparative scale with an excellent 99% yield using an OPME galactose (Gal)-activation and transfer system (OPME 2) containing Streptococcus pneumoniae TIGR4 galactokinase (SpGalK),42 Bifidobacterium longum UDP-sugar pyrophosphorylase (BLUSP),43 PmPpA, and Cvβ3GalT.
Scheme 1.
Preparative-scale sequential one-pot multienzyme (OPME) synthesis of LNT (1).
The efficiency of the sequential OPME process was further demonstrated in a multigram-scale synthesis of LNT. Trisaccharide LNT II was produced from 10 grams of lactose, GlcNAc (1.15 equiv.), and ATP and UTP (1.28 equiv. each) using the OPME GlcNAc-activation and transfer system. When the reaction reached completion as indicated by the complete consumption of lactose, the reaction mixture was concentrated and applied directly without purification to the subsequent OPME Cvβ3GalT-containing galactosylation reaction with additional amounts of Gal (1.20 equiv.), ATP and UTP (1.30 equiv. each). A total consumption of LNT II intermediate was observed. A portion (1/12.5) of the reaction mixture was subjected to purification by a Dowex 1 × 8–200 (formate form) anion exchange column and a Bio-Gel P-2 gel filtration column to produce pure LNT. A yield of 99.3% was determined for the synthesis of LNT from Lac with two OPME systems carried out in sequence. The sequential OPME process without the purification of LNT II intermediate is a highly effective approach to obtain large quantities of LNT in high yields.
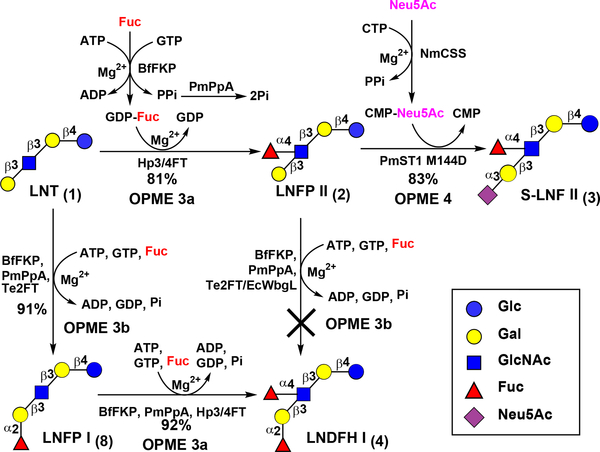
In addition to LNT, the synthesis of fucosylated and/or sialylated HMOSs containing LNT core including LNFP II (2), S-LNF II (3), and LNDFH I (4) (Scheme 2) was explored.
Scheme 2.
Preparative-scale one-pot multienzyme (OPME) synthesis of LNT-containing fucosides (2–4, and 8) from LNT (1).
Lacto-N-fucopentaose II (LNFP II, 2) Galβ3(Fucα4)GlcNAcβ3Galβ4Glc is a pentasaccharide containing a fucose α1–4-linked to the GlcNAc unit in LNT (1). The concentration of LNFP II in human milk was reported to be in the range of 0.14–1.25 g L-1.44 LNFP II was shown to protect human intestinal epithelial cells from E. histolytica-induced cytotoxicity by blocking the binding of the bacterium.45 As shown in Scheme 2, LNFP II (2) was synthesized from LNT using an OPME fucosylation system (OPME 3a) containing a bifunctional Bacteroides fragilis L-fucokinase/GDP-fucose pyrophosphorylase (BfFKP),46 PmPpA, and Helicobacter pylori UA948 α1–3/4-fucosyltransferase (Hp3/4FT).18 When 1.1 equivalents of fucose (Fuc) was used, Hp3/4FT catalyzed the transfer of fucose selectively to the GlcNAc (instead of Glc) residue in LNT to form LNFP II (2) in 81% yield. As we reported previously,18 adding more than two equivalents of Fuc in the reaction system produced difucosylated LNDFH II Galβ3(Fucα4)GlcNAcβ3Galβ4(Fucα3)Glc (7), a hexasaccharide found in human milk at a concentration in the range 0.05 to 0.27 g L-1.44, 47–48 The preference of Hp3/4FT towards fucosylation of GlcNAc-containing disaccharide Galβ3GlcNAcβ motif to the Glc-containing disaccharide Galβ4Glc (Lac) motif in LNT is similar to what was observed previously18 for its preference in the Galβ4GlcNAcβ motif to the Galβ4Glc (Lac) motif in lacto-N-neotetraose (LNnT), a type 2 glycan also found in HMOSs. Sialylfucosyllacto-N-tetraose (S-LNF II, 3) Neu5Acα3Galβ3(Fucα3)GlcNAcβ3Galβ4Glc was subsequently synthesized from LNFP II (2) using an OPME α2–3-sialylation system (OPME 4) containing Neisseria meningitidis CMP-sialic acid synthetase (NmCSS)49 and Pasteurella multocida α2–3-sialyltransferase 1 M144D mutant (PmST1 M144D).50
Lacto-N-difuco-hexaose I (LNDFH I, 4) Fucα2Galβ3(Fucα4)GlcNAcβ3Galβ4Glc, which has both α1–2- and α1–4-linked fucose residues (Scheme 2), has been found in human milk at a concentration of 0.32–1.40 g L-1.44, 47, 51 It presents a terminal Lewis b structure that can bind to H. pylori and has been a candidate for developing potential therapeutics against H. pylori infection.11, 52 Attempts to fucosylate LNFP II (2) using an OPME fucosylation system (OPME 3b) containing Thermosynechococcus elongates α1–2-fucosyltransferase (Te2FT)19 or Escherichia coli O126 α1–2-fucosyltransferase (EcWbgL)53–54 were not successful, indicating that LNFP II (2) was not a suitable acceptor substrate for Te2FT or EcWbgL. Instead, LNDFH I (4) was successfully synthesized by altering the fucosylation sequence of LNT. First, LNT (1) was α1–2-fucosylated at the terminal galactose (Gal) residue using the Te2FT-containing OPME system (OPME 3b) to produce LNFP I Fucα2Galβ3GlcNAcβ3Galβ4Glc (8)19 with an excellent 91% yield. Subsequently, LNFP I was modified with an α1–4-linked fucose at the GlcNAc residue by the Hp3/4FT-containing OPME system (OPME 3a) in excellent (92%) yield. This demonstrated that LNFP I (8) was a suitable acceptor substrate for Hp3/4FT and the GlcNAc was the preferred fucosylation site of Hp3/4FT over the Glc in LNFP I (8).
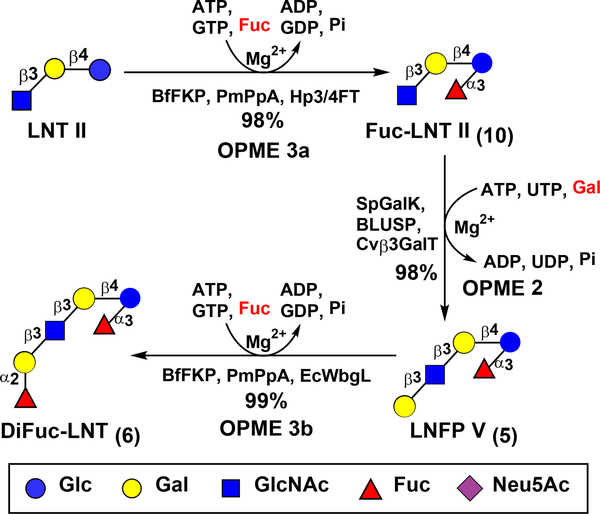
Lacto-N-fucopentaose V (LNFP V, 5) Galβ3GlcNAcβ3Galβ4(Fucα3)Glc (Scheme 3) is another important HMOS. It is a pentasaccharide containing an α1–3-linked fucose at the reducing end glucose (Glc) of LNT. LNFP V, along with other fucosylated HMOSs, was shown to provide protection for infants of secretor mothers from necrotizing enterocolitis (NEC) and sepsis.55 Direct fucosylation of LNT to obtain LNFP V (5) would be challenging due to the lack of a FucT to selectively add Fuc to the Glc of LNT (1). Hence, an alternative approach was carried out. As shown in Scheme 3, LNT II was fucosylated by the Hp3/4FT-containing OPME fucosylation system (OPME 3a) to produce Fuc-LNT II GlcNAcβ3Galβ4(Fucα3)Glc (10) in an excellent 98% yield. The tetrasaccharide was an excellent acceptor for Cvβ3GalT, and LNFP V (5) was synthesized readily in 98% yield using the Cvβ3GalT-containing OPME galactosylation system (OPME 2). LNFP V (5) was found not a suitable acceptor substrate of Te2FT, but a well-tolerated substrate for EcWbgL. Therefore, fucosylation of LNFP V (5) with a Fuc α1–2-linked to the terminal Gal was achieved by EcWbgL-containing OPME system (OPME 3b) for the formation of difucosylated LNT (DiFuc-LNT, 6) Fucα2Galβ3GlcNAcβ3Galβ4(Fucα3)Glc in 99% yield.
Scheme 3.
Preparative-scale sequential OPME synthesis of LNT-containing fucosides (5 and 6) from GlcNAcβ3Galβ4Glc (LNT II).
In conclusion, the newly discovered Cvβ3GalT is a highly efficient catalyst for the synthesis of LNT (1) and LNT-containing fucosylated and/or sialylated HMOSs. LNT (1) was synthesized in a multigram (>10 grams) scale from inexpensive starting materials (lactose, GlcNAc, Gal, ATP, and UTP) using a highly efficient sequential OPME process and without purification of the trisaccharide intermediate LNT II. By designing the order of glycosylation, fucosylated LNT HMOSs with different fucosylation linkages and with fucosylation at different sites were successfully synthesized in high yields. These included monofucosylated HMOSs including LNFP II (2), its sialylated form S-LNF II (3), and LNFP V (5) as well as difucosylated HMOSs such as LNDFH I (4) and its derivative DiFuc-LNT (6). As the Galβ1‒3GlcNAc linkage is common in HMOSs and other human glycans such as sialyl Lewis a and Lewis b, Cvβ3GalT represents a valuable addition to the synthetic glycobiology toolbox. OPME systems are proven again as powerful approaches for synthesizing complex oligosaccharides.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the United States National Institutes of Health (NIH) Common Fund Glycoscience Program grants U01GM120419 and U01GM125288. Bruker Avance-800 NMR spectrometer was funded by NSF grant DBIO-722538.
Footnotes
The authors declare no competing financial interest.
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website at DOI: https://doi.org/10.1021/acscatal.9b03990.
Supplemental figures and experimental details for cloning and characterization of Cvβ3GalT, as well as detailed synthetic procedures, nuclear magnetic resonance (NMR) spectroscopy and high-resolution mass spectrometry (HRMS) data, and NMR spectra of products (PDF)
REFERENCES
- (1).Yu H; Chen X CHAPTER 11: Enzymatic and Chemoenzymatic Synthesis of Human Milk Oligosaccharides (HMOS). in Synthetic Glycomes 2019, pp. 254–280. [Google Scholar]
- (2).Yu RK; Tsai YT; Ariga T; Yanagisawa M Structures, Biosynthesis, and Functions of Gangliosides-An Overview. J. Oleo Sci 2011, 60, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Holgersson J; Lofling J Glycosyltransferases Involved in Type 1 Chain and Lewis Antigen Biosynthesis Exhibit Glycan and Core Chain Specificity. Glycobiology 2006, 16, 584–593. [DOI] [PubMed] [Google Scholar]
- (4).Thurin M; Kieber-Emmons T SA-Lea and Tumor Metastasis: The Old Prediction and Recent Findings. Hybrid Hybridomics 2002, 21, 111–116. [DOI] [PubMed] [Google Scholar]
- (5).Bode L Human Milk Oligosaccharides: Every Baby Needs A Sugar Mama. Glycobiology 2012, 22, 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Rudloff S; Kunz C Milk Oligosaccharides and Metabolism in Infants. Adv. Nutr 2012, 3, 398S–405S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Neu J; Walker WA Necrotizing Enterocolitis. N. Engl. J. Med 2011, 364, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lucas A; Cole TJ Breast Milk and Neonatal Necrotising Enterocolitis. Lancet 1990, 336, 1519–1523. [DOI] [PubMed] [Google Scholar]
- (9).Stuebe A The Risks of Not Breastfeeding for Mothers and Infants. Rev. Obstet. Gynecol 2009, 2, 222–231. [PMC free article] [PubMed] [Google Scholar]
- (10).German JB; Freeman SL; Lebrilla CB; Mills DA Human Milk Oligosaccharides: Evolution, Structures and Bioselectivity as Substrates for Intestinal Bacteria. Nestle Nutr. Workshop Ser. Pediatr. Program 2008, 62, 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gustafsson A; Hultberg A; Sjostrom R; Kacskovics I; Breimer ME; Boren T; Hammarstrom L; Holgersson J Carbohydrate-Dependent Inhibition of Helicobacter pylori Colonization using Porcine Milk. Glycobiology 2006, 16, 1–10. [DOI] [PubMed] [Google Scholar]
- (12).Kunz C; Rudloff S; Baier W; Klein N; Strobel S Oligosaccharides in Human Milk: Structural, Functional, and Metabolic Aspects. Annu Rev Nutr 2000, 20, 699–722. [DOI] [PubMed] [Google Scholar]
- (13).Angeloni S; Ridet JL; Kusy N; Gao H; Crevoisier F; Guinchard S; Kochhar S; Sigrist H; Sprenger N Glycoprofiling with Micro-arrays of Glycoconjugates and Lectins. Glycobiology 2005, 15, 31–41. [DOI] [PubMed] [Google Scholar]
- (14).Kuntz S; Kunz C; Rudloff S Oligosaccharides from Human Milk Induce Growth Arrest via G2/M by Influencing Growth-Related Cell Cycle Genes in Intestinal Epithelial Cells. Br. J. Nutr 2009, 101, 1306–1315. [DOI] [PubMed] [Google Scholar]
- (15).Kobata A Structures and Application of Oligosaccharides in Human Milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci 2010, 86, 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Urashima T; Asakuma S; Leo F; Fukuda K; Messer M; Oftedal OT The Predominance of Type I Oligosaccharides is A Feature Specific to Human Breast Milk. Adv. Nutr 2012, 3, 473s–482s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fushinobu S Conformations of The Type-1 Lacto-N-Biose I Unit in Protein Complex Structures. Acta Crystallogr. F Struct. Biol. Commun 2018, 74, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yu H; Li Y; Wu Z; Li L; Zeng J; Zhao C; Wu Y; Tasnima N; Wang J; Liu H; Gadi MR; Guan W; Wang PG; Chen XH pylori alpha1–3/4-Fucosyltransferase (Hp3/4FT)-Catalyzed One-Pot Multienzyme (OPME) Synthesis of Lewis Antigens and Human Milk Fucosides. Chem. Commun 2017, 53, 11012–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhao C; Wu Y; Yu H; Shah IM; Li Y; Zeng J; Liu B; Mills DA; Chen X The One-Pot Multienzyme (OPME) Synthesis of Human Blood Group H Antigens and A Human Milk Oligosaccharide (HMOS) with Highly Active Thermosynechococcus elongates alpha1–2-Fucosyltransferase. Chem. Commun 2016, 52, 3899–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yu H; Li Y; Zeng J; Thon V; Nguyen DM; Ly T; Kuang HY; Ngo A; Chen X Sequential One-Pot Multienzyme Chemoenzymatic Synthesis of Glycosphingolipid Glycans. J. Org. Chem 2016, 81, 10809–10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Craft KM; Townsend SD Synthesis of Lacto-N-Tetraose. Carbohydr. Res 2017, 440–441, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yu H; Thon V; Lau K; Cai L; Chen Y; Mu S; Li Y; Wang PG; Chen X Highly Efficient Chemoenzymatic Synthesis of beta1–3-Linked Galactosides. Chem. Commun 2010, 46, 7507–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Malekan H; Fung G; Thon V; Khedri Z; Yu H; Qu J; Li Y; Ding L; Lam KS; Chen X One-Pot Multi-Enzyme (OPME) Chemoenzymatic Synthesis of Sialyl-Tn-MUC1 and Sialyl-T-MUC1 Glycopeptides Containing Natural or Non-natural Sialic Acid. Bioorg. Med. Chem 2013, 21, 4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Murata T; Inukai T; Suzuki M; Yamagishi M; Usui AT Facile Enzymatic Conversion of Lactose into Lacto-N-Tetraose and Lacto-N-Neotetraose. Glycoconj. J 1999, 16, 189–195. [DOI] [PubMed] [Google Scholar]
- (25).Henze M; Schmidtke S; Hoffmann N; Steffens H; Pietruszka J; Elling L Combination of Glycosyltransferases and a Glycosynthase in Sequential and One-Pot Reactions for the Synthesis of Type 1 and Type 2 N-Acetyllactosamine Oligomers. ChemCatChem 2015, 7, 3131–3139. [Google Scholar]
- (26).Schmolzer K; Weingarten M; Baldenius K; Nidetzky B Lacto-N-tetraose Synthesis by Wild-Type and Glycosynthase Variants of The beta-N-hexosaminidase from Bifidobacterium bifidum. Org. Biomol. Chem 2019, 17, 5661–5665. [DOI] [PubMed] [Google Scholar]
- (27).Li W; McArthur JB; Chen X Strategies for Chemoenzymatic Synthesis of Carbohydrates. Carbohydr. Res 2019, 472, 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Liu XW; Xia C; Li L; Guan WY; Pettit N; Zhang HC; Chen M; Wang PG Characterization and Synthetic Application of A Novel beta1,3-Galactosyltransferase from Escherichia coli O55:H7. Bioorg. Med. Chem 2009, 17, 4910–4915. [DOI] [PubMed] [Google Scholar]
- (29).Baumgartner F; Conrad J; Sprenger GA; Albermann C Synthesis of The Human Milk Oligosaccharide Lacto-N-Tetraose in Metabolically Engineered, Plasmid-Free E. coli. Chembiochem 2014, 15, 1896–1900. [DOI] [PubMed] [Google Scholar]
- (30).Baumgartner F; Sprenger GA; Albermann C Galactose-Limited Fed-Batch Cultivation of Escherichia coli for The Production of Lacto-N-Tetraose. Enzyme Microb. Technol 2015, 75–76, 37–43. [DOI] [PubMed] [Google Scholar]
- (31).Baumgartner F; Jurzitza L; Conrad J; Beifuss U; Sprenger GA; Albermann C Synthesis of Fucosylated Lacto-N-tetraose Using Whole-Cell Biotransformation. Bioorg. Med. Chem 2015, 23, 6799–6806. [DOI] [PubMed] [Google Scholar]
- (32).Fischoder T; Laaf D; Dey C; Elling L Enzymatic Synthesis of N-Acetyllactosamine (LacNAc) Type 1 Oligomers and Characterization as Multivalent Galectin Ligands. Molecules 2017, 22, 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fischoder T; Cajic S; Reichl U; Rapp E; Elling L Enzymatic Cascade Synthesis Provides Novel Linear Human Milk Oligosaccharides as Reference Standards for xCGE-LIF Based High-Throughput Analysis. Biotechnol. J 2019, 14, e1800305. [DOI] [PubMed] [Google Scholar]
- (34).Fischoder T; Cajic S; Grote V; Heinzler R; Reichl U; Franzreb M; Rapp E; Elling L Enzymatic Cascades for Tailored (13)C6 and (15)N Enriched Human Milk Oligosaccharides. Molecules 2019, 24, 3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Madeira F; Park YM; Lee J; Buso N; Gur T; Madhusoodanan N; Basutkar P; Tivey ARN; Potter SC; Finn RD; Lopez R The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Altschul SF; Gish W; Miller W; Myers EW; Lipman DJ Basic Local Alignment Search Tool. J. Mol. Biol 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- (37).Boix E; Zhang Y; Swaminathan GJ; Brew K; Acharya KR Structural Basis of Ordered Binding of Donor and Acceptor Substrates to The Retaining Glycosyltransferase, alpha-1,3-Galactosyltransferase. J. Biol. Chem 2002, 277, 28310–28318. [DOI] [PubMed] [Google Scholar]
- (38).Li Y; Yu H; Chen Y; Lau K; Cai L; Cao H; Tiwari VK; Qu J; Thon V; Wang PG; Chen X Substrate Promiscuity of N-Acetylhexosamine 1-Kinases. Molecules 2011, 16, 6396–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chen Y; Thon V; Li Y; Yu H; Ding L; Lau K; Qu J; Hie L; Chen X One-Pot Three-Enzyme Synthesis of UDP-GlcNAc Derivatives. Chem. Commun 2011, 47, 10815–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Lau K; Thon V; Yu H; Ding L; Chen Y; Muthana MM; Wong D; Huang R; Chen X Highly Efficient Chemoenzymatic Synthesis of beta1–4-Linked Galactosides with Promiscuous Bacterial beta1–4-Galactosyltransferases. Chem. Commun 2010, 46, 6066–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li Y; Xue M; Sheng X; Yu H; Zeng J; Thon V; Chen Y; Muthana MM; Wang PG; Chen X Donor Substrate Promiscuity of Bacterial beta1–3-N-Acetylglucosaminyltransferases and Acceptor Substrate flexibility of beta1–4-Galactosyltransferases. Bioorg. Med. Chem 2016, 24, 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Chen M; Chen LL; Zou Y; Xue M; Liang M; Jin L; Guan WY; Shen J; Wang W; Wang L; Liu J; Wang PG Wide Sugar Substrate Specificity of Galactokinase from Streptococcus pneumoniae TIGR4. Carbohydr. Res 2011, 346, 2421–2425. [DOI] [PubMed] [Google Scholar]
- (43).Muthana MM; Qu J; Li Y; Zhang L; Yu H; Ding L; Malekan H; Chen X Efficient One-Pot Multienzyme Synthesis of UDP-Sugars Using A Promiscuous UDP-Sugar Pyrophosphorylase from Bifidobacterium longum (BLUSP). Chem. Commun 2012, 48, 2728–2730. [DOI] [PubMed] [Google Scholar]
- (44).Thurl S; Munzert M; Henker J; Boehm G; Muller-Werner B; Jelinek J; Stahl B Variation of Human Milk Oligosaccharides in Relation to Milk Groups and Lactational Periods. Br. J. Nutr 2010, 104, 1261–1271. [DOI] [PubMed] [Google Scholar]
- (45).Bode L; Jantscher-Krenn E Structure-Function Relationships of Human Milk Oligosaccharides. Adv. Nutr 2012, 3, 383s–391s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Yi W; Liu X; Li Y; Li J; Xia C; Zhou G; Zhang W; Zhao W; Chen X; Wang PG Remodeling Bacterial Polysaccharides By Metabolic Pathway Engineering. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chaturvedi P; Warren CD; Altaye M; Morrow AL; Ruiz-Palacios G; Pickering LK; Newburg DS Fucosylated Human Milk Oligosaccharides Vary Between Individuals and Over The Course of Lactation. Glycobiology 2001, 11, 365–372. [DOI] [PubMed] [Google Scholar]
- (48).Kunz C; Rudloff S; Schad W; Braun D Lactose-Derived Oligosaccharides in The Milk of Elephants: Comparison with Human Milk. Br. J. Nutr 1999, 82, 391–399. [DOI] [PubMed] [Google Scholar]
- (49).Yu H; Yu H; Karpel R; Chen X Chemoenzymatic Synthesis of CMP-sialic Acid Derivatives by a One-Pot Two-Enzyme System: Comparison of Substrate Flexibility of Three Microbial CMP-sialic Acid Synthetases. Bioorg. Med. Chem 2004, 12, 6427–6435. [DOI] [PubMed] [Google Scholar]
- (50).Sugiarto G; Lau K; Qu J; Li Y; Lim S; Mu S; Ames JB; Fisher AJ; Chen X A Sialyltransferase Mutant with Decreased Donor Hydrolysis and Reduced Sialidase Activities for Directly Sialylating LewisX. ACS Chem. Biol 2012, 7, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Coppa GV; Pierani P; Zampini L; Carloni I; Carlucci A; Gabrielli O Oligosaccharides in Human Milk During Different Phases of Lactation. Acta Paediatr. Suppl 1999, 88, 89–94. [DOI] [PubMed] [Google Scholar]
- (52).Xu HT; Zhao YF; Lian ZX; Fan BL; Zhao ZH; Yu SY; Dai YP; Wang LL; Niu HL; Li N; Hammarstrom L; Boren T; Sjostrom R Effects of Fucosylated Milk of Goat and Mouse on Helicobacter pylori Binding to Lewis b Antigen. World J. Gastroenterol 2004, 10, 2063–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Yu H; Santra A; Li Y; McArthur JB; Ghosh T; Yang X; Wang PG; Chen X Streamlined Chemoenzymatic Total Synthesis of Prioritized Ganglioside Cancer Antigens. Org. Biomol. Chem 2018, 16, 4076–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Engels L; Elling L WbgL: A Novel Bacterial α1,2-Fucosyltransferase for the Synthesis of 2’-Fucosyllactose. Glycobiology 2014, 24, 170–178. [DOI] [PubMed] [Google Scholar]
- (55).Bering SB Human Milk Oligosaccharides to Prevent Gut Dysfunction and Necrotizing Enterocolitis in Preterm Neonates. Nutrients. 2018, 10, 1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.