Abstract
This study aims to evaluate the association of maternal DNA methylation (DNAm) during pregnancy and offspring birthweight. 122 newborn-mother dyads from the Isle of Wight (IOW) cohort were studied to identify differentially methylated cytosine-phosphate-guanine sites (CpGs) in maternal blood associated with offspring birthweight. Peripheral blood samples were drawn from mothers at 22–38 weeks of pregnancy for epigenome-wide DNAm assessment using the Illumina Infinium HumanMethylation450K array. Candidate CpGs were identified using a course of 100 repetitions of a training-and-testing process with robust regressions. CpGs were considered informative if they showed statistical significance in at least 80% of training and testing samples. Linear mixed models adjusting for covariates were applied to further assess the selected CpGs. The Swedish Born Into Life cohort was used to replicate our findings (n=33).
Eight candidate CpGs corresponding to the genes LMF1, KIF9, KLHL18, DAB1, VAX2, CD207, SCT, SCYL2, DEPDC4, NECAP1, and SFRS3 in mothers were identified as statistically significantly associated with their children’s birthweight in the IOW cohort and confirmed by linear mixed models after adjusting for covariates. Of these, in the replication cohort, three CpGs (cg01816814, cg23153661, and cg17722033 with p-values= 0.06, 0.175, and 0.166, respectively) associated with four genes (LMF1, VAX2, CD207, and NECAP1) were marginally significant. Biological pathway analyses of three of the genes revealed cellular processes such as endocytosis (possibly sustaining an adequate maternal-fetal interface), and metabolic processes such as regulation of lipoprotein lipase activity (involved in providing substrates for the developing fetus). Our results contribute to an epigenetic understanding of maternal involvement in offspring birthweight. Measuring DNAm levels of maternal CpGs may in the future serve as a diagnostic tool recognizing mothers at risk for pregnancies ending with altered birthweights.
Keywords: DNA Methylation, Epigenetics, Epigenome-Wide Association Study, Pregnancy, Birthweight
Introduction
Birthweight is an important indicator of pregnancy outcome and neonatal health. Both low and high birthweight categories have been linked to adverse health outcomes.[1–4] Maternal factors such as cigarette smoking, hyperglycemia, and hypertension have been documented to affect the offspring birthweight.[5] It has been suggested that effects of maternal factors during pregnancy on birthweight are exerted through differential methylation of DNA of offspring exposed to the intrauterine environment.[6] A meta-analysis of epigenome-wide studies on birthweight and neonatal DNA methylation (DNAm) reported that 914 cytosine-phosphate-guanine sites (CpGs) are differentially methylated in association with birthweight.[5] However, due to concurrent measurements of birthweight and DNAm, it remains unclear whether differentially methylated sites represent effects of lower birthweight or risk factors for differences in birthweight.
No study in the literature has yet assessed the associations of maternal DNAm in pregnancy and offspring birthweight. Maternal DNAm may influence birthweight of the offspring by impacting expression of the maternal genes that are important for fetal development. A recent study that investigated DNAm at age 18 years to early pregnancy showed that changes in maternal DNAm may be located on genes belonging to signaling pathways controlling uterus and trophoblast interaction such as cell adhesion and cell division.[7] Considering the critical role of in-utero environment in fetal development, we postulated that differential maternal DNAm during gestation could affect the offspring’s growth and birthweight. In this study, we investigated the association of maternal DNAm during pregnancy and offspring birthweight. In the discovery step, we identified differentially methylated CpGs related to birthweight, using the 3rd generation Isle of Wight (IOW) cohort. Then, in an independent mother-offspring cohort, the Swedish Born Into Life cohort, we tested whether we could replicate our findings. In addition, to assess if maternal DNAm affects the gestational age, not the birthweight, we tested the potential mediating role of gestational age between maternal DNAm and birthweight.
Materials and Methods
Study population
IOW cohort
The IOW cohort was established in 1989 in UK to investigate the natural history of allergic disorders and asthma.[8] The IOW study was approved repeatedly by the local research ethics committee (NRES Committee South Central – Hampshire B, U.K.) and the University of Memphis Institutional Review Board in Memphis, U.S. (FWA00006815). Written consents were obtained from all participants at recruitment and all follow-ups. The IOW cohort consists of three generations (F0-parents of the original birth cohort, F1- birth cohort members, and F2-offspring of F1). The current analysis focuses on adult female IOW study participants (F1) and their offspring (F2). The F2 generation is comprised of 542 newborns from 331 mothers. Excluding subjects with missing data on birthweight, gender, or maternal DNAm, a total of 122 newborns of 114 mothers (8 mothers with 2 newborns) remained for analysis.
Peripheral blood samples were drawn from F1 participants during the first (8–21 weeks) and the second (22–38 weeks) halves of pregnancy for epigenome-wide DNAm assessment. After delivery, umbilical cord blood samples were collected from F2 participants. Maternal anthropometrics (weight, height, BMI) were measured early in pregnancy. Information on F1 such as parity, smoking during pregnancy, and socioeconomic status was ascertained by questionnaires. Smoking during pregnancy was classified as none, light (between 1 and 9 cigarettes per day), and moderate smoking (at or above 10 cigarettes per day). Socioeconomic status is a composite variable derived from three indicators: the British socioeconomic classes based on parental occupation (1–6), number of children in the index child’s bedroom, and family income.[9] The derived variable was categorized into three levels: low, medium, and high, respectively. The proportions of maternal blood cell-types (neutrophil, eosinophil, B cell, CD4+ T cell, natural killer cell) were estimated using the “minfi” R packages [10] based on reference values of cell-type-specific CpGs. [11] Newborns’ gender, gestational age, and birthweight were transcribed from hospital records.
Born into life cohort
The Born into Life cohort focuses on effects of maternal factors and early biomarkers during pregnancy on child’s growth and health outcomes later in life in Sweden. This study is described in a previous publication [12]. The data contribution from this study consisted of 33 women with available DNAm data during late pregnancy (weeks 26–28). Ethical approval was obtained by the Regional Ethics Review Board in Stockholm, Sweden, and written consents were obtained from each subject in the Born into Life cohort. Maternal peripheral blood samples were collected for epigenome-wide DNAm assessment at gestational weeks 26 to 28.
DNAm profiling
DNA from maternal peripheral blood and umbilical cord blood was extracted by a standard salting out procedure.[13] Using the EZ 96-DNA methylation kit (Zymo Research, CA, USA), about 1 μg of DNA was bisulfite-treated for conversion of cytosine to thymine according to the manufacturer’s standard protocol in all samples. DNAm was assessed by the Illumina Infinium HumanMethylation450 Beadchip (Illumina, Inc., CA, USA). A standard protocol was used to process arrays.[14] To control for batch effects, samples were randomly allocated on microarrays. The beadchips were scanned by a BeadStation. We used Bioconductor packages IMA [15] and ComBat [16] for preprocessing the methylation data and removal of batch effect, respectively. Methylation level (β value) was determined for each CpG locus using the Methylation module of BeadStudio software. Beta values indicate the proportions of methylated over the sum of methylated and unmethylated sites and c as a constant to prevent dividing by zero. The M-value was calculated as the log2 ratio of the intensities of methylated sites versus unmethylated sites.[17]
In the replication cohort, DNAm was assessed from peripheral whole blood using DNA extracted at the Karolinska Institutet Biobank. Per sample, an aliquot of DNA (500 ng) underwent bisulfite conversion using the EZ-96 DNA Methylation kit (Zymo Research Corporation, Irvine, USA). Samples were randomized and plated onto 96-well plates and processed with the MethylationEPIC BeadChip, using the standard protocol from the manufacturer (Illumina Inc., San Diego, USA) at the Mutation Analysis Facility, Karolinska Institutet (www.maf.ki.se). This chip measures 866,836 CpG sites across the genome. Methylation data was processed using GenomeStudio Software. The ComBat package was used to remove batch effects.
Statistical analysis
Discovery phase (IOW cohort)
To conduct an epigenome-wide association study (EWAS) identifying maternal CpGs associated with offspring birthweight, the ttScreening R package (v1.5, http://cran.r-project.org/web/packages/ttScreening/) [18] was used. This method removes non-informative CpGs in a course of 100 repetitions of a training-and-testing process with robust regressions. CpGs were considered informative if they showed statistical significance in at least 80% of training and testing samples (selection probability ≥80%). The ttScreening method has the advantage of detecting more truly positives than conventional EWAS methods.[18] For maternal DNAm levels in ttScreening, we used M values due to having a higher power in detecting highly methylated and unmethylated CpG sites.[17] Following the screening, to assess the association of birthweight with methylation of the candidate CpGs, linear mixed models (in SAS 9.4) were applied to adjust for repeated measurements of maternal DNAm for the eight mothers with two offspring. In these models, we also adjusted for potential confounders (newborn’s gender, maternal peripheral blood cell-types, maternal BMI, smoking during pregnancy, and socioeconomic status). Since gestational age was correlated with birthweight (r=0.56, p-value <.0001) and may act as mediator, it was not used as a confounder in the regression models. However, we ran structural equation modeling in SAS 9.4 to examine the relationship between maternal DNAm at the candidate CpGs, birthweight, and gestational age. To assess the variation of DNAm levels of a CpG from age 18 to early and late pregnancy, we used linear mixed models, with DNAm levels as outcome and time as predictor. Multiple testings in all analyses, including EWAS and associations of informative CpGs with birthweight, were adjusted by applying the false discovery rate (FDR) method.[19] An FDR adjusted p-value of <= 0.05 was considered statistically significant.
Replication phase (Born Into Life cohort)
Linear regression models were used to assess the association of candidate CpGs with birthweight in the Born Into Life cohort. Models were adjusted for newborns’ gender. Additional factors such as parity and maternal age at delivery were not significant in the model, hence, not included in the analysis. Further, none of the 33 mothers from this study smoked during pregnancy.
Biological Pathway analysis
Following statistical testing, the study focused on biological pathways to explain the function of the identified CpGs. Function enrichment analyses were conducted using the genes of the discovered CpGs provided in the methylation label file (Infinium MethylationEPIC v1.0 B4 Manifest File). To identify genes linked to the CpGs for which the manifest did not provide any associated gene name, we used SNIPPER (https://csg.sph.umich.edu/boehnke/snipper/) [20] and the University of California Santa Cruz (UCSC) Genome Browser (https://genome.ucsc.edu/) [21]. The chromosome number and map info of the CpGs were queried (using Human GRCh37/hg19) and the nearest genes (maximum distance=250000 SNPs) to the site of the CpG were selected (up to two genes). Once all gene names were obtained, the full list was entered into Toppfun (https://toppgene.cchmc.org/) [22] to identify biological pathways related to these genes. Significant pathways adjusted for multiple testing (false discovery rate p-value <0.05) are presented.
Results
Study characteristics
From the total 542 mother-newborn dyads of IOW, 122 were included in the study. There was no significant difference between characteristics of the population analyzed and the whole F2-IOW cohort subjects (Table 1). Birthweight was normally distributed ranging from 1850 to 4450 grams. Among the 122 newborns, 20 and 19 had birthweights less than 3000 grams and more than 4000 grams, respectively.
Table 1.
Characteristics of Isle of Wight and Born Into Life cohorts comparing included and not included subjects
| F2-Generation of Isle of Wight Cohort (n=542) | Born Into Life Cohort (n= 107) | |||||
|---|---|---|---|---|---|---|
| Analyzed (n=122) | Total (n=542) | P value | Analyzed (n=33) | Total (n=107) | P value | |
| Newborns | 122 | 542 | _ | 33 | 107 | _ |
| Mothers | 114 | 331 | 33 | 107 | ||
| Newborn’s gender | ||||||
| Male | 52% | 55% | 0.7 | 33% | 57% | 0.5 |
| Female | 48% | 45% | 67% | 43% | ||
| Newborn’s birthweight (grams) | 3453.6±491.5 | 3400.0 (710.0) | 0.1 | 3525.0 ± 560.5 | 3531.3 ± 497.8 | 0.9 |
| Maternal age (year) | 23.8±2.4 | 24.3 (5.3) | 0.1 | 32.8±3.8 | 32.5±3.7 | 0.74 |
| Maternal BMI | 25.2 (6.6) | 25.2 (9.0) | 0.8 | 23.1 (3.6) | 23.0 (3.3) | 0.8 |
| Maternal parity | ||||||
| 1 | 62% | 55% | 0.7 | 73% | 66% | 0.8 |
| 2 | 25% | 31% | 27% | 27% | ||
| 3 | 11% | 11% | 5% | 0.8 | ||
| 4 | 2% | 3% | 2% | |||
| Maternal smoking during | 0 | 0 | - | |||
| pregnancy | ||||||
| No smoking | 61% | 64% | 0.7 | |||
| Light smoking | 19% | 16% | ||||
| Moderate smoking | 20% | 20% | ||||
| Socioeconomic status | ||||||
| Low | 17% | 19% | 0.8 | 7% | 9% | 0.7 |
| Medium | 71% | 68% | 89% | 88% | ||
| High | 12% | 13% | 4% | 3% | ||
Data showed as mean± standard deviation for normally distributed and median (interquartile range) for non-normally distributed variables.
The sub-sample from the Born Into Life cohort consisted of 33 mother-newborn dyads. There was no significant difference between the population analyzed and the whole Born into Life cohort in terms of characteristics (Table 1). The average birthweight was 3525 grams.
EWAS (IOW cohort)
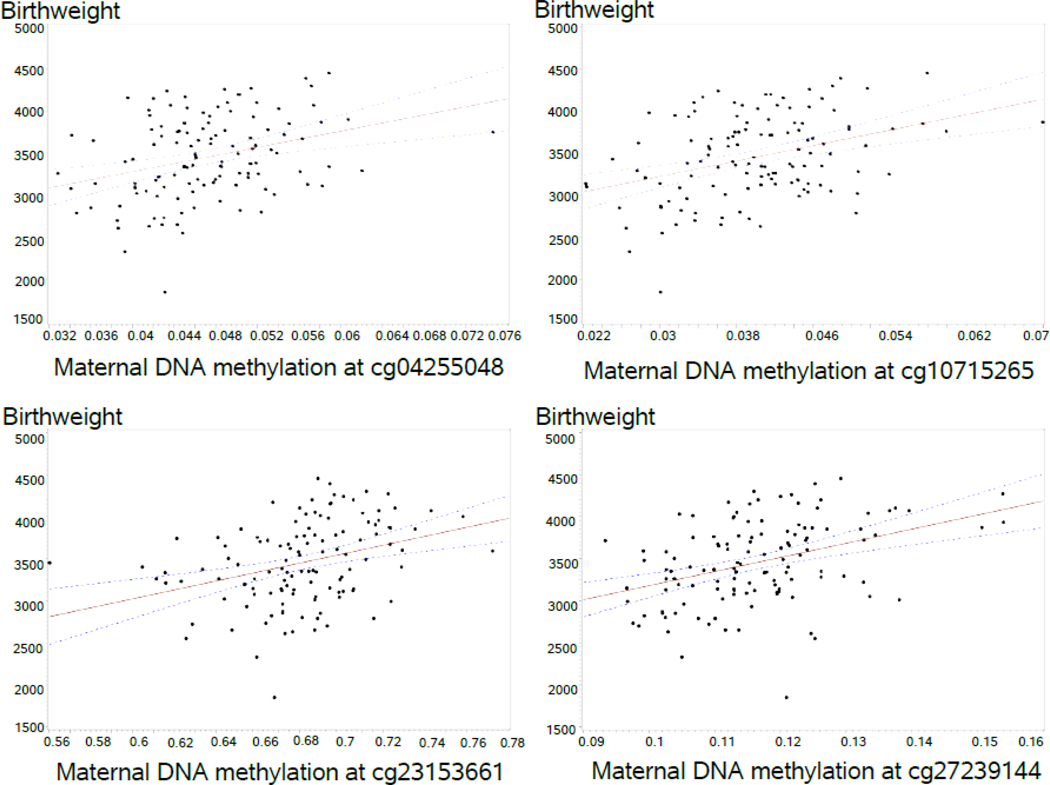
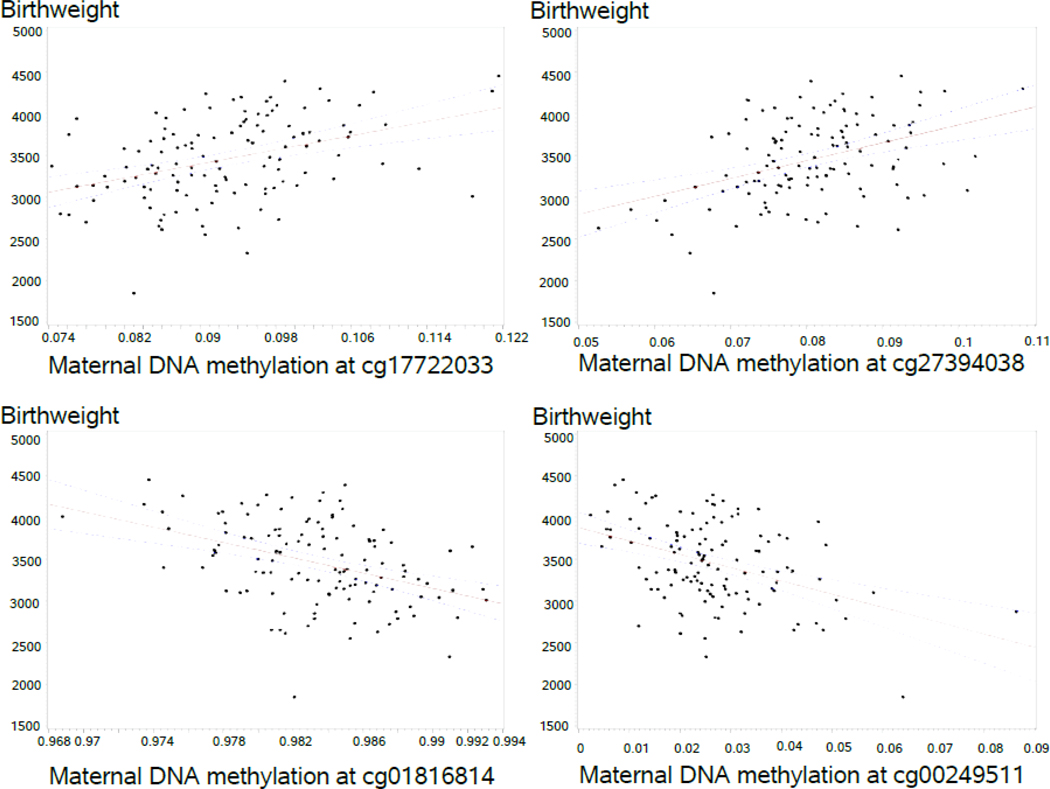
The screening for identifying CpG sites with statistically significant differential DNAm related to birthweight using training and testing datasets yielded 283 CpGs, eight of which had a selection probability of 80% or higher (Table 2). These eight CpGs remained significantly related to birthweight after adjusting for newborn’s gender, maternal peripheral blood cell types, maternal BMI, smoking during pregnancy, socioeconomic status, and multiple births. Among the eight CpGs, two were negatively and six were positively associated with offspring birthweight, respectively (figure 1). Structural equation modeling showed maternal DNAm to affect birthweight both directly and indirectly through gestational age in all candidate CpGs except for cg23153661 and cg 27394038. The latter two CpGs only directly affected birthweight and did not show any significant indirect effect on birthweight through gestational age (supplementary figure 1).
Table 2.
Candidate CpGs in the IOW cohort (n=122) and their replication in the Born Into Life cohort (n=33)
| IOW cohort |
Born into life cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ttscreening | Mixed model | ||||||||
| CpGs | Gene names | Selection Probability | Estimate# M values | P-value | FDR adjusted p-value | Estimate* β values | P-value* | Estimate⸸ β values | P-value⸸ |
| cg01816814 | LMF1 | 89% | −0.00034 | 4.68 E-06 | 0.0004 | −45843 | <.0001 | −38027 | 0.06 |
| cg04255048 | KIF9; KLHL18 | 85% | 0.00015 | 8.83 E-05 | 0.0007 | 25719 | 0.0003 | 23602 | 0.27 |
| cg10715265 | DAB1 | 84% | 0.00025 | 8.18 E-06 | 0.0004 | 23208 | <.0001 | 8991 | 0.39 |
| cg23153661 | VAX2; CD207 | 82% | 0.00016 | 2.50 E-05 | 0.0006 | 6090.19 | <.0001 | 3291 | 0.175 |
| cg00249511 | SCT | 81% | −0.00067 | 4.16 E-06 | 0.0004 | −15925 | <.0001 | −494 | 0.92 |
| cg27239144 | SCYL2; DEPDC4 | 81% | 0.00011 | 7.28 E-05 | 0.0007 | 14176 | 0.0006 | −2247 | 0.79 |
| cg17722033 | NECAP1 | 80% | 0.00012 | 1.85 E-05 | 0.0006 | 23160 | <.0001 | 11840 | 0.166 |
| cg27394038 | SFRS3 | 80% | 0.00015 | 6.37 E-06 | 0.0004 | 19565 | 0.0001 | 6326 | 0.57 |
Controlled for newborns’ gender and maternal peripheral blood cell proportions (CD4, neutrophils, eosinophils, B-lymphocytes, natural killer cells, monocytes)
Adjusted for newborns’ gender, maternal peripheral blood cell proportions (CD4, neutrophils, eosinophils, B-lymphocytes, natural killer cells, monocytes), maternal smoking during pregnancy, socioeconomic status, and repeated measurements in linear mixed models
Adjusted for newborns’ gender
Fig.1.
DNA methylation levels of statistically significant maternal candidate CpGs (ß values) in association with offspring birthweight (grams).
Legend: Solid lines and dashed lines represent regression lines and 95% confidence interval, respectively.
In addition, there was no correlation between maternal and cord blood DNAm levels of candidate CpGs, except for cg10715265, associated with DAB1 gene (r= −0.23, p-value=0.01). Cord blood DNAm levels of the eight candidate CpGs were not associated with birthweight.
The DNAm levels of three CpGs were correlated in early and late pregnancy (cg00249511, rs (Spearman’s rank correlation coefficient) =0.25, p=0.04; cg04255048, rs=0.28, p=0.02; cg23153661, rs=0.29, p=0.02). Comparing the trend of DNAm at the eight candidate CpGs from age 18 to early and to late pregnancy using linear mixed models revealed no time-effect after cell-type adjustment.
Replication (Born-Into-Life cohort)
The directions of the estimates (association of maternal DNAm with offspring birthweight) of the candidate CpGs replicated in the Born Into Life cohort were concordant with those in the IOW cohort, except for one CPG (cg27239144). Of the eight candidate CpGs, three were marginally significant (p≤0.20, n=33) in the replication step (cg01816814, cg23153661, and cg17722033 with p-values= 0.06, 0.175, and 0.166, respectively).
Biological pathway analysis
The eight CpGs identified in the discovery phase were located on or close to 11 genes. Using ToppFun, biological processes related to cellular transport (import into cell, endocytosis, receptor mediated endocytosis), membrane invagination, and forebrain development were identified. Functional analysis of the genes associated with the three CpGs that were marginally significant in the replication, showed enriched biological processes related to endocytosis, protein glycosylation in Golgi, lipoprotein lipase activity positive regulation, and dorsal/ventral axis specification (Table 3).
Table 3.
Biological processes involving gene or gene clusters associated with CpGs identified in IOW and Born Into Life cohorts (replication phase)
| Gene/ gene clusters | Pathways | IDs | FDR adjusted P value |
|---|---|---|---|
| LMF1, CD207, NECAP1 | endocytosis | GO:0006897 | 0.013 |
| membrane invagination | GO:0010324 | 0.013 | |
| vesicle budding from membrane | GO:0006900 | 0.013 | |
| import into cell | GO:0098657 | 0.013 | |
| vesicle organization | GO:0016050 | 0.031 | |
| LMF1, CD207 | receptor-mediated endocytosis | GO:0006898 | 0.029 |
| LMF1 | protein glycosylation in Golgi | GO:0033578 | 0.027 |
| triglyceride-rich lipoprotein particle clearance | GO:0071830 | 0.029 | |
| chylomicron remnant clearance | GO:0034382 | 0.029 | |
| positive regulation of lipoprotein lipase activity | GO:0051006 | 0.029 | |
| positive regulation of triglyceride lipase activity | GO:0061365 | 0.029 | |
| VAX2 | dorsal/ventral axis specification | GO:0009950 | 0.03 |
Discussion
In DNAm measured in maternal blood during gestation, we found eight statistically significant CpGs to be associated with offspring birthweight located on or close to 11 genes. The smaller sample size of the Born Into Life cohort limited the replication and only three CpGs were marginally significant, corresponding to four genes LMF1, NECAP1, VAX2, and CD207.
Biological pathway analysis of three of the four genes enriched processes of endocytosis, membrane invagination, and vesicle organization.
Endocytosis in pregnancy is related to two processes, namely, maternal immune regulation and autophagy. Dendritic cells in uterine mucosa express C-type lectin receptors (CLRs) such as Langerin (encoded by CD207), binding to cell surface carbohydrates, mediating their uptake via endocytosis in the first step of antigen presentation to the immune system.[23–25] CLRs, In addition to function as pathogen recognition receptors, mediate identification of carbohydrate structures on self-glycoproteins promoting self-antigens tolerance.[26] Dendritic cells of the uterine mucosa have been implied in pregnancy maintenance by establishing maternal immune tolerance of fetal tissues.[27] Achieving immune tolerance towards the fetus is required for creating a favorable maternal-fetal interface serving the growth and development of the fetus.[28]
Endocytosis is also involved in autophagy, a lysosome-mediated process maintaining cellular homeostasis by degrading useless or destructive intracellular materials.[29, 30] Autophagy has been implicated in uterovascular changes in pregnancy such as implantation and placentation.[31] Autophagy exists in endometrial stromal cells and epithelial cells and helps constitution of fetal-maternal interface by maintaining cellular homeostasis under physiological or pathological stress. [32] By sustaining a stable fetal-maternal interface, autophagy may be involved in fetal growth. In fact, increased autophagy have been reported in placentas obtained from pregnancies complicated by intra-uterine growth retardation.[30]
In addition to biological pathway analysis of gene clusters, we found potential linkages between three individual genes and pregnancy and fetal growth. The CpG cg01816814 associated with LMF1 gene nearly gained statistical significance in the replication (P value=0.06). LMF1 is an endoplasmic reticulum chaperone required for the post-translational activation of vascular lipases including lipoprotein lipase (LPL), hepatic lipase, and endothelial lipase.[33] The implication of maternal LMF1 gene for offspring birthweight can be explained by its crucial role in regulation of maternal plasma lipids which are required substrates for fetal growth. [34] In the third trimester, the activity of maternal adipose tissue lipoprotein lipase decreases and maternal fat depots break down as a result of increased lipolysis and lipid mobilization.[35] This change in maternal LPL activity leads to physiological maternal hypertriglyceridemia in late pregnancy.[35, 36] Maternal supply of triglycerides have been shown to be correlated with fetal lipid levels and fetal growth.[37] Triglycerides do not cross the placenta; however, placental LPL and endothelial lipase hydrolyze them, liberating fatty acids that can be taken up by the placenta and used by the fetus to grow.[38] Previous studies have reported a positive association between placental LPL enzymatic activity [37] and DNAm of placental LPL gene [39] with birthweight and neonatal adiposity.
SCYL2 gene, in addition to playing a role in positive regulation of endocytosis, is involved in Wnt signaling pathway. Different Wnt signaling components and ligands have been identified in the uterus promoting endometrial changes such as decidualization and endometrial gland formation. [40, 41] Ineffective decidualization have been reported in pregnancies complicated with intrauterine growth retardation. [42]
SCT gene codes for Secretin, a peptide hormone. Murine studies have shown secretin expression by uterine stromal cells from early pregnancy.[43] Secretin levels increases significantly in late pregnancy with highest level at 36 weeks.[44] Our data showed a positive correlation between DNAm of CpG cg00249511, associated with SCT gene, in early and late pregnancy, possibly implying the importance of secretin throughout gestation. Secretin and secretin receptor axis leads to the activation of intracellular secondary messenger system of cAMP, a strong inducer of decidualization.[44] The role of maternal secretin in decidualization could explain its association with fetal development and offspring birthweight. Moreover, secretin has lipolytic effects and is involved in appetite regulation and glucose/insulin homeostasis [45], potentially influencing maternal metabolism and fetal development during pregnancy.
In addition to the maternal effects, several roles of secretin in fetal development, particularly in the developing brain, have been reported.[46] Placenta provides secretin to the developing fetus before it gains the ability to produce its own. [45] Interestingly, neonatal methylation levels of the corresponding receptor gene of SCT, namely, SCTR gene, have been associated with birthweight. [5]
Although correlated, birthweight and gestational age may present different processes. Birthweight is determined by both the duration of gestation and fetal growth rate.[47] Using structural equation modeling, we observed direct and indirect effects of maternal DNAm at six candidate CpGs (cg01816814, cg04255048, cg10715265, cg00249511, cg27239144, and cg17722033) on birthweight through gestational age. This finding suggests that these CpGs influence birthweight by affecting both the duration of gestation and the fetal growth rate. However, the other two CpGs, cg23153661 and cg 27394038 associated with VAX2, CD207, and SFRS3, showed no significant indirect effect on birthweight through gestational age.
Given that the methylation data of the candidate CpGs did not vary significantly with age (18 years, early, and late pregnancy), it is possible that the DNAm is driven by maternal genetic polymorphisms (methylation quantitative trait loci or methQTLs).[48] Hence, there is a need in future studies to assess whether maternal genetic polymorphisms of the linked genes are associated with birthweight of the child.
Our study has some limitations. First, we only evaluated DNAm extracted from maternal peripheral blood samples. Given the substantial differences in the epigenetic markings of distinct cell types, CpG sites other than what we identified may be associated with offspring birthweight in other organs. Second, even though all maternal blood samples were collected in late pregnancy, they were not drawn at the same gestational week. Nonetheless, the gestational week at which the maternal blood was drawn had no association with the DNAm levels of the candidate CpGs after adjusting for blood cell types. Third, although directions of each association among CpGs and birthweight was established, we cannot state that the direction corresponds to gene expression. Lastly, our results were only marginally replicated, presumably due to the small sample size of the Born Into Life cohort. However, for three genes LMF1, SCYL2, and SCT, prior genetic and metabolic findings also support our results. Future studies with larger sample sizes are needed to evaluate all associations described in this study.
Conclusion
Our study (n=122) found eight maternal CpGs differentially methylated in association with offspring birthweight, three of them were replicated with marginal statistical significance in a small sample (n=33). The associated genes were linked to metabolic pathways involved in providing substrates for the developing fetus or in cellular processes such as endocytosis, possibly playing a role in sustaining a sufficient maternal-fetal interface. Overall, our findings contribute to the concept of maternal involvement in offspring birthweight. Additional studies are needed to elucidate the role of maternal DNAm in pregnancy and its association with offspring birthweight and other pregnancy outcomes. This can contribute to the identification of preventive or therapeutic targets for neonatal low and high birthweights. Furthermore, it is necessary to determine which environmental or maternal factors influence the differential maternal DNAm in order to identify targets for interventions reducing the risk of low or high birthweights. Finally, measuring DNAm levels of maternal CpGs may have a diagnostic potential recognizing mothers at risk for pregnancies ending with altered birthweights.
Supplementary Material
Supplementary fig1 Structural equation modeling graphs showing the total effect of maternal DNAm on birthweight, indirect effect of maternal DNAm on birthweight through gestational age, and the direct effects between maternal DNAm, gestational age, and birthweight. Models are adjusted for newborn’s gender, maternal peripheral blood cell types, maternal BMI, smoking during pregnancy, and socioeconomic status. Results are shown as beta values*100 (percentage) *0.001≤P-value <0.05, **P-value<0.001
Acknowledgments
Funding : This work has been supported by National Institute of Allergy and Infectious Diseases [R01 AI091905] and the National Heart, Lung, and Blood Institute [R01HL132321] to W.K and [R01 HL082925] to SHA. The Born into Life cohort was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the Swedish Heart-Lung Foundation and Department of Clinical Sciences, Karolinska Institutet, Danderyd University Hospital, Stockholm, Sweden. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, USA.
Ethical considerations:
The IOW study was approved repeatedly by the local research ethics committee (NRES Committee South Central – Hampshire B, U.K.) and the University of Memphis Institutional Review Board in Memphis, U.S. (FWA00006815). Ethical approval was obtained by the Regional Ethics Review Board in Stockholm, Sweden, for the Born Into Life study. Written consents were obtained from all participants of the IOW and Born Into Life cohorts at recruitment and follow-ups.
Footnotes
Declarations
Consent to participate and for publication: Written consents were obtained from all participants of the IOW and Born Into Life cohorts at recruitment and follow-ups.
Availability of data and material: Data used in this study may be available upon request from the authors with permission from the IOW and Born Into Life cohorts.
Code availability: N/A
Conflicts of interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Asmare G, Berhan N, Berhanu M, Alebel A. Determinants of low birth weight among neonates born inAmhara Regional State Referral Hospitals of Ethiopia: unmatched case control study. BMC Res Notes. 2018;11(1):447. doi: 10.1186/s13104-018-3568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu XF, Li YJ, Sheng YJ, Liu JL, Tang LF, Chen ZM. Effect of low birth weight on childhood asthma: ameta-analysis. BMC Pediatr. 2014;14:275. doi: 10.1186/1471-2431-14-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherman V, Rutherford GW. A meta-analysis of the effect of high weight on asthma. Arch Dis Child.2006;91(4):334–9. doi: 10.1136/adc.2005.080390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palatianou ME, Simos YV, Andronikou SK, Kiortsis DN. Long-term metabolic effects of high birthweight: a critical review of the literature. Horm Metab Res. 2014;46(13):911–20. doi: 10.1055/s-0034-1395561. [DOI] [PubMed] [Google Scholar]
- 5.Kupers LK, Monnereau C, Sharp GC, Yousefi P, Salas LA, Ghantous A et al. Meta-analysis ofepigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat Commun. 2019;10(1):1893. doi: 10.1038/s41467-019-09671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon EJ, Kim YJ. What is fetal programming?: a lifetime health is under the control of in utero health.Obstet Gynecol Sci. 2017;60(6):506–19. doi: 10.5468/ogs.2017.60.6.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, Mukherjee N, Janjanam VD, Arshad SH, Kurukulaaratchy RJ, Holloway JW et al. Consistencyand Variability of DNA Methylation in Women During Puberty, Young Adulthood, and Pregnancy. Genet Epigenet. 2017;9:1179237×17721540. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arshad SH, Holloway JW, Karmaus W, Zhang H, Ewart S, Mansfield L et al. Cohort Profile: The Isle OfWight Whole Population Birth Cohort (IOWBC). Int J Epidemiol. 2018;47(4):1043–4i. doi: 10.1093/ije/dyy023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogbuanu IU, Karmaus W, Arshad SH, Kurukulaaratchy RJ, Ewart S. Effect of breastfeeding duration onlung function at age 10 years: a prospective birth cohort study. Thorax. 2009;64(1):62–6. doi: 10.1136/thx.2008.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al. Minfi: a flexibleand comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England). 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH et al. DNAmethylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smew AI, Hedman AM, Chiesa F, Ullemar V, Andolf E, Pershagen G et al. Limited association betweenmarkers of stress during pregnancy and fetal growth in ‘Born into Life’, a new prospective birth cohort. Acta Paediatr. 2018;107(6):1003–10. doi: 10.1111/apa.14246. [DOI] [PubMed] [Google Scholar]
- 13.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from humannucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol.2009;507:149–63. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Yan L, Hu Q, Sucheston LE, Higgins MJ, Ambrosone CB et al. IMA: an R package for high-throughput analysis of Illumina’s 450K Infinium methylation data. Bioinformatics. 2012;28(5):729–30. doi: 10.1093/bioinformatics/bts013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empiricalBayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 17.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L et al. Comparison of Beta-value and M-valuemethods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJ. An Efficient Approach to Screening Epigenome-Wide Data. Biomed Res Int. 2016;2016:2615348. doi: 10.1155/2016/2615348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach toMultiple Testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 20.Welch RP, Willer CJ, J. SL, Boehnke M. Snipper: a research tool for extracting and searching biologicalannotations on genes near SNPs: University of Michigan; 2013. [Google Scholar]
- 21.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR et al. The UCSC Genome Browserdatabase: 2018 update. Nucleic Acids Res. 2018;46(D1):D762–d9. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis andcandidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–11. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg H, Taylor ME, Razi N, McBride R, Knirel YA, Graham SA et al. Structural basis for langerinrecognition of diverse pathogen and mammalian glycans through a single binding site. J Mol Biol. 2011;405(4):1027–39. doi: 10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg H, Rowntree TJ, Tan SL, Drickamer K, Weis WI, Taylor ME. Common polymorphisms inhuman langerin change specificity for glycan ligands. J Biol Chem. 2013;288(52):36762–71. doi: 10.1074/jbc.M113.528000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valladeau J, Ravel O, Dezutter-Dambuyant C, Moore K, Kleijmeer M, Liu Y et al. Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity. 2000;12(1):71–81. doi: 10.1016/s1074-7613(00)80160-0. [DOI] [PubMed] [Google Scholar]
- 26.Geijtenbeek TB, van Vliet SJ, Engering A, t Hart BA, van Kooyk Y. Self- and nonself-recognition by C-type lectins on dendritic cells. Annu Rev Immunol. 2004;22:33–54. doi: 10.1146/annurev.immunol.22.012703.104558. [DOI] [PubMed] [Google Scholar]
- 27.Blois SM, Kammerer U, Alba Soto C, Tometten MC, Shaikly V, Barrientos G et al. Dendritic cells: keyto fetal tolerance? Biol Reprod. 2007;77(4):590–8. doi: 10.1095/biolreprod.107.060632. [DOI] [PubMed] [Google Scholar]
- 28.Ander SE, Diamond MS, Coyne CB. Immune responses at the maternal-fetal interface. Scienceimmunology. 2019;4(31). doi: 10.1126/sciimmunol.aat6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: Shared machinery for degradation.Bioessays. 2013;35(1):34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 30.Kanninen TT, de Andrade Ramos BR, Witkin SS. The role of autophagy in reproduction fromgametogenesis to parturition. Eur J Obstet Gynecol Reprod Biol. 2013;171(1):3–8. doi: 10.1016/j.ejogrb.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Cao B, Camden AJ, Parnell LA, Mysorekar IU. Autophagy regulation of physiological and pathologicalprocesses in the female reproductive tract. Am J Reprod Immunol. 2017;77(5). doi: 10.1111/aji.12650. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Wang H, Li D, Li M. Role of Endometrial Autophagy in Physiological and Pathophysiological Processes. J Cancer. 2019;10(15):3459–71. doi: 10.7150/jca.31742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterfy M. Lipase maturation factor 1: a lipase chaperone involved in lipid metabolism. BiochimBiophys Acta. 2012;1821(5):790–4. doi: 10.1016/j.bbalip.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehrhardt N, Bedoya C, Peterfy M. Embryonic viability, lipase deficiency, hypertriglyceridemia andneonatal lethality in a novel LMF1-deficient mouse model. Nutr Metab (Lond). 2014;11:37. doi: 10.1186/1743-7075-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrera E, Ortega-Senovilla H. Lipid metabolism during pregnancy and its implications for fetalgrowth. Current pharmaceutical biotechnology. 2014;15(1):24–31. doi: 10.2174/1389201015666140330192345. [DOI] [PubMed] [Google Scholar]
- 36.Herrera E. Lipid metabolism in pregnancy and its consequences in the fetus and newborn. Endocrine.2002;19(1):43–55. doi: 10.1385/endo:19:1:43. [DOI] [PubMed] [Google Scholar]
- 37.Heerwagen MJR, Gumina DL, Hernandez TL, Van Pelt RE, Kramer AW, Janssen RC et al. Placentallipoprotein lipase activity is positively associated with newborn adiposity. Placenta. 2018;64:53–60. doi: 10.1016/j.placenta.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Segura MT, Demmelmair H, Krauss-Etschmann S, Nathan P, Dehmel S, Padilla MC et al. Maternal BMIand gestational diabetes alter placental lipid transporters and fatty acid composition. Placenta. 2017;57:144–51. doi: 10.1016/j.placenta.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Gagne-Ouellet V, Houde AA, Guay SP, Perron P, Gaudet D, Guerin R et al. Placental lipoprotein lipaseDNA methylation alterations are associated with gestational diabetes and body composition at 5 years of age. Epigenetics. 2017;12(8):616–25. doi: 10.1080/15592294.2017.1322254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonderegger S, Pollheimer J, Knofler M. Wnt signalling in implantation, decidualisation and placentaldifferentiation--review. Placenta. 2010;31(10):839–47. doi: 10.1016/j.placenta.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tepekoy F, Akkoyunlu G, Demir R. The role of Wnt signaling members in the uterus and embryoduring pre-implantation and implantation. J Assist Reprod Genet. 2015;32(3):337–46. doi: 10.1007/s10815-014-0409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunk C, Kwan M, Hazan A, Walker S, Wright JK, Harris LK et al. Failure of Decidualization andMaternal Immune Tolerance Underlies Uterovascular Resistance in Intra Uterine Growth Restriction. Frontiers in endocrinology. 2019;10:160. doi: 10.3389/fendo.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Z, Ge YF, Jing J, Wu L, Zhou ZY, Zhu QF et al. [Effect of secretin on the expression of cPLA2 andmPGEs-1 in mouse endometrial stromal cell during early pregnancy]. Sheng Li Xue Bao. 2016;68(6):725–32. [PubMed] [Google Scholar]
- 44.Huang Z, Wang TS, Qi QR, Zuo RJ, Liang XH, Zhao XY et al. Progesterone regulates secretin expressionin mouse uterus during early pregnancy. Reprod Sci. 2014;21(6):724–32. doi: 10.1177/1933719113512527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knox K, Leuenberger D, Penn AA, Baker JC. Global hormone profiling of murine placenta revealsSecretin expression. Placenta. 2011;32(11):811–6. doi: 10.1016/j.placenta.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siu FK, Sham MH, Chow BK. The prenatal expression of secretin receptor. Ann N Y Acad Sci.2006;1070:561–5. doi: 10.1196/annals.1317.081. [DOI] [PubMed] [Google Scholar]
- 47.Kramer MS. The epidemiology of low birthweight. Nestle Nutrition Institute workshop series.2013;74:1–10. doi: 10.1159/000348382. [DOI] [PubMed] [Google Scholar]
- 48.Pan H, Holbrook JD, Karnani N, Kwoh CK. Gene, Environment and Methylation (GEM): a tool suite toefficiently navigate large scale epigenome wide association studies and integrate genotype and interaction between genotype and environment. BMC Bioinformatics. 2016;17:299. doi: 10.1186/s12859-016-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary fig1 Structural equation modeling graphs showing the total effect of maternal DNAm on birthweight, indirect effect of maternal DNAm on birthweight through gestational age, and the direct effects between maternal DNAm, gestational age, and birthweight. Models are adjusted for newborn’s gender, maternal peripheral blood cell types, maternal BMI, smoking during pregnancy, and socioeconomic status. Results are shown as beta values*100 (percentage) *0.001≤P-value <0.05, **P-value<0.001