Abstract
Bile acids have recently emerged as key metabolic hormones with beneficial impacts in multiple metabolic diseases. We previously discovered that hepatic bile acid overload distally modulates glucose and fatty acid metabolism in adipose tissues to exert anti-obesity effects. However, the detailed mechanisms that explain the salutary effects of serum bile acid elevation remain unclear. Here, proteomic profiling identified a new hepatokine, Orosomucoid (ORM) that governs liver-adipose tissue crosstalk. Hepatic ORMs were highly induced by both genetic and dietary bile acid overload. To address the direct metabolic effects of ORM, purified ORM proteins were administered during adipogenic differentiation of 3T3-L1 cells and mouse stromal vascular fibroblasts. ORM suppressed adipocyte differentiation and strongly inhibited gene expression of adipogenic transcription factors such as C/EBPβ, KLF5, C/EBPα, and PPARγ. Taken together, our data clearly suggest that bile acid-induced ORM secretion from the liver blocks adipocyte differentiation, potentially linked to anti-obesity effect of bile acids.
Keywords: bile acid, hepatokine, orosomucoid, adipogenesis, lipogenesis, obesity
INTRODUCTION
The increasing incidence of obesity accelerates the prevalence of type 2 diabetes mellitus, fatty liver disease, and cardiovascular disease [1,2]. The liver is one of the largest internal organs in the human body and plays a central role in energy metabolism. Recent studies revealed that liver-derived bile acids are not only natural surfactants emulsifying dietary lipids, but also important metabolic regulators of glucose and lipid homeostasis [3]. Pharmacological modulation of bile acid signaling pathways generally exhibits metabolic benefits in obesity and diabetes [4,5]. Dietary supplementation with bile acids slows body weight gain in mice and improves metabolic parameters [6]. In addition, we showed that hepatic bile acid accumulation in liver-specific farnesoid X receptor (FXR) and small heterodimer partner (SHP) double knockout (L-DKO) mice is coupled with systemic metabolic improvements in aging-induced obesity. These observations suggest bile acid-induced inter-organ crosstalk prevent obesity and insulin resistance [7].
The liver is an important secretory organ that synthesizes and secretes the majority of the serum proteome including carrier proteins, apolipoproteins, coagulation factors, as well as hormones [8]. To this end, many liver-secreted hormones (i.e., hepatokines) such as fetuin-A/B, retinol-binding protein 4, and selenoprotein P, fibroblast growth factor 21 (FGF21), Tsukushi, and apolipoprotein J link hepatic signaling to systemic metabolic regulation [9,10,11,12]. Among them, FGF21 is known to be associated with hepatic bile acid signaling, which improves lipid and carbohydrate metabolism. FGF21 is induced mainly by peroxisome proliferator-activated receptor alpha (PPARα) [13] and by other nuclear receptors like FXR [14], that respond to hepatic bile acids. However, since hepatic Fgf21 expression in aged L-DKO mice is not altered [7], it cannot explain the metabolic benefit of L-DKO mice and likely requires additional factors that mediates bile acid effects.
In this study, we profiled the serum proteome in global FXR/SHP double knockout (DKO) and L-DKO mice to identify novel mediators of hepatic bile acid signaling. We discovered Orosomucoid (ORM) was abundantly expressed and highly induced by hepatic bile acids in the liver. We find that ORM inhibits adipocyte differentiation through suppression of critical adipogenic transcription factors. Taken together, our studies reveal bile acid regulation of the hepatokine ORM exerts anti-adipogenic activity during adipocyte differentiation. These findings are an important advance in the understanding of how bile acid acids can signal via liver-secreted hormones to coordinate adipocyte function.
MATERIALS AND METHODS
Animal Study
Global FXR/SHP double knockout (DKO) and liver-specific FXR/SHP double knockout (L-DKO) mice were previously described [7]. C57BL/6J WT (BCM Center for Comparative Medicine) and littermate control mice having no Alb-cre transgene were used as controls for DKO and L-DKO, respectively. Mice were maintained on a normal chow (NC) diet (TD.00217, ENVIGO). Dietary bile acid overload mice were generated by feeding 0.5% cholic acid (CA) diet for 1 day (TD.110811, ENVIGO). All mice were humanely euthanized by isoflurane inhalation followed by cervical dislocation. All animal studies and procedures were approved by the Institutional Animal Care and Use Committee of the Baylor College of Medicine.
Cell Culture and Adipocyte Differentiation
Orosomucoid protein isolated from bovine plasma (G3643, Sigma Aldrich) was used for in vitro experiments. 3T3-L1 mouse preadipocytes (SP-L1-F, Zen-Bio) were maintained in DMEM (10–013-CV, Corning) supplemented with 10% bovine calf serum (BCS;16777–206, HyClone) and penicillin-streptomycin (30–002-CI, Corning). Post-confluent 3T3-L1 cells were induced by adding differentiation medium (MDI) containing DMEM/F12 (10–090-CV, Corning), 10% fetal bovine serum (FBS; 35011-CV, Corning), 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; I5879, Sigma-Aldrich), 1 μM dexamethasone (D1756, Sigma-Aldrich), 5 μg/mL insulin (I9278, Sigma-Aldrich) and 0.5 μM rosiglitazone (71740, Cayman Chemical). Three days post-differentiation, medium was replaced with DMEM/F12, 10% FBS, and 5 μg/mL insulin for 2 days and maintained in DMEM/F12, 10% FBS thereafter.
SVF preadipocyte differentiation
The stromal vascular fraction (SVF) was isolated from inguinal white adipose tissue of 8-week-old male mice. Fat pads were digested with Collagenase Type I (17100–017, Gibco) in a 37°C water bath for 1h. After digestion, cells were centrifuged at 700g for 5 min and supernatant was discarded. The cell pellets were resuspended with DMEM/F12 containing 10% FBS and penicillin/streptomycin and passed through 100 μm cell strainer to remove debris. Centrifugation was repeated to wash the pellet and then cells were plated in 24-well plates. Once 80% confluence was reached, the cells were induced to differentiate by adding MDI and maintained as described for 3T3-L1 cells.
Oil Red O staining
Cells were washed with 1X Phosphate-Buffered Saline (PBS; 21040-CV, Corning) once and then fixed with 10% neutral buffered formalin (89370–094, VWR) for 20 min at room temperature. After washing with 1X PBS, fixed cells were pre-incubated with 100% propylene glycol (P4347, Sigma-Aldrich) for 5 min and then stained with Oil Red O solution (I1516, Sigma-Aldrich) for 15 min. Then, cells were washed with 85% propylene glycol once and followed with 1X PBS twice. Cell images were taken using an inverted microscope. To quantify the amount of accumulated lipids, Oil Red O was extracted from the stained cells using 250 μl of 100% isopropanol and two aliquots of 100 μl were transferred to a 96-well assay plate (9017, Corning). The OD values were measured at 520 nm using a plate reader (Multiskan™ FC Microplate Photometer, Thermo Scientific).
Western Blotting
Whole-cell lysates were prepared using RIPA buffer (20mM Tris-HCL pH7.4, 150mM NaCl, 1mM EDTA, 1% Triton-X100, 1% sodium deoxycholate, 0.1% SDS) supplemented with Xpert Protease Inhibitor Cocktail (P3100, GenDEPOT) and Phosphatase Inhibitor Cocktail (P3200, GenDEPOT) for 30 min at 4°C followed by centrifugation at 14,000 rpm for 10 min. Supernatants were denatured in 2x Laemmli Sample buffer (1610737, Bio-Rad Laboratories) at 95°C for 5 min and loaded in 12% Mini-PROTEAN TGX Precast Protein Gels (4561046, Bio-Rad Laboratories). Proteins were transferred to PVDF membrane (162–0177, Bio-Rad Laboratories) by electrophoresis for 30 min at 100V. Primary antibody was incubated overnight at 4°C and followed by secondary anti-rabbit IgG HRP-conjugated antibody (7074, Cell Signaling) incubation for 1h. Immunoreactive bands were visualized by SuperSignal West Pico or Femto (34087 or 34095, Thermo Scientific) reagents using AZURE c600 IMAGING SYSTEM (Azure Biosystems). Detailed information regarding antibodies can be found in Supplementary Table 2.
Real-time Quantitative PCR
Total RNA was isolated by PureXtract RNAsol reagent (R6101, GenDEPOT) and converted into complementary DNA by the qScript Reverse Transcriptase Kit (95047, Quanta Bio). Relative mRNA expression was measured by quantitative PCR using the Light Cycler 480 Real-Time PCR System (Roche) with KAPA SYBR FAST qPCR Master Mix (2X) (KK4611, KAPA Biosystems) and quantified by the comparative cycle threshold method (ΔΔCt method) with normalization to β-actin. Primer information is listed in Supplementary Table 1.
Serum Secretome Analysis
Mouse serum was pre-cleared by Proteome Purify 2 Mouse Serum Immunodepletion resin (MIDR002, RnD Systems) to remove albumin and immunoglobulins. Depleted serum was subjected to Mass Spectrometry-based whole proteome analysis [15,16]. Briefly, trypsinized serum peptides were extracted by 50% acetonitrile/0.1% formic acid and 80% acetonitrile/0.1% formic acid solution. The proteome was profiled by nano-liquid chromatography-tandem mass spectrometry analysis with a nano-LC1000 coupled to a Thermo Q-Exactive (Thermo Fisher Scientific). The relative amount was normalized as the intensity-based absolute quantification algorithm and normalized to the intensity-based fraction of the total (iFOT).
Statistical Analysis
All data are presented as mean ± SEM. Independent t-test was used for comparisons between two groups, otherwise a one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used. Statistical analysis was performed using Prism 8 software (GraphPad Software). A probability value of p < 0.05 was considered statistically significant, unless otherwise indicated.
RESULTS
Bile Acid Overload induces Hepatokine ORM Expression.
Liver-specific farnesoid X receptor (FXR) and small heterodimer partner (SHP) double knockout (L-DKO) in mice results in hepatic bile acid accumulation that is associated with systemic metabolic improvements in aging-induced obesity [7]. To screen for novel hepatokines that mediate the systemic metabolic effects during bile acid accumulation, we performed serum proteomics in DKO and L-DKO mice. We identified 84 and 25 serum proteins that were increased in DKO and L-DKO, respectively (Fig. 1A). Overlap of the elevated proteins revealed 15 proteins in common between DKO and L-DKO (Fig. 1B), consisting of many known liver-secreted proteins (Fig. 1C). Among them, ORM1 and ORM2 were the most abundant in both datasets (Fig. 1D). ORM, also known as alpha1-acid glycoprotein, is an acute phase protein found in blood [17]. While serum ORM is exclusively synthesized by the liver, other tissues such as heart, stomach, lung, and adipose tissue secrete ORM locally [17,18]. ORM was abundant in serum from DKO mice compared to control (Supplementary Fig. S1), consistent with the liver proteomics data.
Figure 1. Differentially Expressed Circulating Proteins in DKO and L-DKO serum.
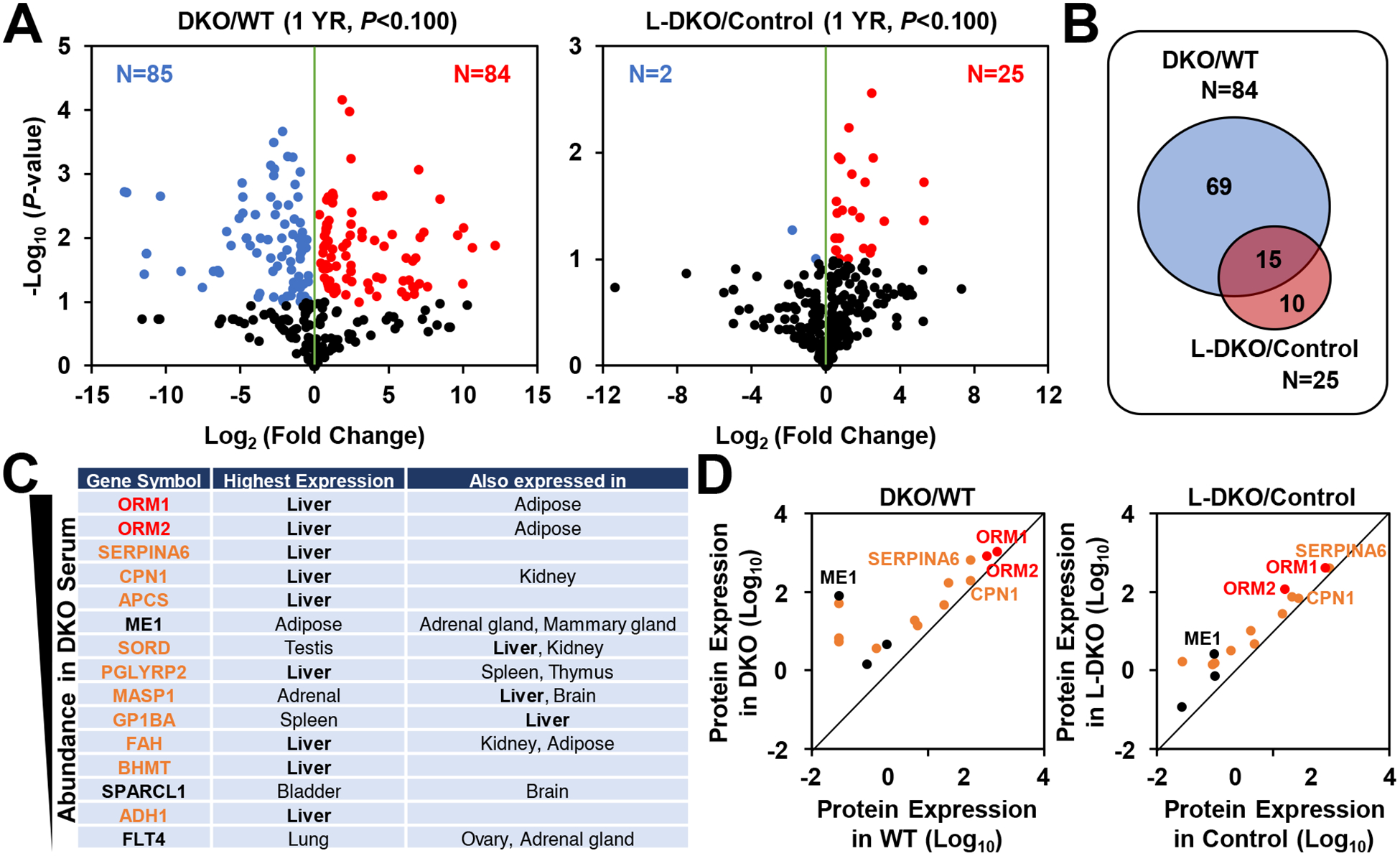
(A) Quantitative proteomic analysis. Volcano plot display of the results from statistical analysis of the serum proteome in 1 year-old DKO and L-DKO serum. (B) Venn diagram showing the distribution of upregulated proteins. (C) List of commonly-upregulated proteins in DKO and L-DKO serum. Among them, ORMs were highly-induced and mainly secreted from the liver. (D) Relative protein expression in DKO and L-DKO. ORMs are the most abundant. Relative protein expression was expressed as iFOT (intensity fraction of total).
To confirm the origin of serum ORM during bile acid accumulation, we quantified mRNA expression in the liver in genetic and dietary bile acid overload. In DKO, hepatic expression of all Orm isoforms were induced (Fig. 2A), whereas L-DKO markedly increased the Orm2 isoform. (Fig. 2B). Notably, these changes were also consistent in the dietary bile acid overload induced by 0.5% cholic acid diet feeding (Fig. 2C). Next, we analyzed Orm expression in adipose tissues, which express low levels of Orm1 [18]. In control mice, Orm1 and Orm2 were highly enriched in the liver compared with white and brown adipose tissues (Fig. 2D). Importantly, bile acid accumulation in L-DKO mice induced Orm2 expression in the liver but had no effect in adipose tissues. Of note, although Orm3 was increased in the liver and adipose tissues, the absolute level of Orm3 was extremely low compared to Orm1 and Orm2 [19] and was undetectable in our serum proteome data. Therefore, these results suggest hepatic bile acid accumulation induces liver-specific expression of ORMs, which might contribute to metabolic functions in other tissues.
Figure 2. Hepatic Orm Induction by Hepatic Bile Acid Overload.
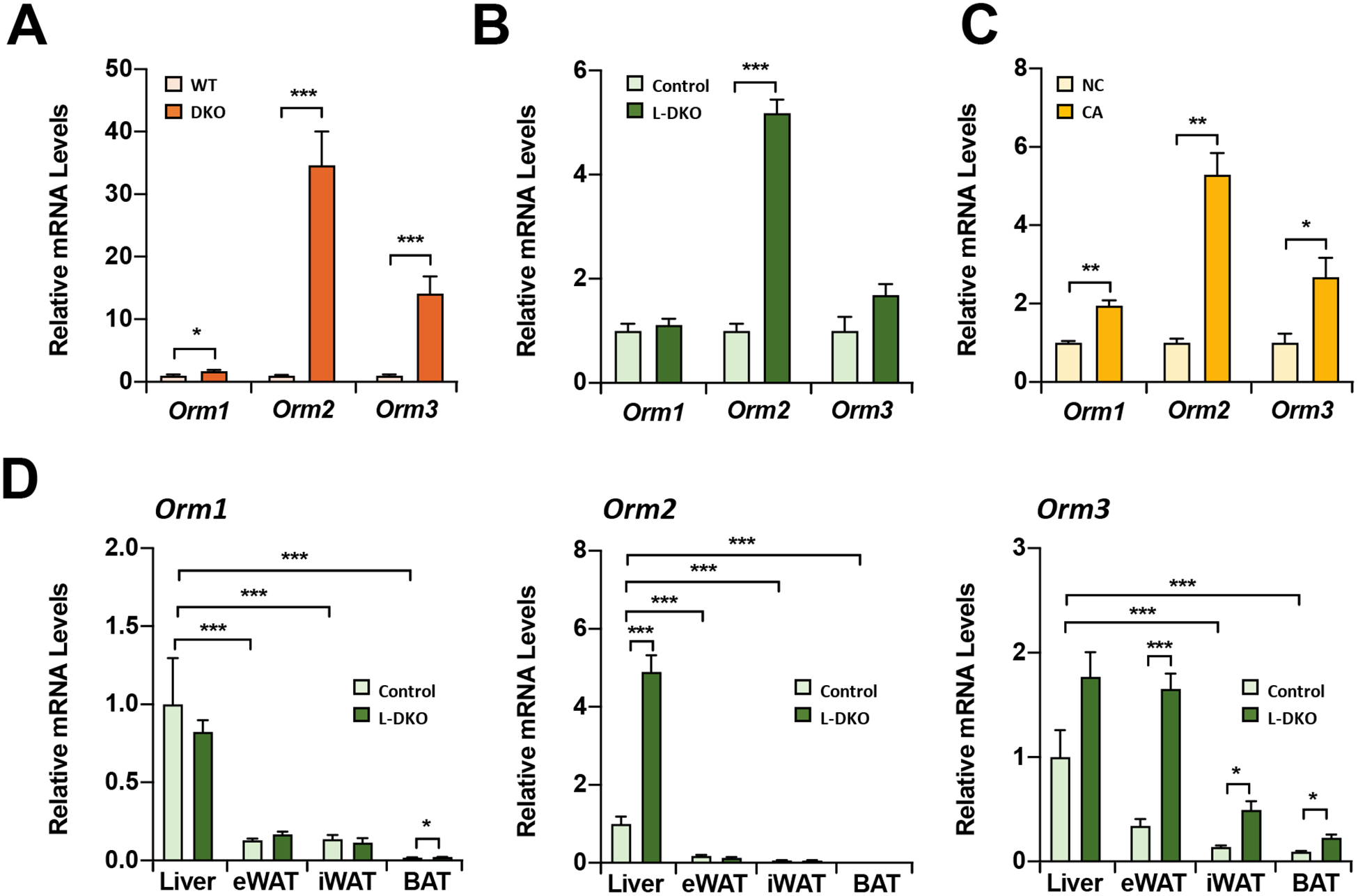
Orm expression in the liver of (A) 1 year-old DKO and (B) 1 year-old L-DKO mice. (C) Hepatic Orm expressions after cholic acid diet (0.5%) feeding for one day (CA). NC, normal chow. (D) The comparison of Orm expressions in the liver and adipose tissues of 1 year-old L-DKO mice. eWAT, epididymal white adipose tissue. iWAT, inguinal WAT. BAT, brown adipose tissue. *p<0.05, **p<0.01, and ***p<0.001
Hepatokine ORM Directly Inhibits Adipogenesis.
Bile acids exert strong anti-obesity effects [6,20]. To investigate whether ORM mediates these effects through modification of adipocyte formation, we exposed preadipocytes to ORM. We utilized 3T3-L1 preadipocytes, the best characterized in vitro model of adipocyte differentiation [21], and treated them with differentiating medium (MDI) in the absence or in the presence of purified ORMs at various concentrations. Oil Red O staining demonstrated decreased lipid accumulation with ORM treatment in a dose-dependent manner, indicating reduced adipogenesis (Fig. 3A). Quantification of intracellular lipid droplets confirmed that ORM treatment dramatically reduced the adipogenic potential of 3T3-L1 cells (Fig. 3B). Furthermore, the anti-adipogenic activity of ORM protein was validated in mouse SVF preadipocytes from inguinal white adipose tissue (Figs. 3C and 3D). Taken together, these results demonstrate that ORM treatment suppresses adipocyte differentiation.
Figure 3. Suppression of Adipogenesis by ORM.
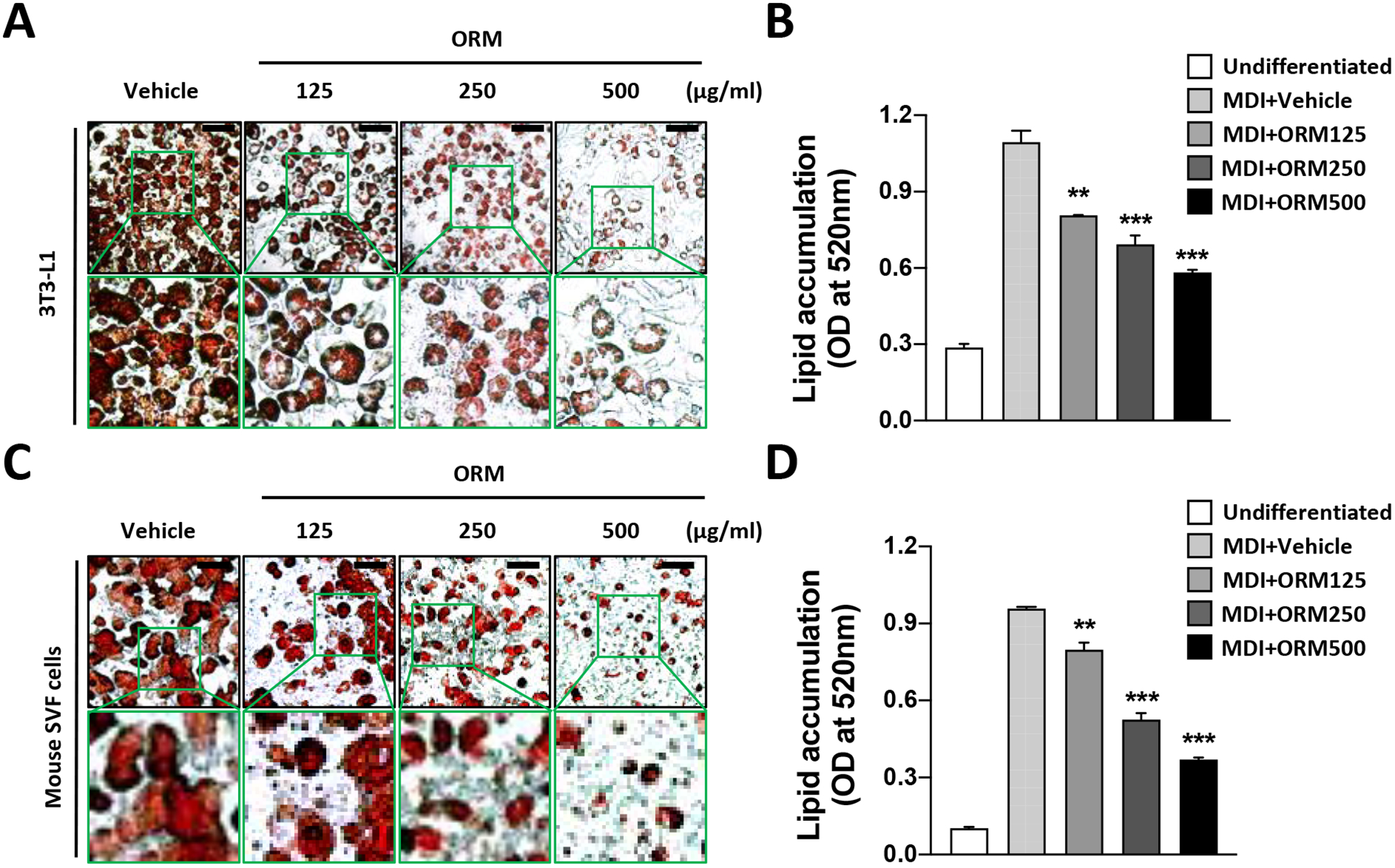
(A) and (B) Differentiation of 3T3-L1 preadipocytes were induced by differentiation medium (MDI) with various concentrations of ORM protein (0, 125, 250, and 500 μg/ml). After 7 days, (A) Oil Red O staining of differentiated adipocytes. (B) Quantification of accumulated lipids by lysis of Oil Red O. (C) and (D) SVF preadipocytes from iWAT were differentiated (MDI) with various concentrations of ORM (0, 125, 250, and 500 μg/ml) for 7 days. (C) Visualization and (D) quantification after Oil Red O staining. Scale bar= 100 μm. *p<0.05, **p<0.01, and ***p<0.001 as compared to vehicle-treated differentiated cells.
Adipogenesis consists of 2 phases, an initial commitment step of preadipocytes (day 0 to day 2) and the subsequent differentiation (day 2 to day 7), which are controlled by temporal expression of multiple transcription factors [22,23]. To test which phase is impacted by ORM, we added purified ORM protein to the differentiation media at various time points as indicated in Supplementary Fig. S2A. Interestingly, ORM suppressed lipid accumulation when administered early in adipogenesis (Supplementary Fig. S2B and C). However, ORM treatment after Day 3 did not inhibit adipogenesis or lipid accumulation. These results suggest that ORM primarily impairs the early commitment steps of adipocyte differentiation.
ORM Coordinately Regulates Adipogenic Transcription Factors.
Adipocyte differentiation is regulated by timely activation and inhibition of multiple transcription factors, including activation of CCAAT/enhancer binding protein beta and delta (C/EBPβ and C/EBPδ) (3) and Kruppel-like factor 4 and 5 (KLF4 and KLF5) in the early phase (day 1 and 2) [24,25], and two master adipocyte transcription factors, C/EBPα and PPARγ to accelerate adipocyte differentiation in the late stage (day 2 to 7) [26]. Conversely, other transcription factors including KLF2, Pref-1, and GATA2/3 inhibit adipogenesis [27,28,29]. To test the impact of ORM on expression of early adipogenic transcription factors, we treated 3T3-L1 preadipocytes with purified ORM during MDI differentiation medium for 2 days. Intriguingly, ORM specifically inhibited the positive regulators Cebpb and Klf5 by day 2 of differentiation (Fig. 4A), although it did not affect anti-adipogenic transcription factors at this time (Fig. 4B). We further examined ORM effects on the two master regulators of adipogenesis, C/EBPα and PPARγ. In accord with the strong effects on differentiation, mRNA (Fig. 4C) and protein levels (Fig. 4D) of C/EBPα and PPARγ were dramatically inhibited by ORM treatment on day 2, with persistent effects until terminal differentiation. Interestingly, ORM treatment to mature adipocytes also significantly suppressed both adipogenic and lipogenic genes (Supplementary Fig. S3). These data indicate that ORM potently suppresses early adipogenic transcription factors C/EBPβ and KLF5, resulting in loss C/EBPα and PPARγ activation, and ultimately inhibiting adipocyte differentiation.
Figure 4. Decreased Expressions of Adipogenic Transcription Factors by ORM.
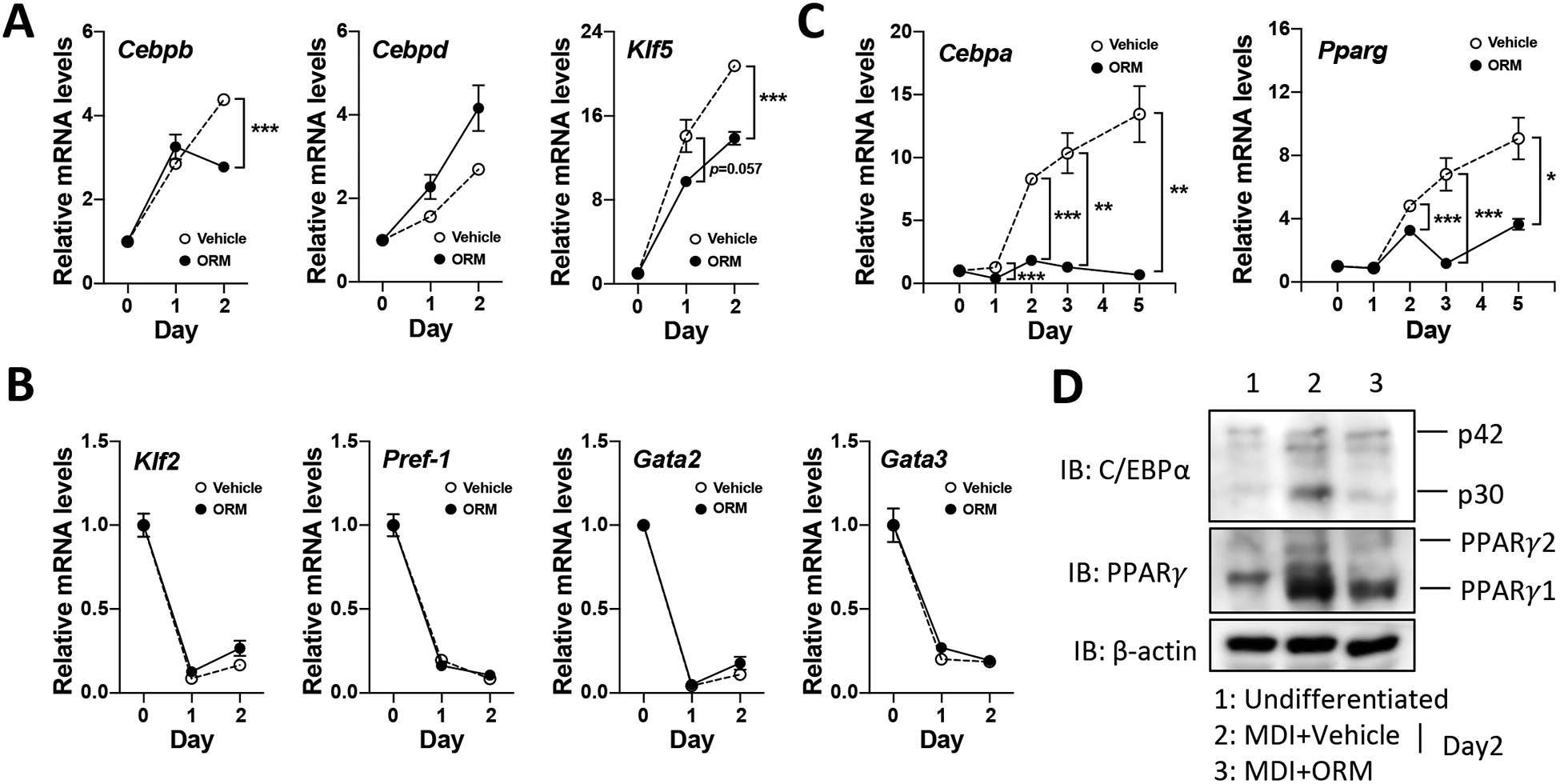
(A) Changes of early adipogenic transcription factor expression by ORM treatment (500 μg/ml) at day 1 and 2. (B) Expression of anti-adipogenic regulators. qPCR was performed measure gene expression. (C) The mRNA expression of master adipogenic genes (Cebpa and Pparg) were analyzed by qPCR (up to day 5). *p<0.05, **p<0.01, and ***p<0.001 as compared to vehicle-treated differentiated cells. (D) The protein level of C/EBPα and PPARγ was examined at day 2 by Western blotting. β-actin was used as a loading control.
DISCUSSION
Bile acids are important signaling molecules that impact glucose and fatty acid metabolism. We previously demonstrated that hepatic bile acid overload in DKO and L-DKO mice has beneficial metabolic effects [7]. Intriguingly, L-DKO fully phenocopied the DKO mice showing anti-obesity and insulin sensitizing effects, uncovering a specific role of hepatic bile acid signaling in the regulation of systemic energy homeostasis, particularly in adipose tissues. These results raise the possibility of undiscovered liver-adipose tissue crosstalk that is mediated by hepatic bile acids.
To identify liver-secreted hormones, we analyzed the serum proteome in DKO and L-DKO and nominated ORM as a novel bile acid-regulated hepatokine. ORM is an abundant plasma protein, accounting for approximately 1% of total plasma protein [30]. In our model, ORM is primarily induced by bile acid overload in the liver, suggesting the liver is the major organ that produces circulating ORM. Although the biological function of ORM has not been clearly defined, it has been suggested that ORM binds to and carries lipophilic drugs, which might affect drug delivery [31]. A limited number of studies suggest a metabolic function of ORM. In muscle, ORM expression is increased by fatigue to enhance exercise endurance through regulation of glycogen metabolism [32,33]. Recently, ORM1 was identified as an adipokine that acts centrally and locally to decrease food intake and adipose tissue inflammation, respectively [18,34]. However, ORM signaling, particularly ORM2, as a mediator of bile acid function has not been studied.
Our results define metabolic functions of ORM as a novel bile acid-regulated hepatokine, acting on adipocyte. During adipocyte differentiation, ORM antagonizes adipogenic signaling by inhibition of adipogenic transcription factors at early stages of adipogenesis. We next asked whether ORM directly affects mitotic clonal expansion, which is one of the earliest events and an essential step for preadipocyte commitment [30]. However, ORM treatment did not affect cell proliferation of 3T3-L1 cells during early adipogenesis (Supplementary Fig. S4), suggesting that ORM may affect another pathway to suppress adipogenic transcription factors. Detailed mechanisms of ORM function during adipogenesis remain unclear.
At the molecular level, ORM activates AMP-activated kinase (AMPK) to increase muscle glycogen accumulation [32]. AMPK is a crucial cellular energy sensor that is activated at low cellular energy level (increasing AMP:ATP and ADP:ATP ratios) or by certain hormones (e.g., adiponectin) [35]. In adipose tissues, AMPK activation promotes energy-producing processes such as fatty acid oxidation and inhibits energy-storing processes like lipid synthesis [36]. Direct AMPK stimulation suppresses 3T3-L1 adipocyte differentiation and reduces adipose tissue mass [37], which is consistent with ORM effects. Therefore, it is likely that ORM effects are partly mediated by AMPK activation. Further studies are needed to determine how ORM acts on AMPK signaling in adipocytes.
In conclusion, we discovered a novel bile acid-regulated hepatokine ORM and characterized its metabolic function in adipocytes. The primary impact of ORM is to decrease C/EBPβ and KLF5 expression at an early stage of adipogenesis and subsequently abolish the expression of key adipogenic transcription factors C/EBPα and PPARγ. Taken together, our data suggest that hepatic bile acid overload induces secretion of the hepatokine ORM to modulate adipocyte differentiation and adipose tissue function.
Supplementary Material
HIGHLIGHTS.
Hepatic bile acid overload stimulates hepatokine orosomucoid secretion.
Hepatokine orosomucoid distally acts on adipocytes to suppress adipogenesis and lipogenesis, potentially exerting anti-obesity effect.
Orosomucoid directly inhibits multiple early-adipogenic transcription factors, interfering C/EBPα and PPARγ activation.
ACKNOWLEDGEMENT
We thank to Dr. Jessica Scott (Baylor College of Medicine) for technical assistance.
FUNDINGS
This work was funded by the Assistant Secretary of Defense for Health Affairs endorsed by the DOD PRMRP Discovery Award (No. W81XWH-18-1-0126 to K.H.K.) and the American Heart Association Career Development Award (19CDA34660196 to K.H.K.). This work was also supported in part by the Baylor College of Medicine Nutrition and Obesity Pilot and Feasibility Fund (to K.H.K. and A.R.C.), the National Institutes of Health (R01DK114356 to S.M.H. and P01DK113954 to D.D.M.), the American Diabetes Association (1-18-IBS-105 to S.M.H.) and the R.P. Doherty Jr.-Welch Chair in Science (Q-0022 to D.D.M.).
Abbreviations
- C/EBP
CCAAT-enhancer-binding proteins
- DKO
FXR/SHP double knockout
- FGF21
fibroblast growth factor 21
- FXR
farnesoid X receptor
- KLF
kruppel-like factor
- L-DKO
liver-specific FXR/SHP double knockout
- ORM
orosomucoid
- PPAR
peroxisome proliferator-activated receptor
- SHP
small heterodimer partner
- WAT
white adipose tissue
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Calle EE, Kaaks R, Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms, Nat Rev Cancer 4 (2004) 579–591. [DOI] [PubMed] [Google Scholar]
- [2].Saxton SN, Clark BJ, Withers SB, Eringa EC, Heagerty AM, Mechanistic Links Between Obesity, Diabetes, and Blood Pressure: Role of Perivascular Adipose Tissue, Physiol Rev 99 (2019) 1701–1763. [DOI] [PubMed] [Google Scholar]
- [3].Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B, Role of bile acids and bile acid receptors in metabolic regulation, Physiol Rev 89 (2009) 147–191. [DOI] [PubMed] [Google Scholar]
- [4].Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, Yoshihara E, Perino A, Jacinto S, Lukasheva Y, Atkins AR, Khvat A, Schnabl B, Yu RT, Brenner DA, Coulter S, Liddle C, Schoonjans K, Olefsky JM, Saltiel AR, Downes M, Evans RM, Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance, Nat Med 21 (2015) 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K, TGR5-mediated bile acid sensing controls glucose homeostasis, Cell Metab 10 (2009) 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J, Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation, Nature 439 (2006) 484–489. [DOI] [PubMed] [Google Scholar]
- [7].Kim KH, Choi S, Zhou Y, Kim EY, Lee JM, Saha PK, Anakk S, Moore DD, Hepatic FXR/SHP axis modulates systemic glucose and fatty acid homeostasis in aged mice, Hepatology 66 (2017) 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kuscuoglu D, Janciauskiene S, Hamesch K, Haybaeck J, Trautwein C, Strnad P, Liver - master and servant of serum proteome, J Hepatol 69 (2018) 512–524. [DOI] [PubMed] [Google Scholar]
- [9].Meex RCR, Watt MJ, Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance, Nat Rev Endocrinol 13 (2017) 509–520. [DOI] [PubMed] [Google Scholar]
- [10].Jung TW, Yoo HJ, Choi KM, Implication of hepatokines in metabolic disorders and cardiovascular diseases, BBA Clin 5 (2016) 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mouchiroud M, Camire E, Aldow M, Caron A, Jubinville E, Turcotte L, Kaci I, Beaulieu MJ, Roy C, Labbe SM, Varin TV, Gelinas Y, Lamothe J, Trottier J, Mitchell PL, Guenard F, Festuccia WT, Joubert P, Rose CF, Karvellas CJ, Barbier O, Morissette MC, Marette A, Laplante M, The Hepatokine TSK does not affect brown fat thermogenic capacity, body weight gain, and glucose homeostasis, Mol Metab 30 (2019) 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Seo JA, Kang MC, Yang WM, Hwang WM, Kim SS, Hong SH, Heo JI, Vijyakumar A, Pereira de Moura L, Uner A, Huang H, Lee SH, Lima IS, Park KS, Kim MS, Dagon Y, Willnow TE, Aroda V, Ciaraldi TP, Henry RR, Kim YB, Apolipoprotein J is a hepatokine regulating muscle glucose metabolism and insulin sensitivity, Nat Commun 11 (2020) 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Potthoff MJ, Kliewer SA, Mangelsdorf DJ, Endocrine fibroblast growth factors 15/19 and 21: from feast to famine, Genes Dev 26 (2012) 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cyphert HA, Ge X, Kohan AB, Salati LM, Zhang Y, Hillgartner FB, Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21, J Biol Chem 287 (2012) 25123–25138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jung SY, Choi JM, Rousseaux MW, Malovannaya A, Kim JJ, Kutzera J, Wang Y, Huang Y, Zhu W, Maity S, Zoghbi HY, Qin J, An Anatomically Resolved Mouse Brain Proteome Reveals Parkinson Disease-relevant Pathways, Mol Cell Proteomics 16 (2017) 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Suh DS, Kwon BS, Hwang SY, Lee NK, Choi KU, Song YJ, Kim KH, Large cell neuroendocrine carcinoma arising from uterine endometrium with rapidly progressive course: report of a case and review of literature, Int J Clin Exp Pathol 12 (2019) 1412–1417. [PMC free article] [PubMed] [Google Scholar]
- [17].Fournier T, Medjoubi NN, Porquet D, Alpha-1-acid glycoprotein, Biochim Biophys Acta 1482 (2000) 157–171. [DOI] [PubMed] [Google Scholar]
- [18].Lee YS, Choi JW, Hwang I, Lee JW, Lee JH, Kim AY, Huh JY, Koh YJ, Koh GY, Son HJ, Masuzaki H, Hotta K, Alfadda AA, Kim JB, Adipocytokine orosomucoid integrates inflammatory and metabolic signals to preserve energy homeostasis by resolving immoderate inflammation, J Biol Chem 285 (2010) 22174–22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Porez G, Gross B, Prawitt J, Gheeraert C, Berrabah W, Alexandre J, Staels B, Lefebvre P, The hepatic orosomucoid/alpha1-acid glycoprotein gene cluster is regulated by the nuclear bile acid receptor FXR, Endocrinology 154 (2013) 3690–3701. [DOI] [PubMed] [Google Scholar]
- [20].Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K, Targeting bile-acid signalling for metabolic diseases, Nat Rev Drug Discov 7 (2008) 678–693. [DOI] [PubMed] [Google Scholar]
- [21].Ruiz-Ojeda FJ, Ruperez AI, Gomez-Llorente C, Gil A, Aguilera CM, Cell Models and Their Application for Studying Adipogenic Differentiation in Relation to Obesity: A Review, Int J Mol Sci 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tong Q, Hotamisligil GS, Molecular mechanisms of adipocyte differentiation, Rev Endocr Metab Disord 2 (2001) 349–355. [DOI] [PubMed] [Google Scholar]
- [23].Gregoire FM, Smas CM, Sul HS, Understanding adipocyte differentiation, Physiol Rev 78 (1998) 783–809. [DOI] [PubMed] [Google Scholar]
- [24].Mota de Sa P, Richard AJ, Hang H, Stephens JM, Transcriptional Regulation of Adipogenesis, Compr Physiol 7 (2017) 635–674. [DOI] [PubMed] [Google Scholar]
- [25].Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R, Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation, Cell Metab 1 (2005) 27–39. [DOI] [PubMed] [Google Scholar]
- [26].Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM, Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity, Mol Cell 3 (1999) 151–158. [DOI] [PubMed] [Google Scholar]
- [27].Smas CM, Sul HS, Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation, Cell 73 (1993) 725–734. [DOI] [PubMed] [Google Scholar]
- [28].Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS, Function of GATA transcription factors in preadipocyte-adipocyte transition, Science 290 (2000) 134–138. [DOI] [PubMed] [Google Scholar]
- [29].Wu J, Srinivasan SV, Neumann JC, Lingrel JB, The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes, Biochemistry 44 (2005) 11098–11105. [DOI] [PubMed] [Google Scholar]
- [30].Tang QQ, Otto TC, Lane MD, Mitotic clonal expansion: a synchronous process required for adipogenesis, Proc Natl Acad Sci U S A 100 (2003) 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Israili ZH, Dayton PG, Human alpha-1-glycoprotein and its interactions with drugs, Drug Metab Rev 33 (2001) 161–235. [DOI] [PubMed] [Google Scholar]
- [32].Qin Z, Wan JJ, Sun Y, Wang PY, Su DF, Lei H, Liu X, ORM Promotes Skeletal Muscle Glycogen Accumulation via CCR5-Activated AMPK Pathway in Mice, Front Pharmacol 7 (2016) 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun Y, Qin Z, Wan JJ, Wang PY, Yang YL, Yu JG, Hu BH, Su DF, Luo ZM, Liu X, Estrogen weakens muscle endurance via estrogen receptor-p38 MAPK-mediated orosomucoid (ORM) suppression, Exp Mol Med 50 (2018) e463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sun Y, Yang Y, Qin Z, Cai J, Guo X, Tang Y, Wan J, Su DF, Liu X, The Acute-Phase Protein Orosomucoid Regulates Food Intake and Energy Homeostasis via Leptin Receptor Signaling Pathway, Diabetes 65 (2016) 1630–1641. [DOI] [PubMed] [Google Scholar]
- [35].Hardie DG, Ross FA, Hawley SA, AMPK: a nutrient and energy sensor that maintains energy homeostasis, Nat Rev Mol Cell Biol 13 (2012) 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Daval M, Foufelle F, Ferre P, Functions of AMP-activated protein kinase in adipose tissue, J Physiol 574 (2006) 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee H, Kang R, Bae S, Yoon Y, AICAR, an activator of AMPK, inhibits adipogenesis via the WNT/beta-catenin pathway in 3T3-L1 adipocytes, Int J Mol Med 28 (2011) 65–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


