Abstract
Purpose
We aimed to assess the concordance of colorectal cancer (CRC)-associated methylated DNA markers (MDMs) in primary and metastatic CRC for feasibility in detection of distantly recurrent/metastatic CRC in plasma.
Experimental Design
A panel of previously discovered CRC-associated MDMs was selected. MDMs from primary and paired metastatic CRC tissue were assayed with quantitative methylation-specific PCR. Plasma MDMs were measured blindly by target enrichment long-probe quantitative amplified signal assays. Random forest modeling was used to derive a prediction algorithm of MDMs in archival plasma samples from primary CRC cases. This algorithm was validated in prospectively collected plasma samples from recurrent CRC cases. The accuracy of the algorithm was summarized as sensitivity, specificity, and area under the curve (AUC).
Results
Of the 14 selected MDMs, the concordance between primary and metastatic tissue was considered moderate or higher for 12 MDMs (86%). At a preset specificity of 95% (91–98%), a panel of 13 MDMs in plasma from 97 CRC cases and 200 controls detected stage IV CRC with 100% (80–100%) sensitivity and all stages of CRC with an AUC 0.91 (0.87–0.95), significantly higher than CEA (AUC 0.72 (0.65–0.79)). This panel in plasma from 40 cases, and 60 healthy controls detected recurrent/metastatic CRC with 90% (76–97%) sensitivity with specificity of 90% (79–96%) and an AUC of 0.96 (0.92–1).The panel was positive in 0.30 (0.19–0.43) of 60 patients with no evidence of disease post-operative CRC patients.
Conclusions
Plasma assay of novel CRC-associated MDMs can reliably detect both primary CRC and distantly recurrent CRC with promising accuracy.
Keywords: DNA methylation, circulating tumor DNA, liquid biopsy
Introduction
Colorectal cancer (CRC) remains one of the most common and fatal cancers in this country despite current screening efforts and advances in therapies [1], which at least is partially attributed to the modest utilization rate of screening for CRC [2]. In addition, nearly half patients undergoing curative resection experience recurrence [3]. Several large studies have shown that comprehensive surveillance with blood and imaging tests can detect pre-symptomatic recurrences and improve the prognosis of metastatic disease where therapy can still be directed toward cure [4,5].
Current surveillance tools have sub-optimal accuracy and high cost. Radiographic modalities may miss focal or diffuse lesions and may fail to reflect total tumor burden [6]. The standard of care biomarker, plasma carcinoembryonic antigen (CEA) suffers from low sensitivity and specificity [7]. Thus, there is a clear need for a noninvasive surveillance tool with better performance characteristics than CEA and at lower cost and more sensitivity than imaging.
Liquid biopsy with mutation-based markers from circulating cell-free DNA (cfDNA) shows strong prognostic value in patients with resected CRC [8–10]. However, this approach is unwieldy for early detection of CRC at screening or surveillance due to the large number of mutational sites that must be assayed [11]. The strategy of customizing targeted markers to those mutations confirmed in each individual’s primary tumor involves high costs of sequencing and tailoring surveillance assays accordingly [10]. Furthermore, there are no guarantees that such targets remain stable or informative in a given patient due to genetic evolution across metastases [12].
Methylated DNA markers (MDMs) offer several advantages over existing tools for screening and surveillance of CRC (Figure 1A). Previous studies showed that MDMs are superior to serum CEA in detecting primary CRC [13]. MDMs, the archetypal epigenetic markers, are also more broadly informative than acquired mutational markers in CRC screening with modest sensitivity [14–16]. Similar to acquired molecular disparities with somatic genetic mutations in CRC [17], some MDMs have been associated with tumor behavior and response to adjuvant chemotherapy reflecting tumor heterogeneity [18]. Our group has previously reported results of a next-generation sequencing approach to the discovery of novel MDMs, strongly associated with primary CRC tissues [19] which we hypothesize would be feasible for liquid biopsy application in early detection of primary CRC and for non-invasive detection of recurrent CRC.
Figure 1:
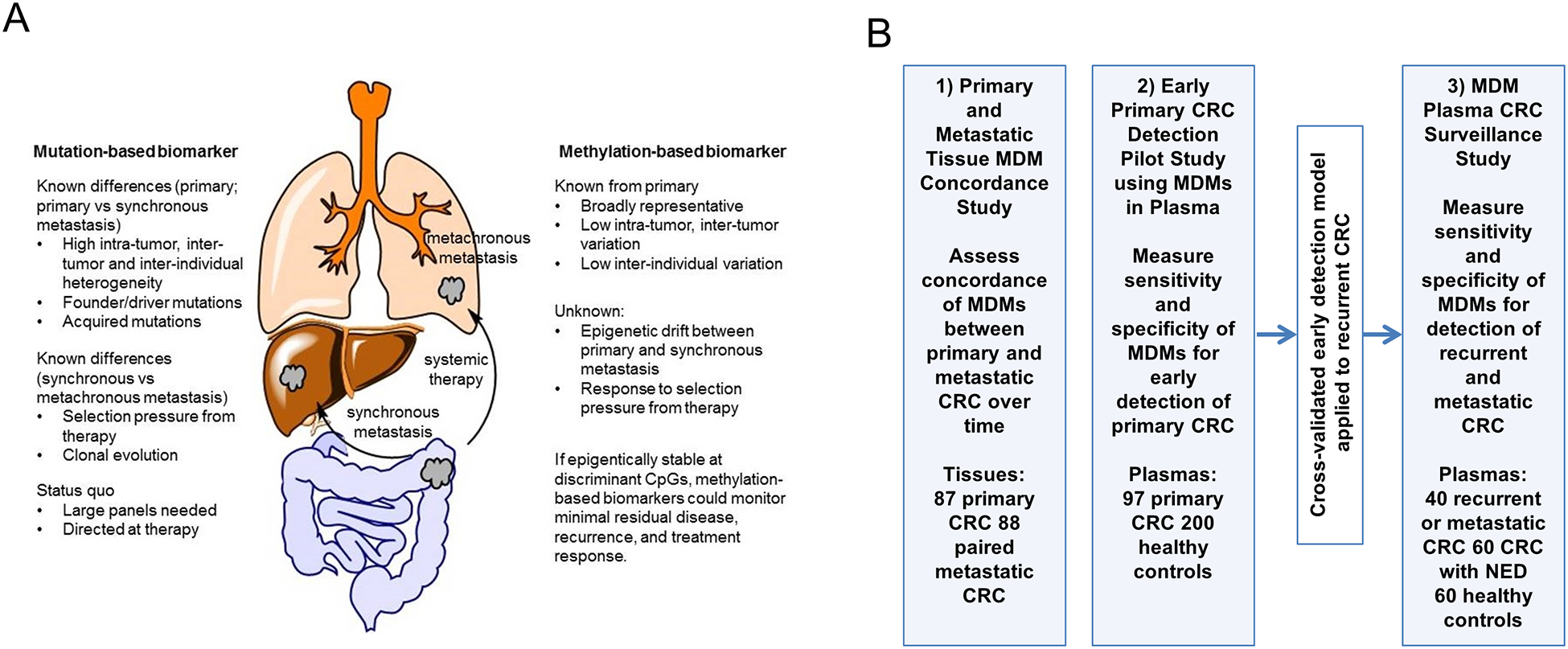
(A) Study rationale for using methylated DNA markers compared to mutation-based biomarker in metastatic colorectal cancer and B) overview of three parallel studies to address primary aims. 1) Primary and Metastatic Tissue MDM Concordance Study to assess concordance of MDMs between primary and metastatic CRC over time; 2) MDM Plasma CRC Early Primary Detection Study to measure sensitivity and specificity of MDMs for early detection of primary CRC; 3) MDM Plasma CRC Surveillance Study to measure sensitivity and specificity of MDMs for detection of recurrent and metastatic CRC. MDM: methylated DNA marker; CRC: colorectal cancer; NED: no radiographic evidence of disease.
Accordingly, the present study has 3 parallel and independent aims. First, we sought to assess concordance of MDM candidates between primary CRC tumors and metastasis, both synchronous and recurrent metachronous disease [Primary and Metastatic Tissue MDM Concordance Study]; high concordance would be required to be useful in the detection of recurrent or minimally residual disease. Second, we assessed the sensitivity and specificity of MDMs for early detection of CRC, when assayed from plasma of treatment naïve case patients and healthy controls [Early CRC Primary Detection Pilot Study] in order to set marker cutoff levels for surveillance study. The model developed to detect treatment naïve primary CRC was then applied in a third case-control study to determine feasibility for identification of previously treated patients with recurrent or metastatic CRC in comparison to those treated for CRC with no radiographic evidence of disease (NED) and healthy controls [MDM Plasma CRC Surveillance Study].
Patients and Methods
All study procedures were conducted after approval from the Mayo Clinic Institutional Review Board in accordance with the United States Common Rule (Department of Health and Human Services Title 45 CFR 46).
Primary and Metastatic Tissue MDM Concordance Study
Surgically resected primary and paired metastatic CRC including synchronous or metachronous metastases were selected from the Mayo Clinic Tissue Registry, following a query assembled from SNOMED pathology diagnostic codes. Synchronous metastases were defined as distant metastases occurring within 3 months, and metachronous metastases as those beyond 3 months relative to initial diagnosis of CRC. Inclusion criteria: written consent recorded and at least 18 years of age; exclusion criteria: history of radiation to the site of tumor being studied and known familial cancer syndrome (e.g. Lynch syndrome, familial adenomatous polyposis). After retrieval of archival formalin-fixed paraffin-embedded tissue specimens, an expert gastrointestinal pathologist (T. T. W.) confirmed the diagnosis, identified tumor location from pathology slides, and estimated tumor purity. Relevant demographic, clinical, and histopathologic information were retrospectively collected on those patients with selected archival tumor specimens. Clinical phenotyping was performed by trained study personnel according to a pre-specified protocol and reviewed for accuracy by a clinician expert (H. X.).
A panel of 14 MDMs (VAV3, CHST2, OPLAH, QKI, PPP2R5C, ARHGEF4, PDGFD, ZNF625, SFMBT2, LRRC4, DOCK10, IKZF1, NDRG4, BMP3) previously identified to be highly discriminant for primary CRC was selected on the basis of high median fold-change of MDM levels 414 (interquartile range: 155–12476) relative to buffy coat and fold-change median of 150 (49–469) relative to normal colon mucosae. Quantitative MSP was used to measure each of the selected MDMs, as previously described [20]. Briefly, DNA was bisulfite treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) and eluted in buffer. One microliter of bisulfite-treated DNA was used as a template for methylation quantification with a fluorescence-based real-time PCR. Primers for each marker were designed to target the bisulfite-modified methylated sequences of each gene (IDT, Coralville IA). A region without cytosine-phosphate-guanine sites in the β-actin gene was used as a reference of bisulfite treatment and DNA input. PCR reactions for tissue DNA samples were performed with SYBR Green master mix (Roche, Germany). All reactions were run on Roche 480 LightCyclers (Indianapolis, IN). Bisulfite-treated CpGenome Universal Methylated DNA (Millipore, Billerica, MA) was used as a positive control, and serially diluted to create standard curves for all plates.
Early Primary CRC Detection Pilot Study using MDMs in Plasma
Archival plasma of 6 mL from healthy controls [Clinicaltrials.gov NCT03628638] and CRC cases and, balanced for age and sex was identified among patients enrolled in an archive of newly diagnosed un-treated cancers [Clinicaltrials.gov NCT03662204] who provided a sample after written informed consent between 09/1/2018 to 03/31/2019. Patients were excluded for prior or concurrent cancer diagnosis defined as any previous cancer diagnosis within the past 5 years (with the exceptions of basal cell or squamous cell skin cancers); or recurrence of the same primary cancer within any timeframe; OR concurrent diagnosis of multiple primary cancers. Case patients were required to be treatment naïve, and samples could not be used if drawn less than 3 days between fine needle aspiration (FNA) of target pathology and blood collection, less than 7 days between biopsy (other than FNA) of target pathology and blood collection or from patients who received IV contrast (e.g. CT and MRI) within 24 hours of blood collection.
Whole peripheral venous blood samples were collected in LBguard tubes (Biomatrica, San Diego CA) which use a proprietary nucleic acid preservative media for maintenance of cell integrity and target DNA analyte recovery, and allow central processing of specimens at room temperature up to 8 days following collection [21].
Plasma samples stored at −80°C were retrieved. DNA from samples was extracted (QIAamp Circulating Nucleic Acid Kit, Qiagen, Germantown MD) and bisulfite-converted. Due to the low abundance of cfDNA in plasma specimens, selected MDMs were assayed and quantified primarily by target enrichment long-probe quantitative amplified signal (TELQAS) assay, which was developed by Exact Sciences (Madison WI); detailed steps of the assay protocol have been previously published [22]. Briefly, a limited number of cycles (12 cycles) multiplex PCR amplification of the MDMs was performed on the bisulfite converted DNA. The PCR products were then diluted 10-fold with a 10 mM Tris-HCl, 0.1 mM EDTA solution; 10 μL of the diluted amplicons were used in triplex LQAS assays in which two MDMs were quantified. MDMs were normalized to a methylated sequence of B3GALT6, measure of total bisulfite converted DNA input. TELQAS reactions were performed on ABI 7500DX equipment (Applied Biosystems, Foster City CA). TELQAS assays for MDMs in this experiment were designed independently of the results from the tissue study; while VAV3, CHST2, QKI, PDGFD, SFMBT2 were in common with the tissue study. Because the studies were conducted non-sequentially, additional MDMs that showed strong association with CRC in pre-clinical experiments were tested. Due to limits of TELQAS multi-plexing capacity and the limited quantity of cell-free DNA in plasma not all MDMs of interest could be studied simultaneously. Therefore, the plasma studies examined additional new candidates JAM3, ZNF671, ZNF568, GRIN2D, DTX1, ANKRD13B, CNNM1, FER1L4. Carcinoembryonic antigen (CEA) testing was performed per manufacturer’s instructions on 10 microliter serum specimens using the Cobas e411 immunoassay analyzer (Roche Diagnostics, Indianapolis IN).
MDM Plasma CRC Surveillance Study
Samples were identified from a prospective longitudinal cohort study of CRC patients from whom blood specimens were obtained using LBgard tubes after written informed consent. The case patient population included those with newly diagnosed or recurrent CRC on active treatment. The disease control patients included those with stage II-IV CRC currently with NED after curative intent therapy. All were followed with CEA and radiographic modalities consistent with routine clinical practice. Age and sex balanced plasma specimens from healthy patients without cancer in a parallel study serve as negative controls. The negative controls were recruited from a 7-county population registry and were required to be up-to-date with CRC screening.
Plasma CEA and the same MDMs used in the MDM Plasma CRC Early Primary Detection Study, were assayed blindly from age/sex-balanced patients including healthy controls, patients with NED, and those with recurrent CRC after primary tumor resection. The cross-validated MDM prediction model from the Early CRC Primary Detection Pilot Study (above) was applied to distinguish those with recurrent or metastatic cancer (cases) from the healthy controls. The prediction accuracy estimate was then applied to the NED disease control group to estimate the percentage of patients who might have minimal residual disease.
Upon receipt, blood samples were immediately spun to separate plasma and buffy coat fractions, which were stored at −80°C. After DNA extraction and bisulfite conversion, selected MDMs were assayed and quantified primarily by TELQAS assay. Four mL of plasma samples were used in this study for MDMs. Ten microliters of plasma was used for CEA analyses as sera were not available for this experiment. Plasma CEA results were compared to available clinical serum values for CRC patients, collected at the same time point.
All assays were performed by laboratory technicians who were blinded to clinical phenotyping and case-control status. Specimen collection, handling, processing and assay methods met REMARK guidelines. All other aspects of study methodology and reporting adhere to the STROBE statement on observational research, as noted in Supplemental Materials.
Statistical analysis
For the Primary and Metastatic Tissue MDM Concordance Study, the level of concordance between MDM levels in primary and metastatic CRC tissue was assessed by the intra-class correlation coefficient (ICC) both overall and stratified by synchronous and metachronous presentation. Briefly, the ICC summarizes the percentage of total variation observed in individual MDM levels that is attributed to differences (non-concordance) between the primary and metastatic CRC tissue. Following the guide of Koo and Li, ICC is considered to indicate ICC<50% poor, 50%≤ICC<75% moderate, 75%≤ICC<90% good, and ICC≥90% excellent concordance relative to the lower bound of the 95% confidence interval [23]. The required sample size was determined based on limiting the width of the 95% confidence interval for the ICC to be no larger than ±10% for the overall comparison. To achieve this level of confidence, and assuming an ICC of 30%, a minimum of 77 patients with paired primary and metastatic tissue was required. In addition to summarizing the ICC with corresponding 95% confidence intervals, the relative difference between primary and metastatic CRC tissue was summarized on the log scale as a median with corresponding 25th and 75th percentiles. All MDMs were standardized to bisulfite-treated β-actin MSP products in each sample.
For the Early Primary CRC Detection Pilot Study, there were a total of 14 MDMs, including B3GALT6, and CEA assayed in serum. Each MDM TELQAS product was standardized to methylated B3GALT6, which was recently shown to be a more reliable normalizing sequence than β-actin in plasma assays [24], using generalized additive models estimated from the training control population and applied to both cases and controls equally within the training data set and the testing data set (i.e., CRC surveillance data). The training data were used to train a random forest (rForest) prediction algorithm [25] (with cross validation) for the endpoint of CRC using all 14 MDMs (with and without continuous CEA values). Briefly, the rForest model consisted of 500 bootstrap random samples of the training data. Each bootstrap sample was used to create a single recursive partition/decision tree resulting in an ensemble of 500 trees (one for each bootstrap sample). Data from an independent sample not used in the development of an individual tree (e.g., an individual is not used in training for roughly 1/3 of the 500 trees) was classified as CRC (Y/N) based on marker levels and the percentage of trees positive for CRC for the independent sample was recorded. After training the model, the cutoff for the percentage of positive trees required to classify someone as CRC (Y/N) was fixed and set to perform predictions at a specificity cutoff of 95%. This model was then applied directly to the CRC surveillance data (test set) and an individual sample was considered positive if the percentage of positive trees exceeded the predefined cutoff from the training set. Sample size justification is based on the recommendation of Riley et al. for the required sample size needed for developing prediction models [25]. Assuming a 2:1 ratio of controls to cases, a total of 14 biomarkers, and a mean absolute prediction error of ±10% for the trained model, a minimum total sample size of 175 patients was required. An additional consideration was to have adequate sample size for the in-silico cross-validation of model sensitivity using bootstrap resampling. To have the 95% confidence interval of the estimated sensitivity of 90% to be no larger than ±10%, a total of 150 patients (50 cases and 100 controls) were required for the test set. Combining these requirements, a total overall study sample size of 300 patients was deemed sufficient for training and cross-validation of the prediction model with an approximate 2:1 ratio of training: testing splits of the data achieved with the bootstrap sampling scheme.
In the MDM Plasma CRC Surveillance Study, sample size was based on comparing the AUC of the trained predictions using MDM levels (with and without CEA) relative to CEA alone. Based on an estimated sensitivity of 63% and specificity of 88%, the anticipated AUC for CEA was 0.75 [7]. In order to detect an AUC of 0.85 or higher for MDM levels with 80% power, a minimum of 64 cases and 64 controls would be required assuming a one-sided test of significance of 5%. With 64 cases and 64 controls, the half-width of the 95% confidence interval for sensitivity and specificity was no larger than ±10% assuming each are greater than 0.85. The models with and without CEA developed in the Early Primary CRC Detection Pilot Study using MDMs in Plasma were applied to predict a cancer diagnosis among recurrent/metastatic CRC cases in comparison to healthy controls. The positivity rate for this model was then assessed in those with NED. ROC analysis was used to evaluate the diagnostic accuracy of trained models and summarized as area under the ROC (AUC) with corresponding 95% confidence intervals. The association of tumor burden measured by Response Evaluation Criteria in Solid Tumors (RECIST) criteria [26] with the diagnostic accuracy of the trained models was evaluated by comparing stratified AUCs.
Continuous variables were summarized as median with interquartile range (IQR) and categorical data were summarized as a percent of group totals. Nonparametric tests were utilized for comparisons between subgroups of patients (e.g., Wilcoxon Rank Sums Test for ordinal data and Fischer’s Exact Test for categorical data).
Results
Figure 1B shows the overview of the three main experiments to test our central hypothesis that novel candidate MDMs are concordant between paired primary and metastatic CRC tissues and that a panel of such MDMs can successfully detect not only primary CRC, but also extra-intestinal recurrent/metastatic CRC in plasma samples.
Primary and Metastatic Tissue MDM Concordance Study
As shown in Supplemental Table 1, the MDM tissue study included 84 patients with paired primary and metastatic CRC of which 56 were synchronous metastasis; 28 were metachronous metastasis. The median age of patients when primary CRC was resected was 60.2 years. Forty-one (49%) patients had neoadjuvant and 56 (67%) patients had adjuvant chemotherapy. Fifty-six (67%) patients had left-sided CRC. All synchronous metastases were to the liver. Metachronous metastases were to the liver in 17 (61%) patients and to the lung in 10 (36%) patients. For the 14 selected MDMs, 1 MDM (7%) had an estimated ICC with a confidence interval greater than ≥ 75% indicating good concordance, 9 (64%) had an ICC ≥ 50% indicating moderate concordance, and 3 (21%) had poor concordance when considering all metastases together (Supplemental Table 2). The levels of 14 selected MDMs measured as median fold change were remarkably similar between paired primary and metastatic CRC for individual MDM (Figure 2A). The levels in the majority of these 14 selected MDMs were comparable when stratified as between primary and synchronous metastatic CRC (Figure 2B), between primary and metachronous metastatic CRC (Figure 2C), between left-side primary and metastatic CRC (Figure 2D), and between right-side primary and metastatic CRC (Figure 2E).
Figure 2:

Differences in tissue methylated DNA marker (MDM) levels between paired primary and metastatic colorectal cancer (CRC). Differences expressed as median and IQR fold change when dividing primary tumor level by paired metastatic tumor level. (A) Overall, (B) between primary and synchronous metastatic CRC; (C) between primary and metachronous metastatic CRC; (D) between left-side primary and metastatic CRC; and (E) between right-side primary and metastatic CRC. Fold change values >1 indicate higher MDM levels in the primary; values <1 indicate higher MDM levels in the metastasis.
In both the Early Primary CRC Detection Pilot Study using MDMs in Plasma and MDM Plasma CRC Surveillance Study, all samples contained least 20,000 strands of B3GALT6 after bisulfite conversion and target amplification. All samples assayed were included in the analysis.
Early Primary CRC Detection Pilot Study using MDMs in Plasma
We then applied selected MDMs to archival plasma samples from patients with stage I-IV primary CRC. Ninety-seven patients with CRC and 200 age- and sex-balanced healthy controls were included. Patients with CRC and healthy controls had comparable age, sex and race distributions (Table 1). Controls were significantly more likely to be current or former smokers. Among patients with CRC, 23 (23.7%) patients had stage IV CRC; 23 (24.7%) patients had stage III CRC; 26 (26.8%) patients had stage II CRC; and 11 (11.3%) patients had stage I CRC. At 95% (91–98%) specificity, the panel of 13 MDMs with CEA had a cross-validated sensitivity of 78% (69–86%) for all stages of CRC; without CEA, the cross-validated sensitivity was 77% (68–85%). Model performance was also evaluated for detecting each stage of CRC. At 95% (91–98%) specificity, the panel of 13 MDMs with or without CEA had cross-validated sensitivities of 64% (31–89%) for stage I CRC; 62% (41–80%) and 65% (44–83%) for stage II CRC, respectively; 79% (58–93%) and 71% (49–87%) for stage III CRC, respectively, and 100% (85–100%) for stage IV CRC. The performance of individual MDMs as well as CEA was also summarized as AUC values as shown in Figure 3. The panel of 13 MDMs was able to detect all stages of CRC with a cross-validated AUC of 0.91 (0.87–0.95). The addition of CEA to the panel did not significantly improve the AUC (0.91 (0.87–0.95) (p=0.5415). The importance of individual MDMs and CEA in contribution to the performance of random forest model in classification of early CRC from healthy controls was shown in Supplemental Figure 2A. Among 13 MDMs, methylated JAM3 and ZNF761 had the most contribution to the performance of overall cross-validated model algorithm. Model accuracy was not impacted by primary tumor location (rectum vs left colon vs right colon).
Table 1:
Baseline characteristics of patients in the Early Primary CRC Detection Pilot Study using MDMs in Plasma
| CRC (n=97) | Healthy controls (n=200) | P value | |
|---|---|---|---|
| Age Median (IQR) | 62.0 (55.0–68.0) | 61.7 (54.8–67.0) | 0.67 |
| Sex | |||
| Female | 37 (38.1%) | 86 (43.0%) | 0.43 |
| Male | 60 (61.9%) | 114 (57.0%) | |
| Race | |||
| White | 82 (90.1%) | 170 (85.0%) | 0.22 |
| Black | 4 (4.4%) | 21 (10.5%) | |
| Other | 11 (11.3%) | 9 (4.5%) | |
| Smoking | |||
| Ever | 51 (52.6%) | 152 (76.0%) | <0.001 |
| Never | 46 (47.4%) | 47 (23.5%) | |
| Unknown | 0 | 1 (0.5%) | |
| Stage | |||
| I | 11 (11.3%) | NA | |
| II | 26 (26.8%) | ||
| III | 24 (24.7%) | ||
| IV | 23 (23.7%) | ||
| Unknown or NA | 13 (13.4%) | ||
| Primary Lesion Site | |||
| Left | 14 (14.4%) | NA | |
| Right | 15 (15.5%) | ||
| Rectum | 45 (46.4%) | ||
| Metastatic | 11 (11.3%) | ||
| Unknown | 12 (12.4%) | ||
| CEA (ng/ml) Median (IQR) | 4.5 (2.3, 15.5) | 2.3 (1.5, 3.6) | <0.001 |
MDM: methylated DNA marker; CRC: colorectal cancer; IQR: interquartile range; CEA: carcinoembryonic antigen; NA: not applicable.
Figure 3:

The performance of individual MDMs, CEA, and a panel of MDMs in plasma to distinguish early CRC from healthy controls in Early Primary CRC Detection Pilot Study using MDMs in Plasma. MDM: methylated DNA marker; CEA: carcinoembryonic antigen; CRC: colorectal cancer; AUC: area under the curve.
MDM Plasma CRC Surveillance Study
We also applied selected MDMs to prospectively collected plasma samples from 160 age/sex-balanced patients including 60 healthy controls, 60 with resected CRC and NED, and 40 recurrent CRC after primary tumor resection (including 10 patients with recurrent CRC after adjuvant therapy and 30 patients with metastatic CRC on active palliative treatment). Patients had a median age of 55 years (IQR: 49–64 years) and median CEA level of 1.7 ng/ml (IQR: 1.1–3.2 ng/ml) (Table 2). Given the high sensitivity and specificity of the cross-validated model from Early Primary CRC Detection Pilot Study using MDMs in Plasma to detect stage IV CRC, we evaluated this model for the detection of recurrent CRC. As a quality control check, the potential drift in distribution of MDMs in Early Primary CRC Detection Pilot Study using MDMs in Plasma and in CRC surveillance study was assessed. The four MDMs (JAM3, ZNF671, SFMBT2, and CHST2) which contributed most to the performance of final model did not significantly drift between studies (Supplemental Figure 3). However, for the Early Primary CRC Detection Pilot Study using MDMs in Plasma, CEA was assayed from serum whereas for the CRC surveillance study CEA was assayed from plasma. A shift in the distribution of CEA was apparent as compared to other assayed markers (Supplemental Figure 4). This could not be explained by a difference in media (serum vs plasma) among patients where a clinically available serum CEA value was available (Supplemental Figure 5). To facilitate comparison and use the same CEA cut-off value from the model developed on Early Primary CRC Detection Pilot Study data, a scaling factor of 1.5 was derived by the ratio of mean CEA observed between controls from each study and then applying this scaling factor to cases and controls within the CRC surveillance study. At the specificity cutoff of 95% from the trained model of MDMs with or without CEA, the sensitivity for detecting recurrent CRC was 90% (76–97%) with an observed specificity of 90% (79–96%). The overall AUC of this MDM model with CEA was 0.96 (0.92–1), similar to the MDM model without CEA (p=0.45), but significantly better than CEA alone with AUC 0.79 (0.69–0.9) (p=0.0015).
Table 2:
Baseline characteristics of patients in MDM Plasma CRC Surveillance Study
| NED (n=60) | Healthy controls (n=60) | Recurrent CRC (n=40) | P value | |
|---|---|---|---|---|
| Age, median (IQR) | 56 (48–63.0) | 56 (48–65) | 53 (50–64) | 0.88 |
| Sex | ||||
| Female | 27 (45.0%) | 27 (45.0%) | 14 (35.0%) | 0.54 |
| Male | 33 (55.0%) | 33 (55.0%) | 26 (65.0%) | |
| Race | ||||
| White | 58 (96.7%) | 49 (81.7%) | 36 (90.0%) | 0.089 |
| Black | 1 (1.7%) | 3 (5.0%) | 2 (5.0%) | |
| Other | 1 (1.7%) | 8 (13.3%) | 2 (5.0%) | |
| Smoking | ||||
| Ever | 27 (45.0%) | 25 (41.7%) | 14 (35.0%) | 0.61 |
| Never | 33 (55.0%) | 35 (58.3%) | 26 (65.0%) | |
| Stage | ||||
| I | 3 (5.0%) | 1 (2.5%) | 0.0625 | |
| II | 5 (8.3%) | 4 (10.0%) | ||
| III | 26 (43.3%) | 8 (20.0%) | ||
| IV | 23 (38.3%) | 26 (65.0%) | ||
| Unknown | 3 (5.0%) | 1 (2.5%) | ||
| Primary Lesion Site | ||||
| Left | 25 (41.7%) | 20 (50.0%) | 0.8668 | |
| Right | 16 (26.7%) | 8 (20%) | ||
| Rectum | 18 (30%) | 12 (30%) | ||
| Unknown | 1 (1.7%) | 0 (0.0%) | ||
| Recurrence Site | ||||
| Liver | 24 (60.0%) | NA | ||
| Lung | 25 (62.5%) | |||
| Lymph nodes | 23 (57.5%) | |||
| Peritoneum | 12 (30% | |||
| Ovary | 4 (10.0%) | |||
| Brain | 2 (5.0%) | |||
| Pelvic Sidewall | 2 (5.0%) | |||
| Bone | 1 (2.5%) | |||
| Adrenal gland | 1 (2.5%) | |||
| Mediastinal | 1 (2.5%) | |||
| RECIST Lesion Burden (cm) Median (IQR) | 4.0 (2.0, 7.6) | NA | ||
| CEA (ng/ml) Median (IQR) | 1.5 (0.9–2.2) | 1.6 (1.1–2.1) | 7.2 (2.2–28.8) | <0.001 |
| Adjusted CEA (ng/ml) Median (IQR) | 2.2 (1.4, 3.3) | 2.4 (1.7, 3.2) | 10.8 (3.3, 43.3) | <0.001 |
NED: no radiographic evidence of disease; MDM: methylated DNA marker; CRC: colorectal cancer; IQR: interquartile range; CEA: carcinoembryonic antigen.
The trained model of MDMs with or without CEA had predicted probabilities of recurrent CRC of 0.32 (0.20–0.45) and 0.30 (0.19–0.43), respectively, for NED patients. (Figure 4) The trained model of MDMs with or without CEA detected recurrent CRC from patients with NED at the immediate previous follow-up with 80% (44–97%) sensitivity and detected metastatic CRC on active palliative treatment with 93% (78–99%) sensitivity. The panel of MDMs with or without CEA detected recurrent CRC liver metastases with 100% (86–100%) sensitivity, lung metastases with 89% (52–100%) sensitivity, and peritoneal/nodal metastases with 57% (18–90%) sensitivity. Lesions with RECIST sum >4 cm were detected with 100% (81–100%) sensitivity and ≤4 cm with 83% (59–96%) sensitivity. The panel of MDMs with or without CEA detected recurrent rectal cancer with 92% (62–100%) sensitivity, left-sided colon cancer with 95% (75–100%) sensitivity, and right-sided colon cancer with 75% (35–97%) sensitivity. As with the early detection study, model accuracy among those with NED or recurrent/metastatic disease was not impacted by primary tumor location (rectum vs left colon vs right colon).
Figure 4:

Performance of the cross-validated model from Early Primary CRC Detection Pilot Study using MDMs in Plasma in patients from MDM Plasma CRC Surveillance Study. (A) The distribution of the probability of CRC being present in the surveillance dataset for each of the patient groups. The blue shaded area indicates where the algorithm derived from training data made positive calls in the test set. (B) ROC curves of MDM models with and without CEA and CEA alone for recurrent CRC versus normal controls with corresponding AUCs and 95% confidence intervals. CRC: colorectal cancer; ROC: receiver operating characteristic; NED: no radiographic evidence of disease; AUC: area under the curve; MDM: methylated DNA marker.
Discussion
In this study, we applied MDMs previously identified by next-generation methylome sequencing on primary tumor tissues to assess concordance of MDMs in independent primary compared to metastatic tissue derived DNA. Next, TELQAS assays with high analytic sensitivity for candidate MDMs were applied for the early detection of primary CRC. This technique was then applied to independent cases, disease controls and healthy controls to demonstrate feasibility for detection of recurrent CRC in patients on or after therapy.
Most importantly, the findings establish that tumor-specific epigenetic alterations are highly concordant in primary and metastatic CRC including synchronous and metachronous metastasis despite potential for clonal selection after the use of neoadjuvant or adjuvant chemotherapy. There appears to be minimal epigenetic drift over time and exposure to therapy. In MDM tissue study, we found that the levels of all the 14 selected MDMs were concordant between left-side primary and metastatic CRC. Further, in contrast to sequencing large or bespoke panels of DNA mutations or sequencing based methylation measurement [27,28], targeted assays for a relatively small group of cross-validated methylation markers appears to show potential clinical utility to detect early-stage disease in the screening setting, or recurrence/metastatic disease. While high specificity was shown relative to healthy controls, nearly a third of patients who were NED after surgery were predicted to have CRC by the panel suggesting possible “minimal residual disease”.
The MDM pilot study for early detection was run to explore accuracy in late stage disease and, accordingly, to select MDM cut-off levels that could be applied in the surveillance study. It should be further emphasized that the MDM pilot study in plasma does not represent a fully optimized assay method for screening and early stage detection, and further studies are underway. However, it is clear from this plasma MDM assay in its current form appears to detect stage IV disease with high accuracy. Previous studies that prospectively assessed the accuracy of several methylation markers from cfDNA with relatively large sample sizes showed sensitivities from 48% to 90% for stage I-IV CRC [14–16]. Using TELQAS assays, we demonstrated that multiple novel MDMs had excellent accuracy in detecting primary CRC at all stages; we also observed increasing sensitivity with increasing stage confirming the expected positive correlation between tumor burden and the quantity of plasma cfDNA [29]. However, these observations on liquid biopsy for early detection of CRC should be viewed with some caution. The case-control design of the present study did not permit estimates of positive predictive value and may bias estimates of sensitivity, especially for early stage disease and the number of stage I cases was relatively small.
Our findings in recurrent/metastatic CRC are supported by others in the literature. Methylated BCAT1/IKZF1 and WIF1/NPY have been shown to be superior to CEA as a prognostic marker for patients with CRC after surgical resection or during adjuvant chemotherapy [13,30–32]. In 397 CRC patients under surveillance after curative management, methylated BCAT1/IKZF1 had only 68% (95% CI 48–84%) sensitivity in predicting CRC recurrence [13]. In the present study after showing high concordance of MDM levels in primary and metastatic CRC and demonstrating high accuracy in detecting primary CRC, we validated the plasma MDM assay in a locked-down model in an independent cohort of patients with extra-intestinal recurrent/metastatic CRC without intact primary in the surveillance setting. As expected, at high specificity cutoff, MDMs with or without CEA were capable of detecting recurrent/metastatic CRC with superior sensitivity, especially in patients with liver or lung metastasis, those with tumor burden greater than 4 cm based on RECIST measurement, and those with left-side primary CRC. The performance of this model appeared robust, despite either prior curative intent treatment of primary CRC or the presence of ongoing palliative chemotherapy.
On the other hand, our current study had limited sample size as it was a cross-sectional analysis of plasma samples collected in the setting of a prospective cohort study. Our model was able to predict the increased probability of the presence of CRC in patients with NED compared to healthy controls. But the number of recurrence events from patients with NED remained low. To further evaluate the ability of these novel MDMs in predicting CRC recurrence and detecting “minimal residual disease” compared to current standard of care, longer follow-up is required with sufficient numbers of events to exclude sub-clinical recurrence in those with NED.
In summary, MDM levels are highly concordant in primary and metastatic CRC. Plasma assay of novel CRC-associated MDMs discovered from primary CRC appear to reliably detect early CRC with extremely high sensitivity for late stage disease. These plasma MDMs in the same model can also detect distantly recurrent/metastatic CRC with promising accuracy. The clinical utility of MDMs for non-invasive post-treatment surveillance and treatment monitoring in CRC warrants further evaluation in longitudinal studies with sufficient follow-up.
Supplementary Material
Statement of translational relevance:
There is a clear need for noninvasive screening and surveillance tools with better performance and lower cost for patients with colorectal cancer (CRC). Methylated DNA markers (MDMs) offer several advantages over existing tools for such purpose. We previously reported the discovery of novel CRC-associated MDMs. In this study, we explored the feasibility of their liquid biopsy application for early detection of primary CRC and for non-invasive detection of recurrent CRC. In contrast to what has been observed with DNA mutation markers, we found that MDM levels were highly concordant in primary and metastatic CRC. Plasma assay of novel CRC-associated MDMs discovered from primary CRC appeared to reliably detect early CRC with intact primary. These plasma MDMs in the same model can also detect distantly recurrent/metastatic CRC with promising accuracy. The clinical utility of MDMs for non-invasive post-treatment surveillance and treatment monitoring in CRC warrants further evaluation in longitudinal studies.
Acknowledgements:
Financial support: This work was supported by CA214679 (to Dr. John Kisiel) and the 2019 Conquer Cancer Foundation of American Society of Clinical Oncology/Boehringer Ingelheim Endowed Young Investigator Award in Gastrointestinal Cancer (to Dr. Hao Xie). Reagents, TELQAS assays and plasma samples for Aim 2 were provided by Exact Sciences (Madison WI).
Footnotes
Conflict of interest statement: Mayo Clinic and Exact Sciences own intellectual property under which Dr. David Ahlquist and Dr. John Kisiel, and Mr. Douglas Mahoney and Mr. William Taylor are listed as inventors; they and Mayo Clinic have rights to receive royalties through a contracted services agreement with Exact Sciences. Drs. Allawi, Kaiser and Lidgard are employees of Exact Sciences.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, et al. Screening of colorectal cancer: present and future. Expert review of anticancer therapy 2017;17:1131–46. [DOI] [PubMed] [Google Scholar]
- [3].Sargent D, Sobrero A, Grothey A, O’Connell MJ, Buyse M, Andre T, et al. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20,898 patients on 18 randomized trials. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009;27:872–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pita-Fernández S, Alhayek-Aí M, González-Martín C, López-Calviño B, Seoane-Pillado T, Pértega-Díaz S. Intensive follow-up strategies improve outcomes in nonmetastatic colorectal cancer patients after curative surgery: a systematic review and meta-analysis. Annals of oncology : official journal of the European Society for Medical Oncology 2015;26:644–56. [DOI] [PubMed] [Google Scholar]
- [5].Primrose JN, Perera R, Gray A, Rose P, Fuller A, Corkhill A, et al. Effect of 3 to 5 years of scheduled CEA and CT follow-up to detect recurrence of colorectal cancer: the FACS randomized clinical trial. JAMA 2014;311:263–70. [DOI] [PubMed] [Google Scholar]
- [6].Akin O, Brennan SB, Dershaw DD, Ginsberg MS, Gollub MJ, Schöder H, et al. Advances in oncologic imaging: update on 5 common cancers. CA: a cancer journal for clinicians 2012;62:364–93. [DOI] [PubMed] [Google Scholar]
- [7].Tan E, Gouvas N, Nicholls RJ, Ziprin P, Xynos E, Tekkis PP. Diagnostic precision of carcinoembryonic antigen in the detection of recurrence of colorectal cancer. Surgical oncology 2009;18:15–24. [DOI] [PubMed] [Google Scholar]
- [8].Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA oncology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tie J, Wang Y, Springer S, Kinde I, Wong H-L, Kosmider S, et al. Serial circulating tumor DNA (ctDNA) and recurrence risk in patients (pts) with resectable colorectal liver metastasis (CLM). JCO 2016;34:e15131–e15131. [Google Scholar]
- [10].Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science translational medicine 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625–34. [DOI] [PubMed] [Google Scholar]
- [12].García-Saenz JA, Ayllón P, Laig M, Acosta-Eyzaguirre D, García-Esquinas M, Montes M, et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC cancer 2017;17:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Young GP, Pedersen SK, Mansfield S, Murray DH, Baker RT, Rabbitt P, et al. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer medicine 2016;5:2763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63:317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Science translational medicine 2020;12. [DOI] [PubMed] [Google Scholar]
- [16].Pedersen SK, Symonds EL, Baker RT, Murray DH, McEvoy A, van Doorn SC, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC cancer 2015;15:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cohen SA, Yu M, Baker K, Redman M, Wu C, Heinzerling TJ, et al. The CpG island methylator phenotype is concordant between primary colorectal carcinoma and matched distant metastases. Clinical epigenetics 2017;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shiovitz S, Bertagnolli MM, Renfro LA, Nam E, Foster NR, Dzieciatkowski S, et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology 2014;147:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Taylor WR, Kisiel JB, Yab TC, Mahoney DW, Smyrk TC, Boardman LA, et al. 109 Discovery of Novel DNA Methylation Markers for the Detection of Colorectal Neoplasia: Selection by Methylome-Wide Analysis. Gastroenterology 2014;146:S–30. [Google Scholar]
- [20].Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer 2012;118:2623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pottekat A, Allawi HT, Boragine GT, Kaiser MW, Sander T, Krueger C, et al. Abstract 4598: A comprehensive assessment of the impact of preanalytical variables on cell free DNA and circulating tumor cells in blood In: Clinical Research (Excluding Clinical Trials): American Association for Cancer Research; 07012018. p. 4598. [Google Scholar]
- [22].Kisiel JB, Dukek BA, V S R Kanipakam R, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology (Baltimore, Md.) 2019;69:1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. Journal of chiropractic medicine 2016;15:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Allawi HT, Giakoumopoulos M, Flietner E, Oliphant A, Volkmann C, Aizenstein B, et al. Abstract 712: Detection of lung cancer by assay of novel methylated DNA markers in plasma In: Clinical Research (Excluding Clinical Trials): American Association for Cancer Research; 07012017. p. 712. [Google Scholar]
- [25].Breiman L Random Forests. Machine Learning 2001;45:5–32. [Google Scholar]
- [26].Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). European journal of cancer (Oxford, England : 1990) 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- [27].Chan KCA, Jiang P, Chan CWM, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proceedings of the National Academy of Sciences of the United States of America 2013;110:18761–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017;168:571–74. Available from: http://www.sciencedirect.com/science/article/pii/S0092867417301150. [DOI] [PubMed] [Google Scholar]
- [29].Donaldson J, Park BH. Circulating Tumor DNA: Measurement and Clinical Utility. Annual review of medicine 2018;69:223–34. [DOI] [PubMed] [Google Scholar]
- [30].André T, Vernerey D, Mineur L, Bennouna J, Desrame J, Faroux R, et al. Three Versus 6 Months of Oxaliplatin-Based Adjuvant Chemotherapy for Patients With Stage III Colon Cancer: Disease-Free Survival Results From a Randomized, Open-Label, International Duration Evaluation of Adjuvant (IDEA) France, Phase III Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36:1469–77. [DOI] [PubMed] [Google Scholar]
- [31].Taieb J, Taly V, Vernerey D, Bourreau C, Bennouna J, Faroux R, et al. Analysis of circulating tumour DNA (ctDNA) from patients enrolled in the IDEA-FRANCE phase III trial: Prognostic and predictive value for adjuvant treatment duration. Ann Oncol 2019;30:v867. [Google Scholar]
- [32].Murray DH, Symonds EL, Young GP, Byrne S, Rabbitt P, Roy A, et al. Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival. Journal of cancer research and clinical oncology 2018;144:1741–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


