Abstract
Psychiatric disorders are associated with accelerated aging and enhanced risk for neurodegenerative disorders. Brain aging is associated with molecular, cellular and structural changes that are robust on the group-level, yet show substantial inter-individual variability. Here we assessed deviations in gene expression from normal age-dependent trajectories, and tested their validity as predictors of risk for major mental illnesses and neurodegenerative disorders. We performed large-scale gene expression and genotype analyses in postmortem samples of two frontal cortical brain regions from 214 control subjects aged 20–90 years. Individual estimates of “molecular age” were derived from age-dependent genes, identified by robust regression analysis. Deviation from chronological age was defined as “delta age”. Genetic variants associated with deviations from normal gene expression patterns were identified by expression quantitative trait loci (cis-eQTL) of age-dependent genes or genome-wide association study (GWAS) on delta age, combined into distinct polygenic risk scores (PRScis-eQTL and PRSGWAS), and tested for predicting brain disorders or pathology in independent postmortem expression datasets and clinical cohorts. In these validation datasets, molecular ages, defined by 68 and 76 age-related genes for two brain regions respectively, were positively correlated with chronological ages (r=0.88/0.91), elevated in bipolar disorder (BP) and schizophrenia (SCZ), and unchanged in major depressive disorder (MDD). Exploratory analyses in independent clinical datasets show that PRSs were associated with SCZ and MDD diagnostics, and with cognition in SCZ and pathology in Alzheimer’s disease (AD). These results suggest that older molecular brain aging is a common feature of severe mental illnesses and neurodegeneration.
Keywords: Aging, molecular age, transcriptome, genetic, cis-eQTL, polygenic risk score
INTRODUCTION
Due to improvements in medical interventions and living conditions, the number of individuals reaching the age of 65 in the United States rose 3-fold in the 20th century, from 4.1 percent in 1900 to 12.4 percent in the year 2000, and may rise above 20% by the middle of this century – roughly 85 million people at current growth rates.1 This increase in life expectancy is paralleled by an increased prevalence of neurodegenerative and neuropsychiatric disorders of late life. Progress has been made in revealing some of the cellular and molecular mechanisms of neurodegenerative diseases, such as Alzheimer’s Disease (AD) and Parkinson’s diseases, and the neurovascular causes of late-life depression and dementia.
“Normal” brain aging and its association with functional changes and late-life brain disorders were long thought to be inescapable, broad-ranging and nonspecific. Recent molecular studies demonstrate that age-dependent molecular changes are conserved, specific and potentially modifiable. Studies by our group2, 3 and others4, 5 have demonstrated that aging of the human brain is associated with continuous and progressive changes in the expression of up to ~10% of all genes, that these changes are conserved across brain regions, and that they affect specific biological pathways, including receptor function, signal transduction, mitochondrial function, cellular defenses, glial/neuronal structure and the extracellular matrix. Moreover, age-dependent genes include numerous genes also associated with brain diseases, wherein age- and disease-related changes in expression are, for the most part, directionally similar. For instance, the expression of brain-derived neurotrophic factor (BDNF) is low in multiple brain disorders, but is also reduced by 50% in healthy individuals aged 65 years or older.6 These parallel trajectories have raised the possibility that deviations from normal age-related gene expression trajectories may contribute to either vulnerability or resilience to various brain-related disorders,7 together providing a compelling rationale for investigating aging and brain disorders simultaneously, and suggesting novel therapeutic approaches. Moreover, people with severe mental illness (particularly SCZ and BP) die at a much younger age, on average, than healthy individuals,8–10 providing further rationale behind looking for mechanisms of accelerated (brain) aging in these illnesses.
We have previously proposed the concept of “molecular age” as an assay to measure biological aging of the brain and to assess individual deviation from chronological age.2, 3 Molecular age is a summary value that describes the predicted chronological age for a specific individual based on regression analysis of expected age-related trajectories for age-dependent gene expression. Other groups have devised similar approaches using either epigenetic marks measured in blood11 or brain structural magnetic resonance imaging,12, 13 with similar results showing progressive, specific and predictive changes. In this report, individual deviations between molecular and chronological ages, defined as delta age, provided a means to identify biological mediators and genetic modulators of normal aging and potentially brain disorders. Specifically, we predicted that biological aging of the brain, i.e. molecular age, would be robust, predictive of chronological age, and significantly elevated in subjects with major mental illnesses, as measured in postmortem brain samples. To extend these studies to patient cohorts, we predicted that common genetic variants associated with changes in either single age-dependent gene expression or with integrated delta age measures would show greater prevalence in psychiatric or neurodegenerative disorders.
To address these questions, we performed large-scale gene expression analyses in two related frontal cortex brain regions, combined with genetic analyses, in a large human postmortem cohort (Figure 1). We applied robust statistical approaches to define age-dependent genes and individual molecular ages in one brain region and used the second region from the same subjects to confirm results. We then identified single nucleotide polymorphisms (SNP) associated with expression changes in age-dependent genes or with delta age, using expression quantitative trait loci (cis-eQTL) or genome-wide association (GWA) methods, respectively, and combined results into polygenic risk scores (PRS). Finally, we used molecular ages and PRS to test our predictions in independent postmortem expression datasets and clinical cohorts (Table 1 and Supplemental Table 1).
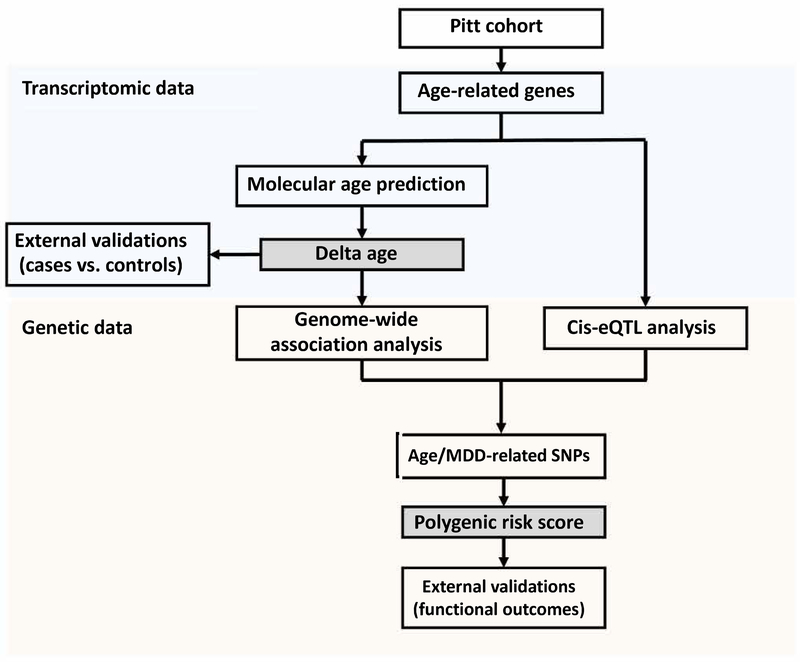
Figure 1. Study flow chart.
Firstly, transcriptomic data from the Pitt cohort (BA47 and BA11) were used to identify age-related genes. A molecular age prediction model was constructed by using top age-related genes. “Delta age” was defined as the difference between molecular and chronological ages, which served as a proxy measure of molecular brain aging. We then tested the robustness of the findings in external cohorts. Secondly, genetic data from the European subjects in same cohorts in was used to identify SNPs associated with brain aging. Two approaches (Genome-wide association and Cis-eQTL analyses) were used to narrow down the potential SNP lists. Polygenic risk scores were defined for each subject as a cumulative score measuring how much genetic risk a subject carries toward premature brain aging. Finally, our proposed risk scores were validated against clinical diagnoses and related functional outcomes.
Table 1.
Cohorts.
| Sample size | Age range | ||||||
|---|---|---|---|---|---|---|---|
| Cohort | Name | Type | Data | Control | Case | 5% | 95% |
| Pitt (cross-validated) | Pitt-aging | Postmortem | Transcriptomics + Genetics | 204 | 0 | 22.0 | 72.9 |
| Pitt-MDD MD2_ACC_F | Pitt-MDD1 | Postmortem | Transcriptomics | 13 | 13 | 26.5 | 64.8 |
| Pitt-MDD MD2_DLPFC_F | Pitt-MDD2 | Postmortem | Transcriptomics | 16 | 16 | 27.1 | 68.6 |
| Pitt-MDD MD2_DLPFC_M | Pitt-MDD3 | Postmortem | Transcriptomics | 14 | 14 | 28.2 | 61.7 |
| Pitt-SCZ PC L5 | Pitt-SCZ1 | Postmortem | Transcriptomics | 32 | 32 | 25.3 | 64.7 |
| Pitt-SCZ MO1 L5 | Pitt-SCZ2 | Postmortem | Transcriptomics | 16 | 16 | 37.7 | 63.4 |
| Lieber SCZ | Lieber | Postmortem + Clinical | Transcriptomics + Genetics | 99 | 88 | 17.2 | 70.3 |
| CommonMind SCZ MSSM | CMMD-SCZ1 | Postmortem + Clinical | Transcriptomics + Genetics | 115 | 85 | 42.0 | 90.0 |
| CommonMind SCZ Pitt | CMMD-SCZ2 | Postmortem + Clinical | Transcriptomics + Genetics | 73 | 36 | 24.4 | 68.6 |
| CommonMind BP MSSM | CMMD-BP1 | Postmortem + Clinical | Transcriptomics + Genetics | 115 | 11 | 41.3 | 90.0 |
| CommonMind BP Pitt | CMMD-BP2 | Postmortem + Clinical | Transcriptomics + Genetics | 73 | 31 | 24.0 | 66.9 |
| ROS-MAP AD | ROSMAP | Postmortem + Clinical | Genetics | 384 | 251 | 76.9 | 90.0 |
| Health ABC | Health ABC | Clinical | Genetics | 1794 | 0 | NA (> 65) | |
| IRL-Grey+HIP fracture studies | IRL | Clinical | Genetics | 254 | 0 | NA (> 65) | |
Sample sizes reflect sample sizes after quality control evaluation.
RESULTS
Age-dependent gene expression and molecular age predictive validity
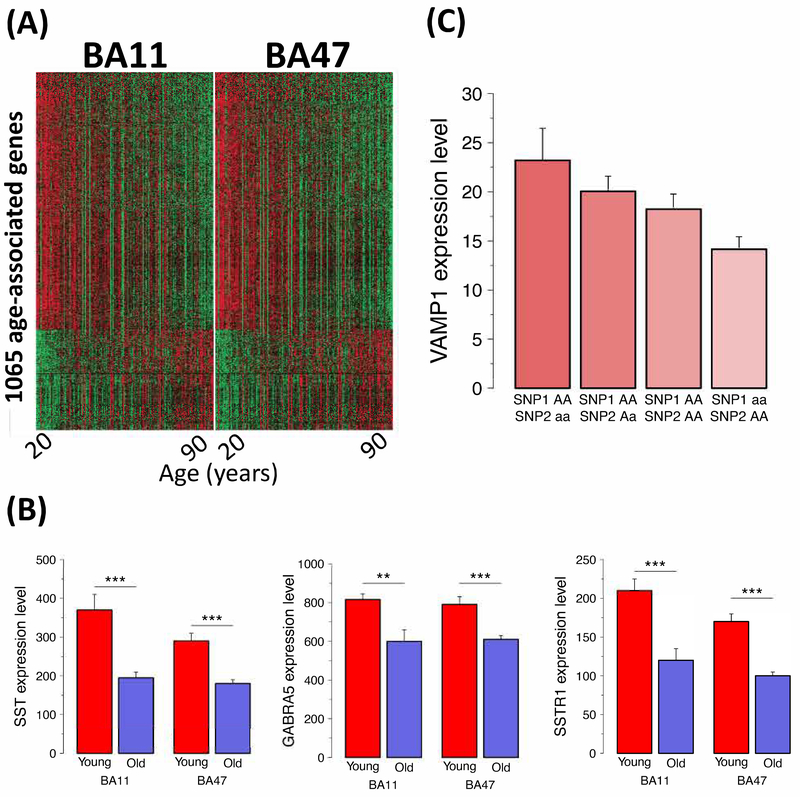
To identify age-related genes, we fitted a robust regression model14 to gene expression data obtained in two closely related regions of the frontal cortex [Brodmann areas (BA) 11 and 47] in a large human postmortem cohort (n=204 after QC, see Supplements; 20–90 year range with even distribution across decades; Supplemental Table 2).15 The model was conducted independently for each gene and each of the two brain regions, taking into account brain and population heterogeneity (see Methods). Supplemental Table 3 reports the number of detected age-associated genes under different levels of false discovery rate (FDR) control in each brain region and their intersections. For instance, at FDR=10−4, 1065 genes were identified as age-dependent in both regions (Figure 2A), being up- or downregulated in older individuals. An analysis of biological pathways represented by these genes revealed high consistency with existing knowledge of brain aging: ~40 of the top 50 affected pathways related to altered synaptic structure (synaptogenesis, axonal guidance) and function, including glutamatergic, GABAergic and multiple neuropeptide (opioid, BDNF, CRH) signaling pathways, and Calcium signaling (Supplemental Table 4). For independent validation of expression changes, we selected three genes that are downregulated with age and that belong to a GABAergic signaling pathway known to be affected by age, i.e. somatostatin (SST), SST receptor 1 (SSTR1) and alpha5-containing GABA-A receptor (GABRA5),6, 16 and confirmed age-dependent changes using qPCR on independent RNA samples (Figure 2B).
Figure 2.
A. Heatmap for 1065 age-associated genes in the Pitt cohort in BA11 and BA47. Red to green describes high to low expression. B. Microarray data validation by qPCR. Robust downregulation of three components of an integrated GABA-related signaling unit with age. Expressions are in arbitrary units. Two-sided p-values are reported. (*, p<0.05; **, p<0.01; ***, p<0.001). C. Cis-eQTL validation by qPCR for VAMP1. The correlation between microarray data and qPCR was 0.73. For validation of VAMP1 cis-eQTL, joint effect between two SNPs (rs2534717 as SNP1 and rs12580729 as SNP2) has been evaluated. There was an observed decrease in VAMP1 expression for subjects carrying homozygous minor alleles from SNP1 in comparison to subjects carrying homozygous major alleles. There was also an observed increase in VAMP1 expression for subjects carrying homozygous alleles from SNP2 in comparison to subjects carrying homozygous major alleles. The correlation between microarray data and qPCR was 0.73 (p<0.001) although two-sample t-tests indicate differences only at trend levels (p<0.1).
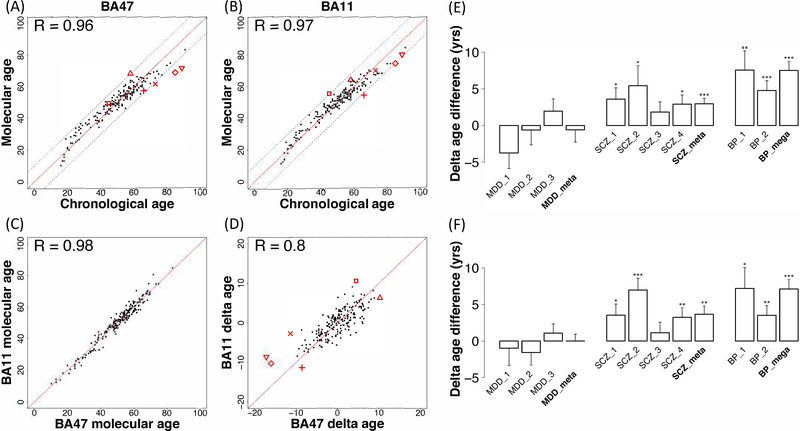
We next defined the molecular ages of individual subjects. We constructed a prediction model using an elastic net approach, based on expression levels of the 1065 age-dependent genes (FDR=10−4; Figure 2A). The derived molecular ages were highly correlated with chronological ages (r=0.96 for BA47 and r=0.97 for BA11, p<10−16 for both regions; Figure 3A–B) and consistent across the two regions (Figure 3C; r=0.98; p<10−16). Accordingly, the differences between molecular age and chronological age, denoted “delta age”, were also correlated across regions (Figure 3D; r=0.80, p<10−16). Similar results were obtained when assessing relative or proportional differences (Supplemental Figure 1; r=0.81; p<10−16). Although the FDR=10−4 identified an approximate number of age-related genes2, 3, 4, 5 (~10%), we also performed a sensitivity analysis to assess the impact of different q-value cut-off for age-related genes included in the molecular age prediction, and determined that it had minimum impact on the results (Supplemental Figures 2–3 and Supplemental Table 5; Supplemental Methods).
Figure 3.
Delta age may index an older or younger brain at the molecular level compared to chronological age. Scatter plot and Pearson’s correlation (r) between chronological and predicted molecular ages in BA47 (A; r=0.96) and BA11 (B; r=0.97). C. Scatter plot and Pearson’s correlation of molecular ages between BA11 and BA47 (r=0.98). D. Scatter plot and Pearson’s correlation of delta ages between BA11 and BA47 (r=0.8). The red symbols indicate brains of distinct subjects with large deviations in molecular level compared to chronological age (absolute delta age greater than 10 in either brain region). Bar plot for delta age comparisons (E: BA47 and F: BA11) in cases versus controls in three brain disorders (MDD, SCZ and BP). MDD_1 to MDD_3 stand for three MDD cohorts from Pittsburgh cohort; SCZ_1 to SCZ_4 are four SCZ studies (two from Pittsburgh cohort, one from Lieber cohort, and one from CommonMind cohort); BP_1 and BP_2 are two studies from CommonMind cohort. See Supplemental Methods and Supplemental Table 7 for detailed naming correspondence. Y-axis is the delta age mean difference in cases over controls in each study plus/minus one standard deviation. *, 0.01–0.05; **, 0.01–0.001; ***, <0.001. Meta-analysis was further applied to summarize results across different studies in MDD and SCZ. Mega-analysis was performed to directly combine two BP studies since both of them are from CommonMind cohorts.
Gene expression-based brain molecular ages in brain disorders
We next hypothesized that delta age may index an older or younger brain at the molecular level compared to chronological age (e.g. red symbols in Figure 3D) and that these deviations may be behaviorally and clinically meaningful. To test this hypothesis, we collected independent postmortem large-scale gene expression studies from control subjects and subjects with MDD, SCZ, BP and AD (Table 1 and Supplemental Table 1; Supplemental Methods for cohort details). Using an elastic net-based approach, 68 and 76 genes with the strongest predictive values were selected from 1065 age-related genes in BA47 and BA11 respectively, and used for the molecular age prediction model (See gene list and weights in Supplemental Table 6).
Since some of the cohorts were not initially designed for assessing progressive age effects across the whole lifespan (e.g., small sample size or uneven distribution across ages), we first tested the potential of the gene panel to predict chronological age using control subjects in each cohort. Of the 24 independent transcriptomic studies we collected (Supplemental Table 7), 17 studies had control sample size less than 40 (category A), 2 studies had large sample size but a very old and narrow age distribution (>95% samples > 65 years; Category B) and 5 studies had large sample size and adequate age distribution (Category C). Successful validation of molecular age prediction was obtained in control samples of all 5 category C studies (i.e. r>0.7 between molecular and chronological ages; Supplemental Table 7), 5 out of 17 category A studies and none of the two category B studies. Molecular ages and delta ages were then calculated using the case subjects of the ten successful validation studies and applying the two separate BA47 and BA11 molecular age prediction models, and compared to control subjects.
Results are depicted in Figure 3E–F. Delta ages of the three MDD cohorts did not significantly differ from chronological ages of control subjects. Three of the five SCZ cohorts showed significantly positive delta ages compared to control subjects using the BA47 model (Supplemental Table 8 - Pitt-SCZ PC L5: p=0.014, 3.59±1.56 years older; Pitt-SCZ MO1 L5: p=0.033, 5.42±2.74 years older), and CommonMind SCZ Pitt (p<3×10−4, 6.34±1.73 years older). Results in the other two SCZ cohorts were similar, but only at trend levels (Lieber SCZ: p=0.096, 1.83±1.40 years older; CommonMind SCZ MSSM: p=0.078, 2.20±1.55 years older). The two BP cohorts showed greatest magnitude of effects using the same BA47 model (CommonMind BP MSSM: p<0.006, 7.56±2.62 years older; CommonMind BP Pitt: p<2×10−4, 4.76±1.36 years older). The results were then summarized across cohorts by meta-analysis using a random effects model for the MDD and SCZ cohorts, and by mega-analysis for BP by directly combining CommonMind MSSM and CommonMind Pitt. These analyses confirmed the absence of molecular age deviation in MDD (−0.59±1.67 years; p, NS) and the older molecular ages for SCZ (2.94±0.77 years older; p<2×10−4) and BP (7.52±1.26 years older; p<4×10−8) patients, compared to control subjects. Almost identical results were obtained using the prediction model derived from the BA11 expression dataset (Supplemental Table 8 and Figure 3F), and similar results were obtained when applying both models to proportional (%), rather than absolute values for delta ages (Supplemental Table 9).
In summary, gene expression-based prediction of molecular brain age is robust and can detect individual deviations from chronological ages. At the group level, SCZ and BD, but not MDD subjects, showed significantly older brain molecular ages compared to control subjects. Results could not be obtained in AD due to the narrow age range of those cohorts.
Genetic-based prediction of clinical diagnostics and functional outcomes
We next sought to construct a SNP-based polygenic risk score (PRS) predictive of delta age in the original expression dataset, and investigate whether PRSs are predictive of disease status or selected functional and pathological measures in independent clinical cohorts (hence bypassing underpowered effects in single cohorts). Diagnostics were available for SCZ, MDD and AD, and functional measures were available for age-related locomotor activity and various aspects of cognition (see cohorts details in Supplemental Methods and functional measures in Supplemental Table 10).
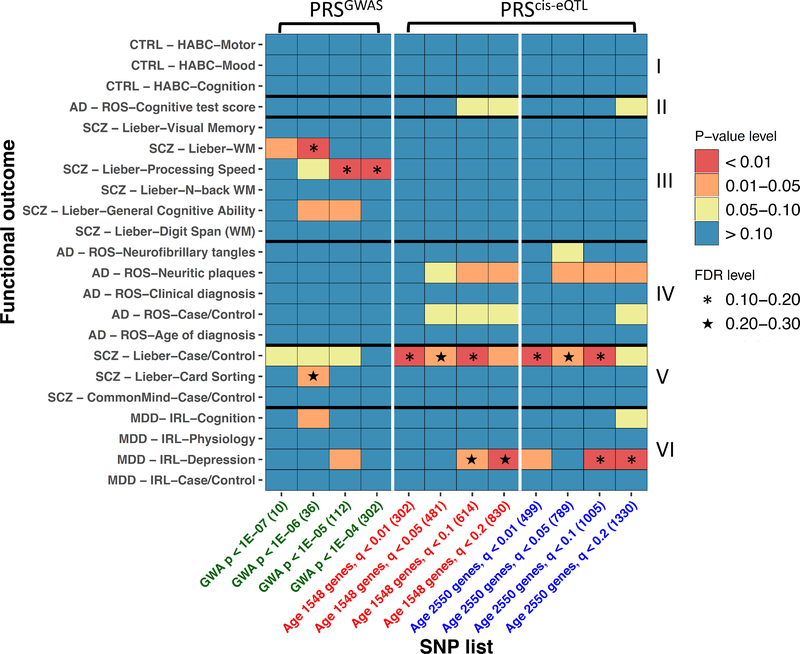
We first used a genome-wide association (GWA) analysis to identify SNPs associated with delta ages independently in BA11 or BA47, as calculated in the previous section using elastic net (with leave-one-out-cross-validation for prediction, see Supplements for details). To account for population admixture, we used the top five principal components as covariates in the regression model (Supplemental Figure 4). The BA11 and BA47 GWA p-values were then meta-analyzed across the two brain regions by adaptively weighted Fisher17 (AW-Fisher; see Supplemental Methods for details) and SNPs were identified under different statistical thresholds (302 SNPs under p<10−4, 112 SNPs under p<10−5, 36 SNPs under p<10−6 and 10 SNPs under p<10−7). See Supplemental Figure 5 for Manhattan plot. We note that although these p-values were overly liberal since they were obtained in two related brain regions, the ranks of these SNP sets are invariant. We therefore tested these associations in independent cohorts in which genetic data was available (Table 1 and Supplemental Table 1) by extracting these SNPs, aggregated them with equal weight to generate four PRSGWAS scores at increasing stringency levels, and assessed for correlations with phenotypes. By controlling for false discovery rate (FDR)=0.2, 3 q-values<0.2 are observed in PRSGWAS (Figure 4) in SCZ Lieber study.
Figure 4.
Heatmap for polygenic risk score associations with clinical diagnostics and functional outcomes in external cohorts (IRL-Grey, ROSMAP, Health ABC, and Lieber center). (A) P-values; (B) FDR. The columns refer to different SNP lists used for calculating polygenic risk scores. The rows refer to different diagnostic or functional outcomes. PRSGWAS scores were associated with SCZ diagnosis and with three aspects of cognitive functions in the Lieber cohort. Rows are classified into broad domains (I) Measures in control cohort, (II) Cognition in AD, (III) Cognition in SCZ, (IV) AD-related diagnosis and pathologies, (V) SCZ diagnostic, and (VI) MDD-related diagnosis and others. PRScis-eQTL scores were associated with late-life depression and SCZ, and marginally with AD. PRScis-eQTL scores were also associated with the presence of neuritic plaques in AD. Only marginal associations were observed with aspects of cognition.
As a complementary approach we applied a cis-expression quantitative trait locus (cis-eQTL) analysis for prioritization of presumably functional variants. For this, we first selected genes relevant to aging in the original gene expression analyses (Supplemental Table 3: 1548 genes at q<10−3; 2550 genes at q<10−2). In addition to genetic PCs, we performed transcriptomic PCA and applied the top PCs to adjust in the regression model to reduce the false positive findings in cis-eQTL analysis 18 (details in Supplemental Methods and Supplemental Figure 6). Cis-eQTL SNPs were identified for those genes in both brain regions, and q-values were derived after AW-Fisher meta-analysis using thresholds with decreasing stringency (q<1%, 5%, 10% and 20%). See Figure 2C and Nikolova et al 19 for qPCR validation of cis-eQTL effects. The cis-eQTL SNPs were then extracted from the respective cohorts, aggregated with equal weight to generate PRScis-eQTL scores at increasing stringency levels for age-related gene and cis-eQTL selection, resulting in 8 PRScis-eQTL scores (Figure 4). The rationale for this approach was that consistent findings across various stringency levels (as shown in Figure 4 Heatmap graphs) would provide a degree of confidence in the results. Assessing the PRScis-eQTL scores for correlations with clinical diagnostic categories revealed significant associations with late-life depression (MDD-IRL cohort) and SCZ (Lieber cohort) with FDR < 0.2, and a marginal association with AD (Figure 4). PRScis-eQTL scores were marginally but concordantly (across different SNP sets) associated with the presence of neuritic plaques in AD. Only marginal associations were observed with aspects of cognition. Overall, 41 comparisons out of 264 (i.e. 12 PRSs by 22 functional outcome) resulted in p-values<0.1 (26.4 false positives expected by chance). Note that the two PRS approaches were not redundant to each other. Indeed, only one SNP overlapped between PRSGWAS (302 SNPs) and PRScis-eQTL (1330 SNPs). This lack of overlap was expected since these two approaches are based on distinct hypotheses and index different biological constructs.
DISCUSSION
In this study, we characterized transcriptional and genetic aspects of molecular aging in the frontal cortex of adults across the adult lifespan. Confirming and extending prior work, we identified a large number of age-related genes showing robust linear effects. Based on these age-dependent genes, we defined the molecular age of individual subjects and confirmed a high correlation with chronological age. We then defined delta age values, based on how far each subject’s molecular age deviates from their chronological age, and confirmed the results in a closely-related brain region in the same cohort. We then validated the functional significance of this metric in several independent cohorts, where we found elevated delta ages in individuals with SCZ as well as BP disorder, suggesting older molecular brain ages in individuals with these severe mental illnesses, compared to chronological age-matched controls. Further demonstrating the functional significance of this construct, we found that two sets of distinct SNPs associated with older molecular age via two distinct mechanisms (GWAS and cis-eQTL) were associated with clinically relevant diagnostic, cognitive and neurological variables across several cohorts. Taken together, these results identify novel genetic contributions of brain aging and provide new insight into age-related brain disorders.
The current findings confirm prior work showing robust and specific age-dependent changes in gene expression2–5 and extend them to a wider age range and additional brain regions. Using age-dependent gene expression trajectories, we defined a novel metric of molecular brain age and a complementary measure, denoted “delta age”, reflecting the discrepancy between this metric and chronological age. Notably, we found that molecular brain age was 2–6 years higher than chronological age in individuals with SCZ across several independent cohorts, while the molecular brain age of subjects with BD was between 4.7 and 7.5 years greater than chronological age across cohorts. No increase in brain aging was noted in subjects with MDD. A limitation of these studies is that correction for several factors was not performed (e.g. psychiatric medications, disease chronicity) since that information was not available for most cohorts. These results are partially consistent with prior work using metrics of neuroanatomical brain age assessed by in vivo structural MRI. Specifically, one prior study found that structural brain age in participants with SCZ was on average 5.5 years older than their chronological age, while BP was associated with a 3.1 year brain age increase 12, with a more recent large-scale study adjusting these estimates to 3.9 and 2 years, respectively20. Notably, the former study also found that the brain age gap was proportional to illness chronicity across disorders. Potential effects of chronicity are also supported by a more recent study, which used similar methods to demonstrate that brain age in individuals with first-episode SCZ was only ~2.6 years older than their chronological age, while early-stage BP was no different from controls.13 The wide range of molecular brain age estimates across SCZ cohorts in our study may reflect differences in pre-mortem disease chronicity, however this information was not available in the majority of the postmortem cohorts. Nonetheless, the overlapping brain age gaps obtained by our current molecular results and prior structural MRI findings in SCZ suggests that the two metrics may reflect related or complementary biological processes and, together, support the notion that SCZ is associated with premature or, possibly, accelerated brain aging.
The gap between molecular brain age and chronological age estimates that we obtained for BD were substantially higher than equivalent metrics reported in structural MRI studies. It is possible that the current cohort had high levels of chronicity, relative to those used in structural MRI studies, which may have driven this discrepancy. Alternatively, it is possible that molecular brain age changes precede macroscale structural brain change detectable by MRI (i.e., molecular brain age may on average be older than structural brain age, particularly in BD). Future studies completing the difficult but not impossible challenge of obtaining structural MRI and postmortem molecular gene expression data in the same individuals would be necessary to test the validity of this conjecture.
Intriguingly, we did not find evidence of elevated molecular brain age in MDD. This result stands in contrast to prior work, which found that MRI-assessed structural brain age was approximately 4 years higher than chronological age in individuals with depression,12 but it is consistent with a more recent study, which found a non-significant increase of only 0.8 years in structural brain age in MDD20. This discrepancy may again be driven by confounding effects of disease chronicity, which strongly drove the structural brain age effect in Koutsouleris et al,12 but were not accounted for by Kaufman or in the current study. It is also possible that the brain age effect sizes in MDD are much smaller or that the disease heterogeneity is greater than those seen in other disorders (e.g., SCZ, BD), thus limiting our power to detect a significant effect.
Our results are interesting to consider in light of well-established, non-overlapping metrics of molecular aging, which rely on age-dependent changes in DNA methylation patterns to estimate tissue-specific epigenetic age. The majority of studies on DNA methylation age use the “epigenetic clock” developed by Horvath et al,11 which estimates epigenetic age based on 353 CpG sites across the entire epigenome. These studies have found no evidence of elevated epigenetic age, relative to chronological age in SCZ, either peripherally or in the brain.21–23 Similarly, another study found no elevated epigenetic age in BD, in the blood or brain.24 However, the latter study did identify a slightly elevated epigenetic age in blood in older, but not younger individuals, once again suggesting the potential confounding effect of chronicity. This effect did not emerge in the brain.24 In contrast, our findings of significantly and robustly elevated molecular brain age in both SCZ and BD suggest that the two metrics likely capture distinct biological processes. A recent study found no effect of MDD on Horvath’s epigenetic clock in brain tissue.25 However, another study combined a larger sample size with sequencing across the entire epigenome to obtain a more detailed estimate of epigenetic age in MDD.26 This study found a small but reliable effect, wherein MDD was associated with 7.68 months older methylation age in peripheral blood and 1.1 years older brain methylation age. This study confirms that accelerated aging effects in MDD are likely small in size; therefore, increased power and precision of estimates obtained through a combination of larger sample sizes and more detailed sampling on the molecular level may be necessary to detect them reliably.
Using an alternate approach to assess epigenetic changes occurring in age-dependent genes, we recently showed in a subset of the present postmortem cohort that epigenetic changes preferentially occur in age-dependent genes,27 hence potentially driving, or at least correlating with molecular brain aging. We further showed that epigenetic changes in age-dependent genes were significantly enriched in subjects with MDD, SCZ, BP and AD, together pointing to a potential mechanism of age-by-disease interactions through preferential age-related methylation changes occurring in disease-associated genes.27
Previous work28, 29 has shown that brain aging is at least partially under genetic control. To confirm this link and probe the potential clinical utility of our novel molecular age construct, we identified SNPs associated with brain aging, through two distinct molecular mechanisms (GWAS and cis-eQTL), aggregated them into two sets of polygenic risk scores (PRSGWAS and PRSciseQTL), and mapped them onto age-related cognitive and clinical outcome variables across several independent cohorts. Several distinct and overlapping phenotypic patterns emerged. Both PRS were higher in individuals with SCZ, suggesting that the difference between molecular and chronological age identified in individuals with SCZ may be at least partially driven by common DNA variations. PRSGWAS was uniquely associated with lower processing speed and lower general cognition, independently of diagnosis, in one of the cohorts. The association with reduced processing speed is in partial agreement with prior proof-of-principle work from our group, showing that genetic variants shifting the expression of an age-dependent gene towards a more old-age-like state are associated with psychomotor slowing even in healthy young adults.19 Finally, PRScis-eQTL was uniquely associated with late-life depression and, marginally, with AD diagnosis. Intriguingly, while there was no link between PRSciseQTL and cognition, PRSciseQTL was associated with neuritic plaques, which are a risk factor for cognitive impairment, particularly in the context of aging.
Although promising, these genetic effects should be interpreted with caution. As a limitation, our sample was relatively underpowered to detect SNPs with significant functional impact. In addition, as our PRS analyses were meant to be exploratory, we did not apply correction for multiple comparisons across the different phenotypic measures and cohorts. While the suggestive effects we identified provide encouraging initial evidence that genetic variants can be used as peripheral indices of molecular brain age, these complex polygenic associations will need to be replicated in future work before they can be used for clinical prediction. For instance, we are planning on testing the predictive values of the various PRSs in the Psychiatric Genomic Consortia for MDD, BPD and SCZ, with a focus on the specific clinical and demographic parameters of the various cohorts included into the consortia databases to ensure adequate power to detect age-related PRS effects. If further refined, the proposed genotype-based predictive model may identify molecular targets to reduce brain aging as a cause or consequence of morbidity in severe mental illness. The model could also provide a screening tool to identify individuals at high risk for accelerated brain aging. Such a tool may offer opportunities for targeted early lifestyle and medical interventions to prevent further age-related decline and offset the risk of age-dependent neuropsychiatric illness in vulnerable groups.
METHODS
Detailed methods are available in the supplements
Human postmortem cohort
Postmortem brains were collected from the Allegheny County Coroner’s Office (Pittsburgh, PA). A committee of experienced clinical scientists examined clinical records, toxicology results and standardized psychological autopsy data. The absence of psychiatric DSM-IV diagnosis was determined by psychopathology, medical and social histories, and history of substance abuse. Individuals were also screened for the absence of neurodegenerative disorders by neuropathological examination. See details in Supplemental Table 2 and Supplements.
Gene expression
Total RNA was extracted from frozen samples and processed for microarray by the Gene Expression & Genotyping Core Facility at Case Western Reserve University. cDNA was hybridized on Affymetrix® Human Gene 1.1 ST arrays (Affymetrix, Santa Clara, CA), covering over 30,000 coding transcripts. Gene expression values were extracted via Expression Console build 1.2.1.20 using RMA method and quantile-normalization to eliminate potential batch effect (Supplemental Figure 7 and 8).
Genetic variants
Total DNA was extracted from fresh frozen brain samples and hybridized onto Genome-Wide Human SNP array 6.0 featuring 909,622 SNPs (Affymetrix, Santa Clara, CA). Genotype calls were generated using Affymetrix Genotyping Console version 4.1.3.
qPCR validation
Subjects were randomly chosen from young (17–38 years old) and old (58–72 years old) age distribution and group-matched for PMI, pH, RNA ratio and RIN. qPCR was performed as previously 30.
Age-related genes detection
A robust regression model 14 was applied to identify age-related genes in a large class of monotonic increasing or monotonic decreasing trajectories. P-values were assessed by fitting the regression model for each gene. Multiple comparisons were corrected by false discovery rate using Benjamini-Hochberg procedure 31.
Molecular age prediction model
The identified age-related genes were combined using elastic net regression, which performed regularization and gene selection. We treated chronological age as a dependent variable and age-related genes and other covariates as independent variables to adjust for potential confounding effects. This method embedded a variable selection procedure that picked up an optimal set of genes to predict chronological ages. To allow the model to automatically select a reasonable number of age-related genes, we started with all 1065 age-related genes (q<10−4 in both brain regions). We defined molecular age as the predicted age from the elastic net model. To avoid over-fitting, we performed a leave-one-out cross-validation (LOOCV) to predict the molecular age for each subject. Delta age was defined as the difference of molecular age and chronological ages.
See Supplements for Genome-wide association (GWA) analysis for delta age and Expression quantitative trait loci (eQTL) analysis of age-related genes
Polygenic risk score
The SNPs were coded as 0, 1, and 2 according to the number of minor alleles. PRSs were defined for each subject by summarizing across multiple SNPs. For GWA, we assigned a weight (1 or −1) to each SNP depending on the sign of GWA effect, and calculated a weighted sum as the PRS. For the cis-eQTL approach, the weight for each SNP was the product of the sign of the cis-eQTL effect and the sign of the age-related gene effect. A weight product equal to 1 indicates older age and −1 indicates younger age.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (MH093723 to E.S. and NIH R01CA190766 for C.L. and G.C.T.), the Campbell Family Mental Health Research Institute (to E.S.). YSN is supported by a NARSAD Young Investigator Award from the Brain & Behavior Research Foundation, a Koerner New Scientist Award, and a Paul Garfinkel Catalyst Award administered by the CAMH Foundation. We thank Kurt Lohman and Yongmei Liu for help with Health ABC analyses.
Data generated as part of the CommonMind Consortium were supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881, AG02219, AG05138, MH06692, R01MH110921, R01MH109677, R01MH109897, U01MH103392, and contract HHSN271201300031C through IRP NIMH. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories, and the NIMH Human Brain Collection Core. CMC Leadership: Panos Roussos, Joseph Buxbaum, Andrew Chess, Schahram Akbarian, Vahram Haroutunian (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Enrico Domenici (University of Trento), Mette A. Peters, Solveig Sieberts (Sage Bionetworks), Thomas Lehner, Stefano Marenco, Barbara K. Lipska (NIMH).
ROSMAP study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute.
The Health ABC study was supported by National Institute on Aging (NIA) Contracts N01-AG-6–2101; N01-AG-6–2103; N01-AG-6–2106; NIA grant R01-AG028050, and NINR grant R01-NR012459. This research was also funded in part by the Intramural Research Program of the NIH, National Institute on Aging and R01 AG028288.
Footnotes
DISCLOSURE. The authors declare no competing interests.
REFERENCES
- 1.Vaupel JW. Biodemography of human ageing. Nature 2010; 464(7288): 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry 2005; 57(5): 549–558. [DOI] [PubMed] [Google Scholar]
- 3.Glorioso C, Oh S, Douillard GG, Sibille E. Brain molecular aging, promotion of neurological disease and modulation by sirtuin 5 longevity gene polymorphism. Neurobiol Dis 2011; 41(2): 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J et al. Gene regulation and DNA damage in the ageing human brain. Nature 2004; 429(6994): 883–891. [DOI] [PubMed] [Google Scholar]
- 5.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A 2008; 105(40): 15605–15610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh H, Lewis DA, Sibille E. The Role of BDNF in Age-Dependent Changes of Excitatory and Inhibitory Synaptic Markers in the Human Prefrontal Cortex. Neuropsychopharmacology 2016; 41(13): 3080–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibille E Molecular aging of the brain, neuroplasticity, and vulnerability to depression and other brain-related disorders. Dialogues Clin Neurosci 2013; 15(1): 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldessarini RJ. Epidemiology of suicide: recent developments. Epidemiol Psychiatr Sci 2019: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laursen TM, Nordentoft M, Mortensen PB. Excess early mortality in schizophrenia. Annu Rev Clin Psychol 2014; 10: 425–448. [DOI] [PubMed] [Google Scholar]
- 10.Gilman SE, Sucha E, Kingsbury M, Horton NJ, Murphy JM, Colman I. Depression and mortality in a longitudinal study: 1952–2011. CMAJ 2017; 189(42): E1304–E1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013; 14(10): R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koutsouleris N, Davatzikos C, Borgwardt S, Gaser C, Bottlender R, Frodl T et al. Accelerated brain aging in schizophrenia and beyond: a neuroanatomical marker of psychiatric disorders. Schizophr Bull 2014; 40(5): 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajek T, Franke K, Kolenic M, Capkova J, Matejka M, Propper L et al. Brain Age in Early Stages of Bipolar Disorders or Schizophrenia. Schizophr Bull 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yohai VJ. High Breakdown-Point and High Efficiency Robust Estimates for Regression. The Annals of Statistics 1987; 15: 642–656. [Google Scholar]
- 15.Chen CY, Logan RW, Ma T, Lewis DA, Tseng GC, Sibille E et al. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc Natl Acad Sci U S A 2016; 113(1): 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lissemore JI, Bhandari A, Mulsant BH, Lenze EJ, Reynolds CF, Karp JF et al. Reduced GABAergic cortical inhibition in aging and depression. Neuropsychopharmacology 2018; 43(11): 2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Tseng GC. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. Ann Appl Stat 2011; 5, NO. 2A: 994–1019. [Google Scholar]
- 18.Liang L, Morar N, Dixon AL, Lathrop GM, Abecasis GR, Moffatt MF et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res 2013; 23(4): 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolova YS, Iruku SP, Lin CW, Conley ED, Puralewski R, French B et al. FRAS1-related extracellular matrix 3 (FREM3) single-nucleotide polymorphism effects on gene expression, amygdala reactivity and perceptual processing speed: An accelerated aging pathway of depression risk. Front Psychol 2015; 6: 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufmann T, van der Meer D, Doan NT, Schwarz E, Lund MJ, Agartz I et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat Neurosci 2019; 22(10): 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA. DNA methylation evidence against the accelerated aging hypothesis of schizophrenia. NPJ Schizophr 2017; 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA. DNA methylation age is not accelerated in brain or blood of subjects with schizophrenia. Schizophr Res 2018; 196: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voisey J, Lawford BR, Morris CP, Wockner LF, Noble EP, Young RM et al. Epigenetic analysis confirms no accelerated brain aging in schizophrenia. NPJ Schizophr 2017; 3(1): 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fries GR, Bauer IE, Scaini G, Wu MJ, Kazimi IF, Valvassori SS et al. Accelerated epigenetic aging and mitochondrial DNA copy number in bipolar disorder. Transl Psychiatry 2017; 7(12): 1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, He Y, Ma X, Chen X. Epigenetic age analysis of brain in major depressive disorder. Psychiatry Res 2018; 269: 621–624. [DOI] [PubMed] [Google Scholar]
- 26.Han LKM, Aghajani M, Clark SL, Chan RF, Hattab MW, Shabalin AA et al. Epigenetic Aging in Major Depressive Disorder. Am J Psychiatry 2018; 175(8): 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney BC, Lin CW, Rahman T, Oh H, Lewis DA, Tseng G et al. DNA methylation in the human frontal cortex reveals a putative mechanism for age-by-disease interactions. Transl Psychiatry 2019; 9(1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revelas M, Thalamuthu A, Oldmeadow C, Evans TJ, Armstrong NJ, Kwok JB et al. Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech Ageing Dev 2018; 175: 24–34. [DOI] [PubMed] [Google Scholar]
- 29.Glorioso C, Sibille E. Between destiny and disease: genetics and molecular pathways of human central nervous system aging. Prog Neurobiol 2011; 93(2): 165–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla R, Prevot TD, French L, Isserlin R, Rocco BR, Banasr M et al. The Relative Contributions of Cell-Dependent Cortical Microcircuit Aging to Cognition and Anxiety. Biol Psychiatry 2019; 85(3): 257–267. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Yosef H. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B 1995; 57(1): 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.