Abstract
Macrophages are among the most common cells in the colorectal cancer microenvironment, but their prognostic significance is incompletely understood. Using multiplexed immunofluorescence for CD68, CD86, IRF5, MAF, MRC1 (CD206), and KRT (cytokeratins) combined with digital image analysis and machine learning, we assessed the polarization spectrum of tumor-associated macrophages in 931 colorectal carcinomas. We then applied Cox proportional hazards regression to assess prognostic survival associations of intraepithelial and stromal densities of M1-like and M2-like macrophages, while controlling for potential confounders, including stage and microsatellite instability status. We found that high tumor stromal density of M2-like macrophages was associated with worse cancer-specific survival, whereas tumor stromal density of M1-like macrophages was not significantly associated with better cancer-specific survival. High M1:M2 density ratio in tumor stroma was associated with better cancer-specific survival. Overall macrophage densities in tumor intraepithelial or stromal regions were not prognostic. These findings suggested that macrophage polarization state, rather than their overall density, was associated with cancer-specific survival, with M1- and M2-like macrophage phenotypes exhibiting distinct prognostic roles. These results highlight the utility of a multimarker strategy to assess the macrophage polarization at single cell resolution within the tumor microenvironment.
Keywords: colorectal neoplasms, immunology, macrophage polarization, immunofluorescence, tumor microenvironment
Introduction
Globally, colorectal cancer is the third most common malignancy and the second most common cause of cancer-related deaths (1). Colorectal cancers elicit variable degrees of antitumor immune response (2), and high densities of tumor-associated T cells are well-established favorable prognostic markers (3,4). However, the immune system may also facilitate tumor progression, as certain inflammatory pathways can enhance angiogenesis, activate tumor cell proliferation, inhibit apoptosis, promote extracellular matrix degradation and tumor invasion, or directly impair an antitumor immune response (5,6).
Macrophages are among the most common non-neoplastic cells in human tumors, including colorectal cancer (7). Macrophages can exhibit broad functional plasticity in response to environmental stimuli, and the concept of “macrophage polarization” has been recognized as a measure of phenotypic state at any given point in space and time (8,9). Traditionally, macrophages have been categorized as either pro-inflammatory M1-like macrophages and anti-inflammatory M2-like macrophages, although it is now generally recognized that a spectrum of phenotypic states exists between these two extremes of macrophage polarization (8,10). In particular, tumor associated macrophages (TAMs), exposed to multiple polarization stimuli in the tumor microenvironment, may exhibit mixed phenotypes that are not readily classified as clearly M1 or M2 (11).
Several studies have evaluated the prognostic significance of TAMs in colorectal cancer and have generally reported an association between higher macrophage densities and better survival (12-16), although contradictory reports exist (17-19). Notably, most of these studies have either evaluated macrophages using only the general macrophage marker CD68 (12,15), or have used single polarization markers to identify M1-like and M2-like populations (13,14,16,18,19), providing a limited view of macrophage phenotypic diversity.
To address this knowledge gap, we utilized multiplex immunofluorescence to identify and quantify macrophages in tumor epithelial and stromal regions and to estimate their polarization state across a M1:M2 spectrum in tissue microarrays (TMAs) of 931 colorectal cancer patients from two U.S.-based large prospective cohort studies. This multiplexed assay enabled us to characterize macrophage polarization by simultaneously measuring the expression of two markers associated with an M1-like antitumorigenic phenotype (CD86, IRF5) and two markers associated with an M2-like pro-tumorigenic phenotype (MAF, MRC1). Our primary hypotheses were that higher density of M1-like polarized macrophages, lower density of M2-like polarized macrophages, and higher M1:M2 density ratio would be associated with lower colorectal cancer-specific mortality. In secondary analyses, we evaluated the relationships between the densities of macrophage subsets and tumor and patient characteristics and assessed the prognostic role of macrophage densities in strata of microsatellite instability (MSI) status. This approach allowed us to define the prognostic significance and clinicopathologic correlates of both macrophage density and polarization in colorectal cancer with greater sophistication and accuracy than prior studies.
Methods
Study population
This study used 4,420 colorectal cancer cases that occurred in two U.S. nationwide prospective cohort studies, the Nurses’ Health Study (NHS, 121,701 women followed since 1976) (20) and the Health Professionals Follow-up Study (HPFS, 51,529 men followed since 1986) (21), including 931 cases with available follow-up data and adequate tissue specimens in TMAs (Table 1). We included both colon and rectal carcinomas in accordance with the colorectal continuum model (22). The medical records related to colorectal cancer were reviewed by study physicians, and tumor, node, metastases (TNM) stage was evaluated according to the American Joint Committee on Cancer (AJCC) criteria. The National Death Index was used to ascertain deaths and to identify unreported lethal colorectal cancer cases. Survival time was defined as the period from the date of colorectal cancer diagnosis to death or the end of follow-up (January 1, 2014 for HPFS; May 31, 2014 for NHS) for those who had not died.
Table 1.
Clinical, pathological, and molecular characteristics of colorectal cancer cases according to overall macrophage density.
| Macrophage density (ordinal quartiles from Q1 to Q4) |
||||||
|---|---|---|---|---|---|---|
| Characteristic* | All cases (N = 931) |
Q1 (N = 232) |
Q2 (N = 233) |
Q3 (N = 233) |
Q4 (N = 233) |
P value† |
| Sex | 0.088 | |||||
| Female (NHS) | 517 (56%) | 117 (50%) | 125 (54%) | 131 (56%) | 144 (62%) | |
| Male (HPFS) | 414 (44%) | 115 (50%) | 108 (46%) | 102 (44%) | 89 (38%) | |
| Mean age ± SD (years) | 68.9 ± 8.9 | 67.2 ± 8.8 | 70.0 ± 8.6 | 69.1 ± 8.9 | 69.4 ± 9.0 | 0.006 |
| Year of diagnosis | <0.001 | |||||
| 1995 or before | 304 (33%) | 107 (46%) | 82 (35%) | 57 (24%) | 58 (25%) | |
| 1996-2000 | 303 (33%) | 60 (26%) | 74 (32%) | 82 (35%) | 87 (37%) | |
| 2001-2008 | 324 (34%) | 65 (28%) | 77 (33%) | 94 (40%) | 88 (38%) | |
| Family history of colorectal cancer in first-degree relative(s) | 0.92 | |||||
| Absent | 738 (80%) | 187 (81%) | 185 (80%) | 183 (79%) | 183 (79%) | |
| Present | 190 (20%) | 44 (19%) | 47 (20%) | 49 (21%) | 50 (21%) | |
| Tumor location | 0.003 | |||||
| Cecum | 161 (17%) | 27 (12%) | 46 (20%) | 35 (15%) | 53 (23%) | |
| Ascending to transverse colon | 301 (32%) | 64 (28%) | 75 (32%) | 78 (34%) | 84 (36%) | |
| Descending to sigmoid colon | 283 (31%) | 85 (37%) | 75 (32%) | 67 (29%) | 56 (24%) | |
| Rectum | 182 (20%) | 56 (24%) | 36 (16%) | 51 (22%) | 39 (17%) | |
| Tumor differentiation | <0.001 | |||||
| Well to moderate | 843 (91%) | 217 (94%) | 221 (95%) | 210 (90%) | 195 (84%) | |
| Poor | 87 (9.4%) | 14 (6.1%) | 12 (5.2%) | 23 (9.9%) | 38 (16%) | |
| AJCC disease stage | 0.81 | |||||
| I | 199 (23%) | 51 (24%) | 52 (24%) | 45 (21%) | 51 (23%) | |
| II | 282 (33%) | 65 (30%) | 67 (31%) | 69 (32%) | 81 (37%) | |
| III | 249 (29%) | 65 (30%) | 59 (28%) | 64 (29%) | 61 (28%) | |
| IV | 134 (16%) | 33 (15%) | 35 (16%) | 39 (18%) | 27 (12%) | |
| MSI status | <0.001 | |||||
| Non-MSI-high | 750 (83%) | 207 (90%) | 197 (86%) | 184 (83%) | 162 (72%) | |
| MSI-high | 154 (17%) | 22 (9.6%) | 31 (14%) | 37 (17%) | 64 (28%) | |
| CIMP status | <0.001 | |||||
| Low/negative | 705 (82%) | 204 (92%) | 184 (85%) | 167 (80%) | 150 (69%) | |
| High | 159 (18%) | 17 (7.7%) | 32 (15%) | 43 (20%) | 67 (31%) | |
| Mean LINE-1 methylation level ± SD (%) | 62.5 ± 9.5 | 61.8 ± 9.9 | 61.6 ± 9.5 | 62.0 ± 8.9 | 64.7 ± 9.5 | 0.001 |
| KRAS mutation | 0.004 | |||||
| Wild-type | 535 (59%) | 122 (54%) | 122 (54%) | 139 (62%) | 152 (68%) | |
| Mutant | 368 (41%) | 106 (46%) | 104 (46%) | 86 (38%) | 72 (32%) | |
| BRAF mutation | <0.001 | |||||
| Wild-type | 771 (85%) | 206 (90%) | 203 (89%) | 187 (83%) | 175 (77%) | |
| Mutant | 139 (15%) | 24 (10%) | 25 (11%) | 39 (17%) | 51 (23%) | |
| PIK3CA mutation | 0.54 | |||||
| Wild-type | 712 (84%) | 186 (87%) | 170 (81%) | 178 (84%) | 178 (84%) | |
| Mutant | 136 (16%) | 29 (13%) | 39 (19%) | 35 (16%) | 33 (16%) | |
| Neoantigen load | 0.10 | |||||
| Q1 (lowest) | 104 (25%) | 28 (27%) | 27 (26%) | 27 (26%) | 22 (21%) | |
| Q2 | 104 (25%) | 32 (31%) | 31 (30%) | 24 (23%) | 17 (16%) | |
| Q3 | 104 (25%) | 21 (20%) | 28 (27%) | 26 (25%) | 29 (28%) | |
| Q4 (highest) | 104 (25%) | 23 (22%) | 18 (17%) | 27 (26%) | 36 (35%) | |
Percentage indicates the proportion of patients with a specific clinical, pathologic, or molecular characteristic among all patients or in strata of macrophage density in overall region.
To compare categorical data between the ordinal categories of macrophage density, the chi-square test was performed. To compare continuous variables, an analysis of variance was performed.
Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; HPFS, Health Professionals Follow-up Study; LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability; NHS, Nurses’ Health Study; SD, standard deviation.
Formalin-fixed paraffin-embedded tissue samples were collected, and hematoxylin and eosin (H&E)-stained tissue sections were reviewed by a single study pathologist (S.O.) to confirm the diagnosis and categorize tumor grade as high (≤50% gland formation) or low (>50% gland formation). Four histologic patterns of lymphocytic reactions, Crohn’s-like lymphoid reaction, peritumoral lymphocytic reaction, intratumoral periglandular reaction, and tumor-infiltrating lymphocytes (TILs), were scored using a 4-tiered scale (23). DNA was extracted from formalin-fixed paraffin-embedded tumor blocks using QIAmp DNA Mini Kit (Qiagen, Valencia, CA, USA) (24) to evaluate MSI and CpG island methylator phenotype (CIMP) status (25), KRAS, BRAF, and PIK3CA mutations (26), and neoantigen load (Supplemental Table S1). TMAs were constructed to include 1-4 cores (diameter 0.6 mm) from each tumor, selected to best represent overall morphology (27). Patients who received any type of neoadjuvant therapy prior to resection were excluded from the TMA set. The study was conducted in accordance with the U.S. Common Rule. All participants gave written informed consent for the study. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health (Boston, MA, USA), and those of participating registries as required.
Multiplex immunofluorescence to evaluate macrophage polarization
We designed a 7-plex immunofluorescence panel to characterize macrophage polarization, incorporating a previously-validated tyramide signal amplification technique to maximize sensitivity (Fig. 1; Supplemental Table S2) (28,29). Based on the published literature (8,10,31,11-14,16,18,19,30), a broad variety of target antigen candidates were screened with available antibody clones to evaluate suitability for panel inclusion, related to the level of evidence supporting association with macrophage polarization states in human tissue and antibody clone performance in formalin-fixed paraffin-embedded tissue specimens. Candidate antibodies were first evaluated using standard immunohistochemistry to assess their degree of expression pattern correspondence with prior studies and their biologically expected distribution using a bank of colorectal cancer tissue samples and normal lymphoid tissue controls. Correspondence was assessed both at a tissue level and at the level of subcellular localization. Once contextual specificity was assured, single-plex immunofluorescence was then performed, combining the selected antibody clones with Opal fluorophores (Akoya Biosciences, Hopkinton, MA, USA) to further evaluate performance and to assess signal-to-noise ratio. Based on this testing, we compiled a multiplex immunofluorescence panel consisting of a pan-macrophage marker (CD68), two M1 phenotype markers [CD86 (32,33), IRF5 (10,31)], two M2 phenotype markers [MRC1 (10,34), MAF (30,35,36)], cytokeratin, and DAPI (Supplemental Table S2). We optimized the sequence of the antibodies and their concentrations, as well as the fluorophore pairing and concentration, confirming similar staining patterns in standard immunohistochemistry and multiplex immunofluorescence (Supplemental Fig. S1). Immunofluorescence staining was performed using an automated Leica Bond RX Research Stainer (Leica Biosystems, Buffalo, IL, USA), as described in Supplemental Fig. S2.
Figure 1.
Quantitative multiplexed evaluation of macrophage polarization, location, and density in colorectal cancer tissue microarrays. (A) Multiplex immunofluorescence image of a representative tumor core. (B-D) Machine learning based image processing, including tissue category classification (B), cell segmentation (C), and cell type classification (D) to identify macrophages in tumor intraepithelial and stromal regions. (E) Examples of macrophages with M1-like, M2-like, and mixed expression phenotypes. (F) Assignment of polarization state for each macrophage across the M1:M2 spectrum using a scaled, four marker index. Plot shows the distribution of M1:M2-index values for all macrophages (N=367,264) across 931 tumors. Macrophages present within the two-sided 30% tails of the graph were classified as M1-like or M2-like for the main analysis. (G) Distribution of overall macrophage density and M1:M2 density ratio across 931 tumors. Scale bars indicate 100 μm (A) or 10 μm (E).
The immunofluorescence slides were scanned using the Vectra 3.0 Automated Quantitative Pathology Imaging System (Akoya Biosciences) equipped with a 20× objective and configured to capture images at discrete intervals from 420 nm to 720 nm (Supplemental Fig. S3). We used slides stained with a single fluorophore to generate a spectral library, enabling the unmixing of spectrally overlapping fluorophores, as well as a negative control slide processed with no fluorophores to profile background autofluorescence. Utilizing the captured spectral data and the fluorophore spectral library, the TMA core images were decomposed into individual fluorophore components using the inForm software package (Akoya Biosciences). Representative images were then processed with pathologist-trained machine learning algorithms implemented using the inForm software package to perform tissue category segmentation (tumor epithelium, stroma, other; using KRT expression to delineate epithelial areas), cell segmentation (using the DAPI signal to identify nuclei), and cell type classification [macrophage, tumor cell, other; using a combination of cellular morphology, CD68 expression (macrophages), and KRT expression (tumor cells) to distinguish these phenotypes] (Fig. 1A-D; Supplemental Fig. S3).
Data were exported from inForm at the single cell level for each core and further processed using the R statistical programming language version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria). We inspected the consistency of the data across the TMAs using violin plots of cell densities and marker intensities. Additionally, we used t-Distributed Stochastic Neighbor Embedding (tSNE) to project high dimensional, cell level fluorophore intensity data into two-dimensional space for visual inspection of potential heterogeneity in fluorophore intensities across all TMAs. For each CD68+ macrophage, an M1:M2 polarization index was calculated based on the scaled (0-1) mean intensities (CD86, MRC1: cytoplasmic; IRF5, MAF: nuclear) of the four polarization markers using the formula “(CD86×IRF5)/(MRC1×MAF)” (Fig. 1). In this approach, macrophages with a high index value are more strongly M1-polarized, while macrophages with a low index value are more strongly M2-polarized. As estimates of the prevalence of M1-like and M2-like macrophages in colorectal cancer vary significantly according to the markers and methods used for their detection (13,14,16), we opted to examine the distribution of the M1:M2-index across all macrophages within the 931 tumors and to categorize equal percentages of macrophages as M1-like or M2-like. For the main analyses, macrophages with M1:M2-index values within the highest 30% were considered M1-like, while those with M1:M2-index values within the lowest 30% were considered M2-like. These data were then combined into aggregate measures at the level of individual cores and individual tumors, with densities of M1-like and M2-like macrophages measured separately. To evaluate the robustness of this approach, we also evaluated a range (10-50%) of M1:M2-index cut points, in addition to the 30% cut-off, for classifying macrophages as either M1-like or M2-like. For a secondary analysis, we defined macrophage populations (CD86+, IRF5+, MAF+, MRC1+) based on visually estimated fluorophore signal intensity cut-off points for each polarization marker. The code used to calculate the M1:M2 index and assign macrophage phenotypes is freely available (https://github.com/MPE-Lab).
Statistical analysis
The statistical analyses were conducted using SAS software (version 9.4, SAS Institute, Cary, NC, USA). All P values were two-sided. Our primary hypothesis tested relationships between the intraepithelial and stromal densities of M1-like and M2-like macrophages, as well as the M1:M2 density ratio, and cancer-specific mortality using multivariable adjusted Cox proportional hazards regression models. We used the stringent two-sided α level of 0.005 (37). All other analyses represented secondary analyses, where we interpreted our data cautiously, in addition to using the stringent α level of 0.005.
We used the chi-square test and Spearman’s rank correlation test to assess the relationships between macrophage densities and clinicopathologic features, as appropriate. Cumulative survival probabilities were estimated using the Kaplan-Meier method and were compared using the log-rank test. Univariable and multivariable Cox proportional hazards regression models were used to estimate the hazard ratios (HRs) and confidence intervals (CIs) of cancer-specific survival according to ordinal macrophage density or density ratio quartiles (Q1-Q4). We assessed the statistical interaction between macrophage densities or density ratio (low vs. high) and MSI status (high vs. non-high) or year of diagnosis (1995 or before vs. 1996-2000 vs. 2001-2008) in relation to cancer-specific survival using the Wald test for the cross-product in multivariable-adjusted Cox regression models. We utilized the inverse probability weighting (IPW) method, using covariate data from the 4,420 incident colorectal cancer cases with or without tumor tissue, to adjust for selection bias due to tissue availability (38). A detailed description of the statistical methods is shown in Supplemental Table S3.
Results
Multiplex immunofluorescence assay to evaluate macrophage density and polarization
Multiplex immunofluorescence staining to evaluate macrophage density and polarization was completed across all cohort TMAs as a single batch, yielding data for 2,942,057 tumor cells and 367,024 macrophages in 931 colorectal carcinomas. Cell densities (Supplemental Fig. S4) and marker intensities (Supplemental Fig. S5) were consistent across TMAs. T-SNE analysis of single cell-level marker intensities did not show evidence of TMA-level batch effect (Supplemental Fig. S6), suggesting that the assay had performed in a technically uniform manner across all TMAs and that the data were suitable for aggregate analysis.
Data from 1,682 cores from 931 tumors (average 1.8 cores/tumor, range 1-4) were processed to calculate macrophage densities. Macrophages were present in all tumors, and their density was higher in tumor stromal regions (median 866/mm2, IQR 485-1,414/mm2) than within tumor intraepithelial regions (median 163/mm2, IQR 79-323/mm2). Intraepithelial and stromal macrophage densities showed moderate positive correlation (Spearman rank correlation coefficient (rho)=0.66). Each macrophage was also assigned a position on macrophage polarization spectrum via an M1:M2-index based on joint measurement of two M1 (CD86, IRF5) and two M2 (MAF, MRC1) markers (Fig. 1). When examining the extreme 30% ends of this spectrum, the densities of M1-like and M2-like macrophages were not strongly correlated (intraepithelial: Spearman rho=0.09; stromal: Spearman rho=−0.16).
The Spearman rho between macrophage densities in two randomly chosen cores of tumors with two or more cores were 0.57 and 0.60 for intraepithelial and stromal macrophages, 0.53 and 0.59 intraepithelial and stromal M1-like macrophages, and 0.61 and 0.65 for intraepithelial and stromal M2-like macrophages (Supplemental Fig. S7), indicating moderate core-to-core correlation.
Survival analyses according to macrophage densities and M1:M2 density ratio
During the median follow-up time of 12.2 years (IQR 8.7-16.3 years) for censored cases, there were 567 all-cause deaths, including 288 colorectal cancer-specific deaths. Kaplan-Meier analysis demonstrated an association between higher stromal macrophage density and better cancer-specific survival (Log-rank P=0.001) (Supplemental Fig. S8). However, when adjusted for potential confounders in multivariable Cox regression models, overall macrophage densities in tumor intraepithelial region (Ptrend=0.37), tumor stroma (Ptrend=0.33), or overall region (Ptrend=0.73) were not significantly associated with cancer-specific survival (Table 2, Supplemental Table S4).
Table 2.
Macrophage density, M1-like macrophage density, M2-like macrophage density, and M1:M2 macrophage density ratio in tumor intraepithelial and stromal regions and patient survival with inverse probability weighting (IPW).
| Colorectal cancer-specific survival |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|
| No. of cases |
No. of events |
Univariable HR (95% CI)* |
Multivariable HR (95% CI)*,† |
No. of events |
Univariable HR (95% CI)* |
Multivariable HR (95% CI)*,† |
|
| Tumor intraepithelial region | |||||||
| Macrophage density | |||||||
| Q1 | 232 | 79 | 1 (referent) | 1 (referent) | 149 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 75 | 0.95 (0.68-1.34) | 1.05 (0.75-1.47) | 139 | 1.05 (0.79-1.39) | 1.03 (0.78-1.38) |
| Q3 | 233 | 72 | 0.97 (0.68-1.37) | 0.96 (0.67-1.37) | 141 | 1.06 (0.79-1.42) | 1.01 (0.74-1.38) |
| Q4 | 233 | 62 | 0.72 (0.51-1.02) | 0.83 (0.55-1.25) | 138 | 0.92 (0.69-1.23) | 0.92 (0.67-1.28) |
| Ptrend‡ | 0.096 | 0.37 | 0.64 | 0.65 | |||
| M1-like macrophage density | |||||||
| Q1 | 232 | 88 | 1 (referent) | 1 (referent) | 152 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 70 | 0.79 (0.56-1.11) | 0.82 (0.59-1.16) | 143 | 0.93 (0.70-1.24) | 0.92 (0.69-1.23) |
| Q3 | 233 | 79 | 0.84 (0.61-1.17) | 0.87 (0.62-1.23) | 143 | 0.93 (0.70-1.24) | 0.93 (0.68-1.26) |
| Q4 | 233 | 51 | 0.51 (0.35-0.73) | 0.71 (0.49-1.05) | 129 | 0.76 (0.57-1.02) | 0.87 (0.63-1.18) |
| Ptrend‡ | 0.001 | 0.12 | 0.092 | 0.40 | |||
| M2-like macrophage density | |||||||
| Q1 | 232 | 72 | 1 (referent) | 1 (referent) | 144 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 73 | 1.02 (0.71-1.45) | 0.98 (0.69-1.40) | 139 | 1.09 (0.81-1.45) | 1.03 (0.76-1.40) |
| Q3 | 233 | 70 | 1.13 (0.80-1.60) | 1.23 (0.86-1.76) | 141 | 1.26 (0.96-1.67) | 1.25 (0.92-1.69) |
| Q4 | 233 | 73 | 1.06 (0.75-1.50) | 1.07 (0.74-1.54) | 143 | 1.08 (0.80-1.44) | 1.03 (0.75-1.41) |
| Ptrend‡ | 0.61 | 0.48 | 0.41 | 0.57 | |||
| M1:M2 density ratio | |||||||
| Q1 | 226 | 80 | 1 (referent) | 1 (referent) | 146 | 1 (referent) | 1 (referent) |
| Q2 | 226 | 72 | 0.92 (0.65-1.29) | 1.20 (0.84-1.71) | 135 | 1.00 (0.75-1.33) | 1.12 (0.83-1.51) |
| Q3 | 227 | 76 | 0.93 (0.66-1.31) | 0.98 (0.69-1.39) | 142 | 0.91 (0.67-1.22) | 0.95 (0.70-1.29) |
| Q4 | 226 | 48 | 0.48 (0.32-0.71) | 0.59 (0.40-0.89) | 127 | 0.70 (0.52-0.95) | 0.72 (0.53-0.98) |
| Ptrend‡ | <0.001 | 0.013 | 0.019 | 0.028 | |||
| Tumor stromal region | |||||||
| Macrophage density | |||||||
| Q1 | 232 | 80 | 1 (referent) | 1 (referent) | 155 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 83 | 1.05 (0.75-1.46) | 1.30 (0.91-1.85) | 156 | 1.09 (0.82-1.44) | 1.12 (0.83-1.52) |
| Q3 | 233 | 71 | 0.93 (0.66-1.31) | 1.12 (0.79-1.60) | 129 | 0.93 (0.70-1.24) | 0.94 (0.69-1.28) |
| Q4 | 233 | 54 | 0.61 (0.42-0.88) | 0.81 (0.54-1.21) | 127 | 0.77 (0.58-1.02) | 0.78 (0.56-1.08) |
| Ptrend‡ | 0.008 | 0.33 | 0.043 | 0.091 | |||
| M1-like macrophage density | |||||||
| Q1 | 232 | 90 | 1 (referent) | 1 (referent) | 160 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 88 | 0.93 (0.68-1.28) | 1.01 (0.74-1.38) | 152 | 1.03 (0.78-1.36) | 1.14 (0.86-1.52) |
| Q3 | 233 | 61 | 0.58 (0.41-0.83) | 0.70 (0.48-1.03) | 134 | 0.69 (0.52-0.93) | 0.73 (0.53-1.00) |
| Q4 | 233 | 49 | 0.45 (0.31-0.66) | 0.69 (0.47-1.01) | 121 | 0.70 (0.53-0.94) | 0.85 (0.62-1.16) |
| Ptrend‡ | < 0.001 | 0.019 | 0.002 | 0.070 | |||
| M2-like macrophage density | |||||||
| Q1 | 232 | 66 | 1 (referent) | 1 (referent) | 142 | 1 (referent) | 1 (referent) |
| Q2 | 233 | 63 | 0.96 (0.66-1.39) | 1.15 (0.80-1.67) | 136 | 1.11 (0.82-1.48) | 1.31 (0.95-1.79) |
| Q3 | 233 | 85 | 1.55 (1.10-2.18) | 1.90 (1.32-2.73) | 157 | 1.59 (1.20-2.11) | 1.88 (1.38-2.57) |
| Q4 | 233 | 74 | 1.21 (0.84-1.73) | 1.48 (1.01-2.17) | 132 | 1.15 (0.85-1.55) | 1.30 (0.93-1.81) |
| Ptrend‡ | 0.061 | 0.005 | 0.089 | 0.024 | |||
| M1:M2 density ratio | |||||||
| Q1 | 231 | 88 | 1 (referent) | 1 (referent) | 154 | 1 (referent) | 1 (referent) |
| Q2 | 232 | 74 | 0.82 (0.58-1.15) | 1.01 (0.72-1.42) | 135 | 0.99 (0.74-1.32) | 1.11 (0.83-1.48) |
| Q3 | 232 | 74 | 0.77 (0.56-1.08) | 0.82 (0.58-1.15) | 148 | 0.91 (0.68-1.21) | 0.93 (0.69-1.25) |
| Q4 | 231 | 50 | 0.46 (0.31-0.67) | 0.58 (0.39-0.86) | 126 | 0.64 (0.47-0.87) | 0.67 (0.48-0.92) |
| Ptrend‡ | < 0.001 | 0.005 | 0.005 | 0.010 | |||
IPW was applied to reduce bias due to the availability of tumor tissue after cancer diagnosis (see “Statistical Analysis” subsection for details).
The multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long-interspersed nucleotide element-1 methylation level. A backward elimination with a threshold P of 0.05 was used to select variables for the final models.
Ptrend value was calculated across the ordinal quartiles of each macrophage density or M1 to M2 density ratio within tumor intraepithelial and stromal regions in the IPW-adjusted Cox regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio; IPW, inverse probability weighting.
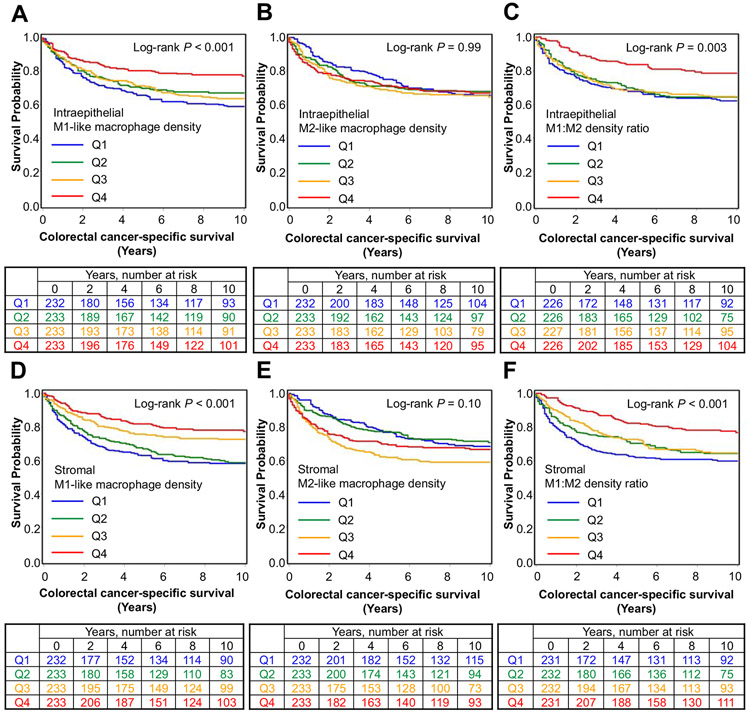
As our primary analysis, we examined the prognostic role of macrophages with either M1-like or M2-like polarization as defined by the most extreme 30% ends of the M1:M2-index (Fig. 2; Table 2). We found that higher stromal densities of M1-like macrophages were associated with a tendency towards longer cancer-specific survival (Q4 vs. Q1: HR 0.69, 95% CI 0.47-1.01, Ptrend=0.019), while higher stromal densities of M2-like macrophages were associated with shorter cancer-specific survival (Q4 vs. Q1: HR 1.48, 95% CI 1.01-2.17, Ptrend=0.005) in multivariable Cox regression models. These results suggested that the relatively weak survival associations for overall macrophage densities might be due to the fact that this population was composed of discrete subpopulations with opposing survival effects. Accordingly, higher stromal M1:M2 density ratio, reflecting M1-like macrophage polarization irrespective of overall macrophage density, was associated with longer cancer-specific survival (Q4 vs. Q1: multivariable adjusted HR 0.58, 95% CI 0.39-0.86, Ptrend=0.005) (Table 2, Table 3). As a secondary analysis, we examined the prognostic value of macrophage populations defined by positivity for single polarization markers. We found no significant associations between the densities of these populations and survival at the α level of 0.005 (Supplemental Table S5).
Figure 2.
Inverse probability weighting-adjusted Kaplan-Meier survival curves of colorectal cancer-specific survival and corresponding number at risk tables according to the intraepithelial (A-C) and stromal (D-F) densities of M1-like and M2-like macrophages and M1:M2 density ratio. P values were calculated using the log-rank test.
Table 3.
M1:M2 macrophage density ratio in tumor intraepithelial and stromal regions and colorectal cancer-specific survival with inverse probability weighting (IPW). Final multivariable Cox regression models with all variables.
| M1:M2 density ratio in intraepithelial region |
M1:M2 density ratio in stromal region |
|
|---|---|---|
| Multivariable HR (95% CI)*,† |
Multivariable HR (95% CI)*,† |
|
| M1:M2 macrophage density ratio | ||
| Q1 | 1 (referent) | 1 (referent) |
| Q2 | 1.20 (0.84-1.71) | 1.01 (0.72-1.42) |
| Q3 | 0.98 (0.69-1.39) | 0.82 (0.58-1.15) |
| Q4 | 0.59 (0.40-0.89) | 0.58 (0.39-0.86) |
| Age (per 10-year increase) | 1.33 (1.12-1.57) | 1.29 (1.08-1.53) |
| Year of diagnosis (per 5-year increase) | 0.80 (0.70-0.92) | 0.78 (0.69-0.89) |
| Tumor differentiation | ||
| Well to moderate | 1 (referent) | 1 (referent) |
| Poor | 2.13 (1.40-3.22) | 2.36 (1.54-3.63) |
| AJCC disease stage | ||
| I-II | 1 (referent) | 1 (referent) |
| III-IV | 5.79 (4.22-7.93) | 5.66 (4.15-7.70) |
| MSI status | ||
| Non-MSI-high | 1 (referent) | 1 (referent) |
| MSI-high | 0.31 (0.16-0.59) | 0.28 (0.15-0.53) |
| LINE-1 methylation level (per 30% decrease) | 1.61 (1.06-2.47) | Did not remain in this model |
| BRAF mutation | ||
| Wild-type | 1 (referent) | 1 (referent) |
| Mutant | 2.00 (1.29-3.08) | 1.84 (1.18-2.87) |
IPW was applied to reduce bias due to the availability of tumor tissue after cancer diagnosis (see “Statistical Analysis” subsection for details).
The multivariable Cox regression model initially included sex, age, year of diagnosis, family history of colorectal cancer, tumor location, tumor differentiation, disease stage, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and long-interspersed nucleotide element-1 methylation level. A backward elimination with a threshold P of 0.05 was used to select variables for the final models.
Abbreviations: AJCC, American Joint Committee on Cancer; CI, confidence interval; HR, hazard ratio; IPW, inverse probability weighting, LINE-1, long-interspersed nucleotide element-1; MSI, microsatellite instability.
While we initially selected a 30% cut-point for defining macrophage polarization in order to balance the total number of macrophages assigned to either the M1-like or M2-like categories against the strength of macrophage polarization at a single cell level, we sought to evaluate the robustness of these results by testing a range of different cut-points from 10-50% (Supplemental Fig. S9-S11). While a 50% cut-point assigns all identified macrophages into either an M1-like or M2-like category, it also resulted in both categories containing macrophages with widely varying degrees of polarization. Conversely, a 10% cut-off results in smaller populations of more uniformly polarized macrophages. These analyses indicated that the favorable prognostic tendency of stromal M1-like polarized macrophages remained quite consistent across the range of cut-points (Supplemental Fig. S9), whereas the adverse prognostic tendency of stromal M2-like macrophages was limited to the extreme 10% to 30% most strongly polarized macrophages (Supplemental Fig. S10). High stromal M1:M2 density ratio was associated with lower colorectal cancer-specific mortality within the entire range of M1:M2-index cut-points (Ptrend≤0.008 in multivariable adjusted models) (Supplemental Fig. S11).
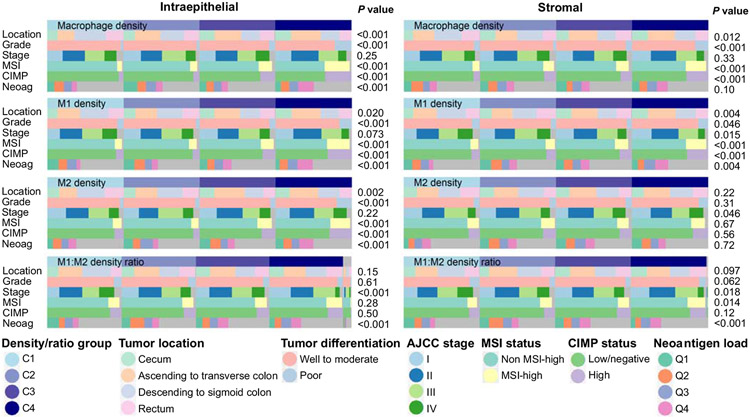
Relationships with tumor and patient characteristics
As secondary analyses, we examined the associations between macrophage densities and tumor and patient characteristics (Table 1, Fig. 3). MSI status was a strong determinant of overall macrophage densities; tumors with high-level microsatellite instability (an MSI-high phenotype) had higher densities of both intraepithelial and stromal macrophages (P<0.001). Moreover, higher intraepithelial and stromal macrophage densities were observed in tumors harboring other characteristics commonly associated with an MSI-high phenotype, including high tumor grade and a CIMP-high phenotype (P<0.001 for both). Higher intraepithelial macrophage densities were also associated with proximal tumor location and higher neoantigen load (P<0.001 for both). Tumors that were diagnosed in more recent years were associated with higher macrophage densities (P<0.001) and was also associated with older patient age (P<0.001), CIMP-high phenotype (P=0.004), and high LINE-1 methylation level (P<0.001) (Supplemental Table S6).
Figure 3.
Heatmap depicting relationships between clinicopathologic features and macrophage densities in tumor intraepithelial region and tumor stroma. The plots show the distribution of clinicopathologic features according to ordinal categories (C1-C4) of macrophage densities. P values are based on the comparison of categorical data between the ordinal categories of macrophage density/ratio using the Chi-square test. Abbreviations: AJCC, American Joint Committee on Cancer; CIMP, CpG island methylator phenotype; MSI, microsatellite instability.
When examining macrophages defined by the most extreme 30% cut-offs for the polarization index, higher stromal densities of M1-like macrophages (but not M2-like macrophages) were associated with an MSI-high phenotype (P<0.001) and higher neoantigen load (P=0.004), whereas in the tumor epithelial region, higher densities of both M1-like and M2-like macrophages were associated with MSI-high phenotype, CIMP-high phenotype, high neoantigen load, and poor differentiation (P<0.001 for all) (Fig. 3). Taken together, these results suggested that, compared to non-MSI-high tumors, MSI-high tumors were characterized by more pronounced macrophage infiltration adjacent to tumor cells, and stronger M1-like polarization of stromal macrophages, which may reflect the higher immunogenicity of these tumors (39).
We also evaluated whether the densities of differentially polarized macrophages were associated with histologic lymphocytic reaction patterns (Supplemental Fig. S12). We found that higher intraepithelial and stromal M1-like macrophage densities were associated with more intense lymphocytic reactions of all four types (TILs, intratumoral periglandular reaction, peritumoral reaction, Crohn’s-like reaction) (P<0.001 for all), whereas intraepithelial M2-like macrophage density showed significant, but relatively weaker, association with TILs (P<0.001). Stromal M2-like macrophage density was not significantly associated with any of the four lymphocytic reaction patterns (P>0.24).
Overall macrophage densities were not significantly associated with disease stage (Fig. 3). When macrophage polarization was evaluated, stromal M1-like macrophages tended to be more common in lower stage tumors (P=0.015), while M2-like macrophages tended to be more common in higher stage tumors (P=0.046), although these findings did not reach statistical significance at the stringent α level of 0.005.
Survival analyses stratified by MSI status and year of diagnosis
Given that we identified a strong association between macrophage densities and MSI status, we performed survival analyses stratified by MSI-status in order to further characterize the relationship between these tumor features (Supplemental Table S7). We found that high stromal densities of overall macrophages (Pinteraction=0.011) and M1-like macrophages (Pinteraction=0.057) tended to be more strongly associated with better survival in MSI-high tumors compared to non-MSI-high tumors, although these findings did not reach statistical significance at α level of 0.005 and need to be interpreted cautiously because of low numbers of events in patients with MSI-high tumors. There were no significant differences in survival associations for macrophage densities stratified by year of diagnosis (Supplemental Table S8).
Discussion
We performed quantitative, multiplexed analysis of macrophage polarization and density in colorectal cancers from patients in two large U.S. nationwide prospective cohort studies. This analysis revealed that the overall macrophage population had heterogenous prognostic significance, with M1-like and M2-like phenotypes exhibiting opposite effects. Higher M1:M2 density ratio, capturing the degree of M1-like macrophage polarization regardless of overall density, was associated with lower colorectal cancer-specific mortality, independent of potential confounding factors, including tumor stage and tumor molecular features.
During the past few decades, there have been significant conceptual and experimental advances in the understanding of macrophage biology (8). Single-cell RNA sequencing studies increasingly show that the macrophage population exhibits more heterogeneity than previously appreciated, particularly in inflammatory conditions such as cancer (40). These studies and others have demonstrated that macrophages exist along a spectrum of polarization states, with M1-like pro-inflammatory and M2-like anti-inflammatory phenotypes representing the extremes of this spectrum (8,9,33,40). However, there is a lack of specific markers that can cleanly separate these populations (8,10), necessitating multimarker approaches for the detection of differentially polarized macrophage subsets. Developments in multiplex immunofluorescence have enabled simultaneous visualization of multiple protein targets in a single tissue section (41). Although the number of markers in these assays is limited compared to some other methods such as bulk RNA sequencing or single-cell sequencing, a key benefit is the ability to examine macrophages in the spatial context of the tumor microenvironment across hundreds of specimens.
The multiplex immunofluorescence panel utilized in this study included two markers associated with an M1-like phenotype and two associated with an M2-like phenotype. Of these markers, IRF5 is a key transcription factor promoting inflammatory M1-like macrophage polarization, including induction of IL12A and IL12B as well as repression of IL10 in macrophages (31). Transcription factor MAF is associated with an M2-like homeostatic phenotype and is downregulated in the IFNG-driven M1-like phenotype (35). CD86 is an important co-stimulatory signal for antigen presentation to T cells and is commonly associated with an M1-like phenotype (32,42,43). MRC1 (CD206) is a scavenger receptor contributing to multiple cellular functions, including the endocytosis of mannosylated inflammatory glycoproteins (44), and is often found to be highly expressed in M2-like macrophages (10). Our four-marker approach had a balanced distribution of M1 and M2 markers, as well as transcription factors and cell surface markers that were spatially distinct at a subcellular level, enabling us to study macrophage polarization spectrum via an M1:M2-index based on marker signal intensities within each individual macrophage.
We found that overall macrophage density was not significantly associated with survival in multivariable Cox regression models. However, this result was likely due to the fact that the overall population was comprised of M1-like cells with a favorable prognostic effect and M2-like cells with an adverse effect. Notably, high M1:M2 density ratio in tumor stroma, representing M1-like polarization of tumor stromal macrophages, was associated with lower cancer-specific mortality independent of other tumor or patient features, including tumor stage, MSI status, and KRAS, BRAF, and PIK3CA mutation status. This finding suggested that macrophage polarization state had prognostic value beyond common tumor molecular and clinical features. Interestingly, a study suggested that high macrophage density in colorectal cancer could predict benefit from 5-fluorouracil adjuvant therapy (45); further studies are warranted to assess whether this effect is dependent on macrophage polarization.
Prior studies have identified numerous potential mechanisms underlying the diverse roles for M1-like and M2-like macrophages in the tumor microenvironment (46,47). Briefly, antitumorigenic M1-like macrophages may phagocytose tumor cells and efficiently present antigen to lymphocytes, thereby enhancing the antitumor immune response, while pro-tumorigenic M2-like macrophages have been implicated in anti-inflammatory effects (46). Supporting the divergent roles of M1-like and M2-like macrophage populations in dictating the status of the antitumor lymphocytic response, we found that higher density of M1-like but not M2-like macrophages in tumor stroma positively correlated with the degree of four histologic lymphocytic reaction patterns (TILs, intratumoral periglandular reaction, peritumoral reaction, Crohn’s-like reaction).
The progression of colorectal cancer is frequently associated with a systemic inflammatory response, characterized by the release of inflammatory mediators into the circulation, thereby supporting tumor growth and dissemination by numerous mechanisms including promotion of tumor cell proliferation and shaping pre-metastatic niches to promote subsequent metastasis (48,49). We found that a lower intraepithelial M1:M2 density ratio was associated with advanced disease stage. While a previous study did not indicate strong correlations between CD68+ macrophage densities in tumor tissue and serum cytokine levels (50), future studies could evaluate whether the local polarization of macrophages might reflect or contribute to a systemic inflammatory response.
Two major forms of genomic instability, namely chromosomal instability and microsatellite instability, are recognized in colorectal carcinoma (51). MSI-high tumors are characterized by a deficient mismatch repair system, leading to a hypermutated phenotype and production of immunogenic neoantigens (39). This often results in a prominent lymphocytic infiltrate in these tumors, which is associated with favorable clinical outcome (3,4,23,52). We found that MSI status was an important determinant of macrophage densities in the colorectal cancer microenvironment, as MSI-high tumors had higher intraepithelial macrophage densities regardless of polarization, and higher densities of M1-like but not M2-like macrophages in tumor stroma. These results suggest that these immunogenic tumors are characterized by M1-polarization of stromal macrophages, as well as close contact between macrophages and tumor cells. However, there is also variation within the MSI-high tumors, and a case report describes high M2-like macrophage infiltration in an immune checkpoint blockade resistant patient with a MSI-high tumors (53). Although part of the prognostic effect of macrophage infiltrates observed in our univariable analyses was attributable to associations between MSI status and macrophage densities, multivariable and MSI-stratified Cox regression models indicated that macrophage polarization harbored prognostic value independent of MSI status.
Several important limitations need to be considered in the interpretation of these results. First, the study was based on TMAs, and macrophage densities and polarization in small tissue samples may not perfectly represent the entire tumor. However, we examined an average of 1.8 cores per tumor and observed reasonable core-to-core correlation between macrophage densities, supporting the validity of this approach. Moreover, TMAs enabled us to examine a large number of samples stained uniformly as one batch. Automated computer-based image analysis ensured that all the images were processed in a uniform manner, reducing the effects of analytical imprecision. Second, we tested multiple hypotheses in this observational study. However, we had specific primary hypotheses, and we used a stringent α level of 0.005 to reduce the risk of type I statistical error (37). Third, although our 7-plex immunofluorescence panel is capable of more comprehensively characterizing macrophage polarization than single- or double-marker assays, the number of included polarization markers was still limited. We recognize the need for further validation of our approach in other cohorts as well as examination of additional markers to further define polarization stages and identify CD68- macrophage populations. Fourth, as the estimates of the prevalence of M1-like and M2-like macrophages in colorectal cancer vary significantly according to the markers and methods used for their detection (13,14,16), we decided to examine the 30% extreme ends of the polarization spectrum in the main analyses. We acknowledge that this may not reflect the true distribution of the anti- and pro-inflammatory macrophage phenotypes in the colorectal cancer microenvironment. However, sensitivity analyses supported the robustness of the results across a wide range of cut-points to define M1-like (10-50%) and M2-like (10-30%) phenotypes. Fifth, patients in the cohort were diagnosed and treated over several decades, a time period long enough that clinical practice, including staging accuracy and treatment strategies, may have changed. Notably, macrophage densities were lower and CIMP-high tumors were less prevalent in patients diagnosed in 1995 or before compared to those diagnosed afterwards. However, there was no significant association between year of diagnosis and tumor cell density or cytokeratin fluorophore signal intensity, suggested that macrophage density differences over time were not due to time-dependent loss of antigenicity in tissue samples. Taken together, these results raised the possibility of evolution in the molecular epidemiology of colorectal cancer during the study period. We also adjusted for year of diagnosis in the multivariable survival models, mitigating its potential effect on the main results. Additionally, there were no significant differences in survival associations for macrophage densities stratified by year of diagnosis. Finally, treatment data were unavailable for our cohorts. However, it is unlikely that macrophage densities or polarization influenced the treatment decisions, as such data were not available during treatment periods. A major strength of this study was the availability of a rich molecular pathological epidemiology database, providing comprehensive data regarding tumor molecular characteristics and clinicopathologic features. The patients were recruited from a large number of hospitals across the U.S., supporting the generalizability of the findings.
In conclusion, we measured macrophage M1:M2 polarization spectrum in colorectal cancer microenvironment using a quantitative, multiplexed immunofluorescence assay. We found that macrophage polarization rather than absolute overall density was associated with colorectal cancer mortality, with M1-like and M2-like macrophages showing opposite effects. These findings support the utility of a multimarker approach for identifying biologically and prognostically relevant macrophage subpopulations in the tumor microenvironment.
Supplementary Material
Synopsis.
Macrophage polarization state, rather than their overall density, in the colorectal cancer microenvironment is associated with cancer-specific survival independent of potential confounding factors, with M1-like and M2-like macrophage phenotypes exhibiting distinct prognostic roles.
Acknowledgements
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. Portions of this research were conducted on the O2 High Performance Compute Cluster, supported by the Research Computing Group, at Harvard Medical School. See http://rc.hms.harvard.edu for more information. This work was supported by U.S. National Institutes of Health (NIH) grants (P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; U01 CA167552 to W.C. Willett and L.A. Mucci; P50 CA127003 to C.S.F.; R01 CA118553 to C.S.F.; R01 CA169141 to C.S.F.; R01 CA137178 to A.T.C.; K24 DK098311 to A.T.C.; R35 CA197735 to S.O.; R01 CA151993 to S.O.; K07 CA190673 to R.N.; R03 CA197879 to K.W.; R21 CA222940 to K.W. and M.G.; and R21 CA230873 to K.W. and S.O.); by Cancer Research UK Grand Challenge Award (UK C10674/A27140 to M.G. and S.O.); by Nodal Award (2016-02) from the Dana-Farber Harvard Cancer Center (to S.O.); by the Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17 to C.S.F. and M.G.), Stand Up to Cancer is a division of the Entertainment Industry Foundation. The SU2C research grant is administered by the American Association for Cancer Research, a scientific partner of SU2C; and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.H. was supported by fellowship grants from the Uehara Memorial Foundation and the Mitsukoshi Health and Welfare Foundation. S.A.V. was supported by grants from the Finnish Cultural Foundation and Orion Research Foundation sr. J.B. was supported by a grant from the Australia Awards-Endeavour Scholarships and Fellowships Program. K.F. was supported by a fellowship grant from the Uehara Memorial Foundation. K.A. was supported by grants from Overseas Research Fellowship from Japan Society for the Promotion of Science (JP201860083). K.W. was supported by an Investigator Initiated Grant from the American Institute for Cancer Research (AICR). A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. J.A.M. research is supported by the Douglas Gray Woodruff Chair fund, the Guo Shu Shi Fund, Anonymous Family Fund for Innovations in Colorectal Cancer, Project P fund, and the George Stone Family Foundation. M.G. was supported by a Conquer Cancer Foundation of ASCO Career Development Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- AJCC
American Joint Committee on Cancer
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- DNA
deoxyribonucleic acid
- HPFS
Health Professionals Follow-up Study
- HR
hazard ratio
- IPW
inverse probability weighting
- IQR
interquartile range
- LINE-1
long-interspersed nucleotide element-1
- MSI
microsatellite instability
- NHS
Nurses’ Health Study
- RNA
ribonucleic acid
- SD
standard deviation
- TAM
tumor-associated macrophage
- TIL
tumor-infiltrating lymphocyte
- TMA
tissue microarray
- TNM
tumor, node, metastasis
Footnotes
Disclosure of Potential Conflicts of Interest: M.C.L., J.B., S.O., and J.A.N. are listed as co-inventors on a provisional application for a patent titled “System for and Method of Discovering Spatially-Derived Signatures of Tumor-Immune Cell Interactions through Tumor-Immune Partitioning and Clustering” regarding novel methods for characterizing immune cell distributions in solid tumors that has been filed through Partners Healthcare. J.L.G. is a consultant for GlaxoSmithKline, Codagenix, Array BioPharma, and Verseau Therapeutics; and receives research support from GlaxoSmithKline, Eli Lilly and Array BioPharma for the study of the breast tumor microenvironment. A.T.C. previously served as a consultant for Bayer Healthcare and Pfizer Inc.. C.S.F. previously served as a consultant for Agios, Bain Capital, Bayer, Celgene, Dicerna, Five Prime Therapeutics, Gilead Sciences, Eli Lilly, Entrinsic Health, Genentech, KEW, Merck, Merrimack Pharmaceuticals, Pfizer Inc, Sanofi, Taiho, and Unum Therapeutics; C.S.F. also serves as a Director for CytomX Therapeutics and owns unexercised stock options for CytomX and Entrinsic Health. J.A.M. received institutional research funding from Boston Biomedical. J.A.M. has also served as an advisor/consultant to Ignyta, Array Pharmaceutical, and Cota. M.G. receives research funding from Bristol-Myers Squibb and Merck. R.N. is currently employed by Pfizer Inc. She contributed to this study before she was employed by Pfizer Inc. This study was not funded by any of these commercial entities. No other conflicts of interest exist. The other authors declare that they have no conflicts of interest.
Use of Standardized Official Symbols: We use HUGO (Human Genome Organization)-approved official symbols (or root symbols) for genes and gene products, including BRAF, CD68, CD86, IFNG, IL10, IL12A, IL12B, IRF5, KRAS, KRT, MAF, MRC1, PIK3CA; all of which are described at www.genenames.org. Gene symbols are italicized whereas symbols for gene products are not italicized.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018;67:1168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander PG, McMillan DC, Park JH. The local inflammatory response in colorectal cancer - Type, location or density? A systematic review and meta-analysis. Cancer Treat Rev. 2020;83:101949. [DOI] [PubMed] [Google Scholar]
- 4.Pagès F, Mlecnik B, Marliot F, Bindea G, Ou F, Bifulco C, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–39. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 6.Shalapour S, Karin M. Pas de Deux: Control of Anti-tumor Immunity by Cancer-Associated Inflammation. Immunity. 2019;51:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kather JN, Halama N. Harnessing the innate immune system and local immunological microenvironment to treat colorectal cancer. Br J Cancer. 2019;120:871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541–66. [DOI] [PubMed] [Google Scholar]
- 9.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. [DOI] [PubMed] [Google Scholar]
- 10.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerriero JL. Macrophages: The Road Less Traveled, Changing Anticancer Therapy. Trends Mol Med. 2018;24:472–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forssell J, Oberg A, Henriksson ML, Stenling R, Jung A, Palmqvist R. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res. 2007;13:1472–9. [DOI] [PubMed] [Google Scholar]
- 13.Edin S, Wikberg ML, Dahlin AM, Rutegard J, Oberg A, Oldenborg P-AA, et al. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7:e47045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koelzer VH, Canonica K, Dawson H, Sokol L, Karamitopoulou-Diamantis E, Lugli A, et al. Phenotyping of tumor-associated macrophages in colorectal cancer: Impact on single cell invasion (tumor budding) and clinicopathological outcome. Oncoimmunology. 2016;5:e1106677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaput N, Svrcek M, Aupérin A, Locher C, Drusch F, Malka D, et al. Tumour-infiltrating CD68+ and CD57+ cells predict patient outcome in stage II-III colorectal cancer. Br J Cancer. 2013;109:1013–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ålgars A, Irjala H, Vaittinen S, Huhtinen H, Sundström J, Salmi M, et al. Type and location of tumor-infiltrating macrophages and lymphatic vessels predict survival of colorectal cancer patients. Int J Cancer. 2012;131:864–73. [DOI] [PubMed] [Google Scholar]
- 17.Herrera M, Herrera A, Domínguez G, Silva J, García V, García JM, et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013;104:437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q, Chang W, Mao Y, He G, Zheng P, Tang W, et al. Tumor-associated macrophages as prognostic and predictive biomarkers for postoperative adjuvant chemotherapy in patients with stage II colon cancer. Clin Cancer Res. 2019;25:3896–907. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Wen X, Bae JM, Kim JH, Cho NY, Kang GH. The distribution of intratumoral macrophages correlates with molecular phenotypes and impacts prognosis in colorectal carcinoma. Histopathology. 2018;73:663–71. [DOI] [PubMed] [Google Scholar]
- 20.Colditz GA, Manson JE, Hankinson SE. The Nurses’ Health Study: 20-year contribution to the understanding of health among women. J women’s Heal. 1997;6:49–62. [DOI] [PubMed] [Google Scholar]
- 21.Ascherio A, Rimm EB, Giovannucci EL, Colditz GA, Rosner B, Willett WC, et al. A prospective study of nutritional factors and hypertension among US men. Circulation. 1992;86:1475–84. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haruki K, Kosumi K, Li P, Arima K, Väyrynen JP, Lau MC, et al. An integrated analysis of lymphocytic reaction, tumour molecular characteristics and patient survival in colorectal cancer. Br J Cancer. 2020;122:1367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. [DOI] [PubMed] [Google Scholar]
- 28.Tóth ZE, Mezey E. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J Histochem Cytochem. 2007;55:545–54. [DOI] [PubMed] [Google Scholar]
- 29.Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget M-A, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep. 2017;7:13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barros MHM, Hauck F, Dreyer JH, Kempkes B, Niedobitek G. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One. 2013;8:e80908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–8. [DOI] [PubMed] [Google Scholar]
- 32.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–96. [DOI] [PubMed] [Google Scholar]
- 33.Xue J, Schmidt S V, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Sime W, Juhas M, Sjölander A. Crosstalk between colon cancer cells and macrophages via inflammatory mediators and CD47 promotes tumour cell migration. Eur J Cancer. 2013;49:3320–34. [DOI] [PubMed] [Google Scholar]
- 35.Kang K, Park SH, Chen J, Qiao Y, Giannopoulou E, Berg K, et al. Interferon-γ Represses M2 Gene Expression in Human Macrophages by Disassembling Enhancers Bound by the Transcription Factor MAF. Immunity. 2017;47:235–250.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers E-J, Berk R, et al. Redefine statistical significance. Nat Hum Behav. 2018;2:6–10. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Nevo D, Nishihara R, Cao Y, Song M, Twombly TS, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33:381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov. 2018;8:730–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, Schultze JL. The Myeloid Cell Compartment-Cell by Cell. Annu Rev Immunol. 2019;37:269–93. [DOI] [PubMed] [Google Scholar]
- 41.Parra ER, Francisco-Cruz A, Wistuba II. State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers (Basel). 2019;11:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerriero JL. Macrophages: Their Untold Story in T Cell Activation and Function. Int Rev Cell Mol Biol. 2018;342:73–93. [DOI] [PubMed] [Google Scholar]
- 43.Gensel JC, Kopper TJ, Zhang B, Orr MB, Bailey WM. Predictive screening of M1 and M2 macrophages reveals the immunomodulatory effectiveness of post spinal cord injury azithromycin treatment. Sci Rep. 2017;7:40144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Pomares L The mannose receptor. J Leukoc Biol. 2012;92:1177–86. [DOI] [PubMed] [Google Scholar]
- 45.Malesci A, Bianchi P, Celesti G, Basso G, Marchesi F, Grizzi F, et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. Oncoimmunology. 2017;6:e1342918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang M, McKay D, Pollard JW, Lewis CE. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018;78:5492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aras S, Zaidi MR. TAMeless traitors: macrophages in cancer progression and metastasis. Br J Cancer. 2017;117:1583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuomisto AE, Mäkinen MJ, Väyrynen JP. Systemic inflammation in colorectal cancer: Underlying factors, effects, and prognostic significance. World J Gastroenterol. 2019;25:4383–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolan RD, Lim J, McSorley ST, Horgan PG, McMillan DC. The role of the systemic inflammatory response in predicting outcomes in patients with operable cancer: Systematic review and meta-analysis. Sci Rep. 2017;7:16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Väyrynen JP, Kantola T, Väyrynen SA, Klintrup K, Bloigu R, Karhu T, et al. The relationships between serum cytokine levels and tumor infiltrating immune cells and their clinical significance in colorectal cancer. Int J Cancer. 2016;139:112–21. [DOI] [PubMed] [Google Scholar]
- 51.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Väyrynen JP, Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, et al. Prognostic Significance of Immune Cell Populations Identified by Machine Learning in Colorectal Cancer Using Routine Hematoxylin and Eosin-Stained Sections. Clin Cancer Res. 2020;26:4326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurjao C, Liu D, Hofree M, AlDubayan SH, Wakiro I, Su M-J, et al. Intrinsic Resistance to Immune Checkpoint Blockade in a Mismatch Repair-Deficient Colorectal Cancer. Cancer Immunol Res. 2019;7:1230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.