Abstract
Purpose:
Several biomarkers of response to immune checkpoint inhibitors (ICIs) show potential but are not yet scalable to the clinic. We developed a pipeline that integrates deep learning on histology specimens with clinical data to predict ICI response in advanced melanoma.
Experimental design:
We used a training cohort from New York University and a validation cohort from Vanderbilt University. We built a multivariable classifier that integrates neural network predictions with clinical data. A receiver operating characteristic (ROC) curve was generated and the optimal threshold was used to stratify patients as high versus low risk for progression. Kaplan-Meier curves compared progression free survival (PFS) between the groups. The classifier was validated on two slide scanners (Aperio AT2 and Leica SCN400).
Results:
The multivariable classifier predicted response with area under the curve (AUC) 0.800 on images from the Aperio AT2 and AUC 0.805 on images from the Leica SCN400. The classifier accurately stratified patients into high versus low risk for disease progression. Vanderbilt patients classified as high risk for progression had significantly worse PFS than those classified as low risk (P = 0.02 for the Aperio AT2; P = 0.03 for the Leica SCN400).
Conclusions:
Histology slides and patients’ clinicodemographic characteristics are readily available through standard of care and have the potential to predict ICI treatment outcomes. With prospective validation, we believe our approach has potential for integration into clinical practice.
Introduction
Immune checkpoint inhibitors (ICIs) produce durable clinical response for a subset of patients with advanced melanoma [1–4]. However, treatment is often complicated by immune-related toxicity, which may necessitate permanent discontinuation of immunotherapy or lead to lifelong secondary conditions [5]. Thus, a major challenge in the management of metastatic melanoma is optimizing patient selection for checkpoint blockade. Several recent attempts to predict ICI response showed potential, but rely on biomarkers that lack scalability, demand high availability of resources, or still require extensive validation of their utility for clinical decision-making [6–10].
Visual microscopic assessment of hematoxylin and eosin (H&E) stained tissue remains standard of care for diagnosing melanoma and staging disease severity. Yet, despite the ready availability of histologic specimens, conventional light microscopy plays a limited role in prognosticating treatment outcomes [11–13]. This could be due to the practical constraint that human evaluations are time consuming and highly subjective. More likely, the phenotypic information that provides insight to drug-responsiveness may be unapparent to human observers. We asked whether machine learning algorithms could be trained to identify prognostically important features of melanoma tissue histology. Within the field of dermatopathology, deep convolutional neural networks (DCNNs) have proven efficacious at computer vision tasks such as image classification [14, 15]. This form of machine learning distinguished malignant melanoma from benign nevi with a level of accuracy comparable to that of a dermatologist [16]. In a separate investigation, our group developed a DCNN pipeline that reliably discriminated between malignant and normal lung tissue and accurately predicted the most commonly mutated genes found within lung tumors [17].
In this study, we aim to develop a streamlined approach to pre-treatment prognostication by leveraging information immediately available through routine clinical care. We adapt our machine learning framework to whole slide image (WSI) analysis of metastatic melanoma tissue. Our hypothesis is that a DCNN can learn to break down WSIs into component features, detect non-obvious patterns, and correlate those patterns with the likelihood of response to immunotherapy. We then integrate patient clinicodemographic variables to create a multifactorial paradigm for accurately predicting immunotherapy response.
Materials and Methods
Patient population
In this analysis, the training cohort consisted of 121 patients who received treatment at New York University (NYU) Perlmutter Comprehensive Cancer Center between 2004 and 2018. The independent validation cohort included 30 patients who were treated at Vanderbilt University Ingram Cancer Center between 2010 and 2017. The NYU patients had been prospectively enrolled with written informed consent in the institutional review board (IRB) approved (#10362) Interdisciplinary Melanoma Cooperative Group (IMCG) database at NYU Langone Health. The IMCG study maintains protocol driven follow-up for melanoma patients treated at Perlmutter Comprehensive Cancer Center and stores tissue specimens for research purposes, which are connected with the clinicopathologic database.
We included patients with metastatic disease who had lymph node (LN) and/or subcutaneous tissue (ST) resected before treatment with first-line anti-CTLA-4, anti-PD-1, or combination anti-CTLA-4 plus anti-PD-1 therapy. Clinical decisions regarding the selection of a treatment regimen were made independent of this study. Response was assessed via imaging done in 3-month intervals following treatment initiation or sooner as dictated by changes in clinical status. Treatment outcomes were classified according to the revised Response Evaluation Criteria in Solid Tumors (RECIST) guideline version 1.1. Response was recorded as progression of disease (POD) or “response,” which included complete response (CR) and partial response (PR). We excluded patients with stable disease in order to focus on extremes of outcome for this proof-of-principle study. Progression free survival was defined as the time from the first dose of immunotherapy until disease progression or death. For the NYU cohort, best response was noted at a median of 3.4 months after treatment [interquartile range (IQR) = 4.7]. Median overall follow-up was 14.0 months (IQR = 34.6). For Vanderbilt patients, best response was noted at a median of 2.3 months (IQR = 1.2). The median overall follow-up was 28.5 months (IQR = 32.5). We adhered to the reporting recommendations for tumor marker prognostic studies (REMARK) guidelines [18].
Image processing
Tissue was obtained through either excisional biopsy or surgical resection of metastases. Formalin-fixed paraffin embedded (FFPE) H&E stained slides were scanned with at least 20X magnification using an Aperio AT2 slide scanner (Leica Microsystems, Wetzlar, Germany). There were 302 slides from NYU patients and 40 slides from Vanderbilt patients. For validation purposes, we scanned 39 slides from 29 patients in the Vanderbilt cohort using a Leica SCN400 machine (Leica Microsystems, Wetzlar, Germany). We partitioned the WSIs into non-overlapping 299×299 pixel tiles at 0.5 μm/pixel resolution (equivalent to 20x magnification) or 1μm/pixel resolution (equivalent to 10x). Background coverage was defined as pixels with an average gray level above 220 (8 bits coded images). Tiles with >75% background coverage were removed.
DCNN architecture and development
In this study, we developed two DCNN classifiers, which we refer to as the Segmentation Classifier and the Response Classifier (Supplemental Figure 1). We utilized the pipeline previously described by our group [17], which relies on Tensorflow and the Inception v3 architecture developed by Google [19]. Inception v3 served as a foundation architecture and was fully re-trained in this study. The jobs were run on NYU Langone Health’s distributed memory high-performance computing cluster Big Purple using either Cray CS-Storm 500NX GPU stations or Cray CS500 CPU stations (2.4 GHz, 384 to 768 GB/node), where it takes approximately 5 seconds to pre-process 500 tiles and another 5 seconds to obtain their probability from a trained network.
Segmentation Classifier
We aimed to predict clinical outcomes based on the analysis of tumor regions within the sampled tissue. In order to do so, we first developed a classifier that could selectively distinguish tumor from the surrounding microenvironment. Given that our dataset included lymph node and subcutaneous tissue, we trained the classifier to identify connective tissue and extra-tumor lymphocyte clusters in addition to tumor compartments. Using Aperio ImageScope (Leica Biosystems, Wetzlar, Germany), our board-certified pathologist colleague manually annotated 153 slides coming from a subset of 72 NYU patients. The manual annotations of the 3 regions of interest (ROI) served as labels for each tile within the delineated region. The labeled slides were divided into training, validation, and test sets (70%, 15%, and 15% of the data, respectively). To prevent overlaps between sets, the slides from a given patient were kept together. The classifier generated a set of 3 probabilities (for normal, lymphocyte, tumor) for each entire tile. During the segmentation step, each tile was assigned the label with the highest probability. In order to determine performance accuracy, the area under the curve (AUC) was calculated using segmentations done by the pathologist as the ground truth. After the Segmentation Classifier was trained and tested, we applied it to segment all of the remaining tiles from the NYU and Vanderbilt datasets. In the NYU dataset, the median area of tumor tiles per slide was 0.7cm2 (IQR 1.0cm2). In the Vanderbilt dataset, the median area of tumor tiles per slide was 1.2cm2 (IQR 0.8cm2). The minimum amount of tissue used for a patient in this study was 2.2mm2 of tumor tiles.
Response Classifier
The NYU training cohort consisted of 1,265,166 tumor ROI tiles from 302 slides. There were 173 slides from metastatic LN and 129 from metastatic ST. We included multiple slides per patient to augment training by increasing the total number of tiles. Most patients had either 1 or 2 slides included in the study (n=57 and n=31, respectively). In order to mitigate skewed training, we limited the number of slides to ≤10 per patient. We optimized the Response Classifier using a five-fold cross validation approach that involved randomly splitting the full set of tiles labeled as tumor ROI into five balanced subsets. Four of the subsets (80%) were used as a training set and the remaining 20% were used for testing (Supplemental Figure 2). We repeated this process five times until all tiles were used in the test set once (Supplemental Table 1). During the five-fold cross validation runs, we noted that color normalization with Reinhard’s method led to higher and more consistent average area under the curve (AUC) than normalization with Vahadane’s method or with no normalization [20, 21]. After identifying additional optimal hyper-parameters through the five-fold cross validation, we retrained on the full NYU dataset using those same parameters, which were as follows: color normalized tiles, batch size of 400, 15 epochs per decay (num_epochs_per_decay parameter in Inception v3), and training for 175,000 iterations. Data augmentation is integrated into Inception v3 (see distort_image function in image_processing.py), and includes color distortion, image distortion and flipping. Since training is a stochastic process and to further check the magnitude of its uncertainty, the final network was trained a total of 5 times. We then tested the fully trained model on the independent cohort from Vanderbilt. Importantly, the Vanderbilt dataset was balanced with only 1 – 2 slides per patient thus mitigating the possibility of performance inflation.
To analyze features used by the classifier to make its decisions, we followed the protocol developed by Kim et al (2020) [22]. First, we performed Class Activation Mapping (CAM) to identify regions within each tile that the neural network uses to generate predictions [23, 24]. To do this, we analyzed a set of tiles that were classified as POD with high probability (POD probability above 0.75; 136,109 “POD” tiles) and another set of tiles classified as Response with high probability (POD probability below 0.25; 51,220 “Response” tiles). The results of our CAM analyses suggested that cell nuclei are important to the algorithm’s predictions. We then used CellProfiler to identify whether there is variation in the characteristics of the nuclei assigned POD versus Response. We started by segmenting the nuclei, then measured the shape and quantity of the segmented objects, and then analyzed whether these features were associated with the prediction assigned to the tile.
Statistical analysis
The DCNN yielded a probability value for each tile for every class of interest. For the Segmentation Classifier, the classes were tumor, lymphocyte, and connective tissue compartments. For the Response Classifier, the classes were response and progression of disease. We averaged the probabilities of each tile from the patients’ slides in order to assign a final probability to each patient. We investigated the relationship between per slide performance accuracy and the amount of time between tissue resection and treatment initiation. We also evaluated the relationship between per slide performance accuracy and the number of tumor tiles. For both, the mean squared error of the DCNN prediction was used as a measure of accuracy. We then performed the Shapiro test for normality and calculated Spearman’s correlation coefficient and its significance. For our CellProfiler analyses, Student’s t-test was used to compare the area, density, and eccentricity of cell nuclei in tiles labeled POD versus Response. We then performed multivariable logistic regressions that combined the Response Classifier output with conventional clinical characteristics to predict treatment outcomes for the NYU training cohort. The candidate predictors included age, gender, histological subtype, treatment category, disease stage, lactate dehydrogenase, Eastern Cooperative Oncology Group (ECOG) performance status, the number of metastatic sites, and the log-transformed tumor mutation burden (TMB). TMB was defined as the total number of non-synonymous somatic mutations and synonymous mutations (single nucleotide variants and small insertions/deletions) per mega-base of the coding regions examined. The mutation count was calculated using customized pipelines based on the LoFreq assay [25]. The results of univariable analyses are shown in Supplemental Table 2. We performed backward stepwise selection to select the final multivariable model. The least significant variables were removed one at a time until all of the variables left in the model were significant. The linear combination of the selected model predictors weighted by regression coefficients was defined as the risk score and applied to the Vanderbilt test cohort. The overall function is:
Logit(p) = −0.4970 + (2.0966*DCNN output) + (1.2522*ECOG) – (1.8908*Anti-CTLA-4 and Anti-PD-1 status) – (1.2013*Anti-PD-1 status).
The probabilities calculated by the neural network and logistic regression classifiers were used to generate receiver operating characteristic (ROC) curves. Prognostic potential was reported as area under the curve values with corresponding 95% confidence intervals (CIs). Using AUC as the metric, we compared the ability of each variable and combination of variables to discriminate outcomes. We also compared the value importance of the variables using the absolute value of their Z-scores. After validation of the DCNN and logistic regression models on the Vanderbilt dataset, we identified the coordinates for the optimal threshold on the ROC Curves from the NYU training dataset. We then determined the corresponding prediction probability scores, which were set as the assay cutoff value. Vanderbilt patients who scored above the cutoff point were classified as high risk for progression; those who scored below the cutoff point were classified as low risk. We generated Kaplan-Meier curves to compare progression free survival of the high and low risk groups. The level of significance was set at P < 0.05. Analyses were performed using R software (http://www.R-project.org/) or scikit-learn.
Results
Patient characteristics
Baseline demographic characteristics were generally well balanced between the training cohort from NYU and the independent validation cohort from Vanderbilt (Table 1). However, there were differences in the treatments and outcomes of the two cohorts. The majority of the NYU population received anti-CTLA-4 monotherapy whereas most patients from Vanderbilt were treated with anti-PD-1 agents (63.6% and 53.3%, respectively). Compared to the NYU cohort, a lower proportion of patients from Vanderbilt suffered POD (50% vs 64.5%, respectively). Deidentified clinical and demographic characteristics for each patient are shown in Supplemental File 1.
Table 1.
Baseline clinical and demographic characteristics of the patients.
| NYU | Vanderbilt | |||
|---|---|---|---|---|
| n | 121 | 30 | ||
| Age | Mean (SD) | 59.82 (15.46) | 60.12 (12.8) | |
| Gender | Male | n (%) | 80 (66.1) | 21 (70.0) |
| Female | n (%) | 41 (33.9) | 9 (30.0) | |
| ECOG score | 0 | n (%) | 87 (71.9) | 11 (36.6) |
| 1 | n (%) | 23 (19.0) | 17 (56.7) | |
| 2 | n (%) | 5 (4.1) | 1 (3.3) | |
| 3 | n (%) | 0 (0) | 1 (3.3) | |
| Unknown | n (%) | 6 (5.0) | 0 (0.0) | |
| Histologic type | Superficial Spreading | n (%) | 17 (14.0) | 11 (36.6) |
| Nodular | n (%) | 39 (32.2) | 8 (26.7) | |
| Other | n (%) | 15 (12.4) | 3 (10.0) | |
| Unclassified | n (%) | 50 (41.3) | 8 (26.7) | |
| Stage at treatment initiation | Stage IIIB | n (%) | 5 (4.1) | 0 (0) |
| Stage IIIC | n (%) | 14 (11.6) | 0 (0) | |
| Stage IV | n (%) | 102 (84.3) | 30 (100.0) | |
| Immunotherapy treatment category | Anti-CTLA-4 | n (%) | 77 (63.6) | 4 (13.3) |
| Anti-PD-1 | n (%) | 26 (21.5) | 16 (53.3) | |
| Combination | n (%) | 18 (14.9) | 10 (33.3) | |
| Best response | CR | n (%) | 24 (19.8) | 5 (16.7) |
| PR | n (%) | 19 (15.7) | 10 (33.3) | |
| POD | n (%) | 78 (64.5) | 15 (50.0) | |
| Time to best response (months) | Median (IQR) | 3.4 (4.7) | 2.3 (1.2) | |
| Alive status | Alive | n (%) | 56 (46.3) | 16 (53.3) |
| Dead | n (%) | 65 (53.7) | 14 (46.7) | |
| Time to last follow-up (months) | Median (IQR) | 14.0 (34.6) | 28.5 (32.5) | |
IQR, interquartile range
Training and validation of the tissue Segmentation Classifier
The neural network distinguished tumor, lymphocyte, and connective tissue compartments with robust accuracy. In metastatic lymph nodes, the Segmentation Classifier identified tumor ROI with area under the curve (AUC) 0.961 (95% CI: 0.959–0.963), lymphocyte ROI with AUC 0.962 (95% CI: 0.960–0.965), and connective tissue ROI with AUC 0.969 (95% CI: 0.967–0.971). In metastatic subcutaneous tissue, the Segmentation Classifier identified tumor ROI with AUC 0.957 (95% CI: 0.950–0.963), lymphocyte ROI with AUC 0.886 (95% CI: 0.867–0.904), and connective tissue ROI with AUC 0.984 (95% CI: 0.977–985). ROC curves are shown in Fig. 1a, and Fig. 1b shows representative images of segmentations done by our pathologist co-investigator and the neural network classifier.
Figure 1.
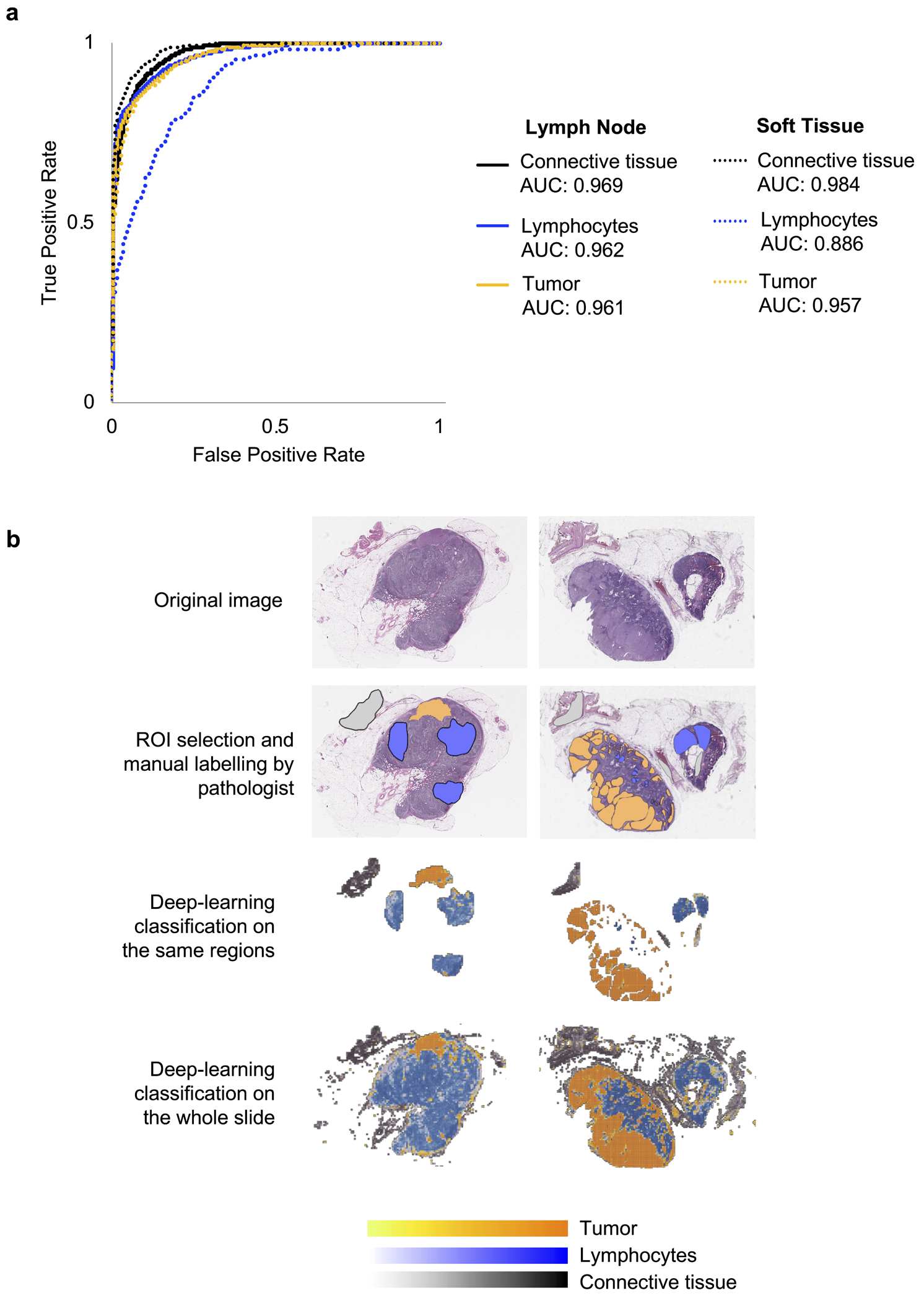
Training of a Segmentation Classifier to distinguish tumor, lymphocyte, and connective tissue compartments. (a) Performance of the classifier was measured in terms of area under the curve (AUC) of the receiver operating characteristic (ROC) curve. The model performed with robust accuracy and was equally efficacious when applied to lymph node and subcutaneous tissue samples. (b) Representative images of the computational workflow. In the first row, there are two WSIs of H&E stained tissue from lymph nodes infiltrated with melanoma. In the subsequent rows, the images show manual annotation for the three regions of interest (ROI) by our pathologist coinvestigator, then training of the neural network classifier on the annotated regions, and finally, application of the classifier to the whole slide images.
Development of predictive models for immunotherapy response using a DCNN
While developing the Response Classifier, we identified the optimal learning conditions through a series of 5-fold cross validations. Using this approach, the selected model predicted response with micro AUC 0.685 (95% CI: 0.593–0.777) and average macro AUC of 0.721 (95% CI: 0.468–0.9331) on the five NYU subsets left aside for testing (Supplemental Figure 2). After validating the model with optimal parameters, we re-trained using the entire NYU cohort as the training dataset. The fully trained model performed with AUC 0.691 (95% CI: 0.597–0.786) [Supplemental Figure 3]. Next, we tested the fully trained classifier on the Vanderbilt cohort. The validation process was independently repeated 5 times in order to check the impact of the stochastic process of learning (Supplemental Tables 3 and 4). The model performed with an average AUC of 0.707 (95% CI: 0.518–0.896) on the test slides scanned with the Aperio AT2. When applied to the test slides scanned with the Leica SCN400, the model performed with an average AUC of 0.667 (95% CI: 0.463–0.870) [Figure 2]. Of note, neural network predictions were better when applied to lymph node than soft tissue. For Aperio AT2 stained slides, the neural network had AUC 0.857 [0.654 – 1.060] on LN and 0.583 [0.312 – 0.855] on ST. For Leica SCN400 scanned slides, the DCNN had AUC 0.738 [0.464 – 1.012] on LN and AUC 0.609 [0.326 – 0.893] on ST (Supplemental Table 5).
Figure 2.
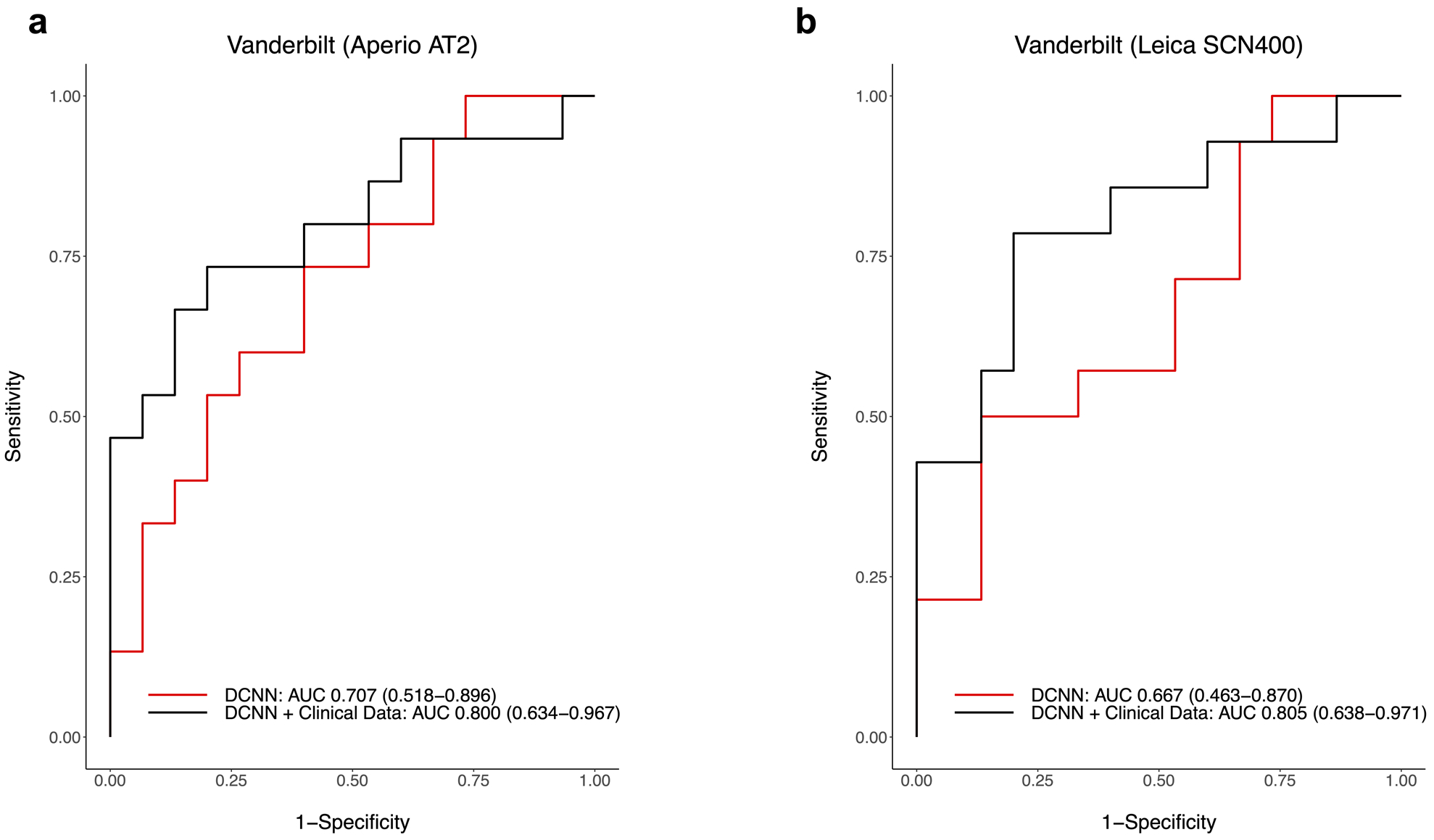
Performance of the neural network and logistic regression classifiers was measured in terms of area under the curve (AUC) of the receiver operating characteristic (ROC) curve. Validation was done on the independent cohort from Vanderbilt University. Slides were scanned with two different scanners as an additional validation measure. ROC Curves are shown for prediction of best response on slides scanned with the (a) Aperio AT2 and (b) Leica SCN400 scanners.
Of note, the predictions reported above were made by analysis of images scanned at 20x magnification. Performance was comparable to predictions made on the same images scanned at 10x magnification (Supplemental Table 6). In the training dataset, we observed a weak negative association between prediction accuracy and the amount of time from tissue resection to treatment initiation (r = −0.16; p = 0.01). When we applied our model to the test cohorts, we found that there was a weak positive, but insignificant association between prediction accuracy and the amount of time between tissue resection and initiation of treatment (for slides scanned with Aperio AT2, r = 0.17 and p = 0.28; for slides scanned with the Leica SCN400, r = 0.08 and p = 0.62). Finally, we found that there was a weak negative, but insignificant association between DCNN prediction accuracy and the number of tiles used (for the NYU dataset, r = −0.07 and p = 0.23; for slides scanned with the Aperio AT2, r = −0.04 and p = 0.83; for slides scanned with the Leica SCN400, r = −0.17 and p = 0.30). The results of Class Activation Mapping are shown in Supplemental Figure 4. Overlaying the original tile images with the image of the CAM analyses revealed that cell nuclei play an important role in the decision to classify POD or response. The results of our CellProfiler analyses are shown in Supplemental Figure 5 [22]. Tiles labeled POD appear to be denser in number of nuclei and with larger nuclei than tiles labeled as Response (p < 0.0001 for both).
Multivariable logistic regression incorporating clinical variables augments prediction accuracy
In order to select from the candidate predictors, we performed multivariable logistic regressions that combined the neural networks’ outputs with conventional clinical characteristics. Among all of the parameters considered, the optimal multivariable logistic regression classifier combined the Response Classifier prediction with patients’ ECOG performance status (as a linear continuous score) and immunotherapy treatment (as a categorical variable) [Supplemental Table 7]. Other candidate predictors including tumor mutation burden did not have significant prognostic value in the multivariable analysis (results of univariable analysis are shown in Supplemental Table 2). The multivariable classifier achieved AUC 0.793 (95% CI: 0.713–0.874) on the NYU training cohort used to establish the regression model (Supplemental Figure 3). When tested on the Vanderbilt dataset, the classifier achieved an average AUC of 0.800 (95% CI: 0.634 – 0.967) on images from the Aperio AT2 scanner and an average AUC of 0.805 (95% CI: 0.638 – 0.971) on images from the Leica scanner (Figure 2). Model performance was better when applied to lymph node versus soft tissue. For Aperio AT2 scanned slides, the multivariable classifier performed with AUC 0.929 [0.787 – 1.070] on LN and AUC 0.708 [0.450 – 0.966] on ST. For Leica SCN400 scanned slides, the classifier performed with AUC 0.881 [0.693 – 1.069] on LN and AUC 0.734 [0.468 – 1.001] on ST. Model performance was consistent across 5 independent test runs for images scanned with each of the Aperio AT2 and Leica SCN400 scanners (Supplemental Tables 3 and 4). There was a trend towards higher AUC with the addition of each variable to the model, although the confidence intervals overlapped (Supplemental Table 8). The variable importance values were 1.99 for the DCNN, 2.25 for baseline ECOG, 2.39 for treatment with anti-PD-1, and 3.03 for treatment with anti-CTLA-4 plus anti-PD-1 (Supplemental Figure 6 and Supplemental Table 9).
An integrated approach can be used to stratify patients into high versus low risk for disease progression
After validating the DCNN and multivariable logistic regression models on the independent dataset from Vanderbilt, we identified the coordinates for the optimal threshold on the ROC curves from the NYU training set. The prediction probability score at the optimal threshold point was then set as a cutoff for stratifying Vanderbilt patients into two groups: high risk for disease progression or low risk for progression. For the multivariable classifier, the sensitivity and specificity at the optimal threshold were 64% and 84%, respectively. When using the predictions generated by the multivariable classifier, Vanderbilt patients were stratified into groups with significantly different progression free survival outcomes (P=0.02 for Aperio AT2 scanned slides; P=0.03 for Leica SCN400 scanned slides) [Figure 3]. The confusion matrices are shown in Supplemental Table 10 and 11. For Aperio scanned slides, the model performed with a sensitivity and specificity of 73% and 80%. For Leica scanned slides, the model performed with a sensitivity and specificity of 79% and 80%.
Figure 3.
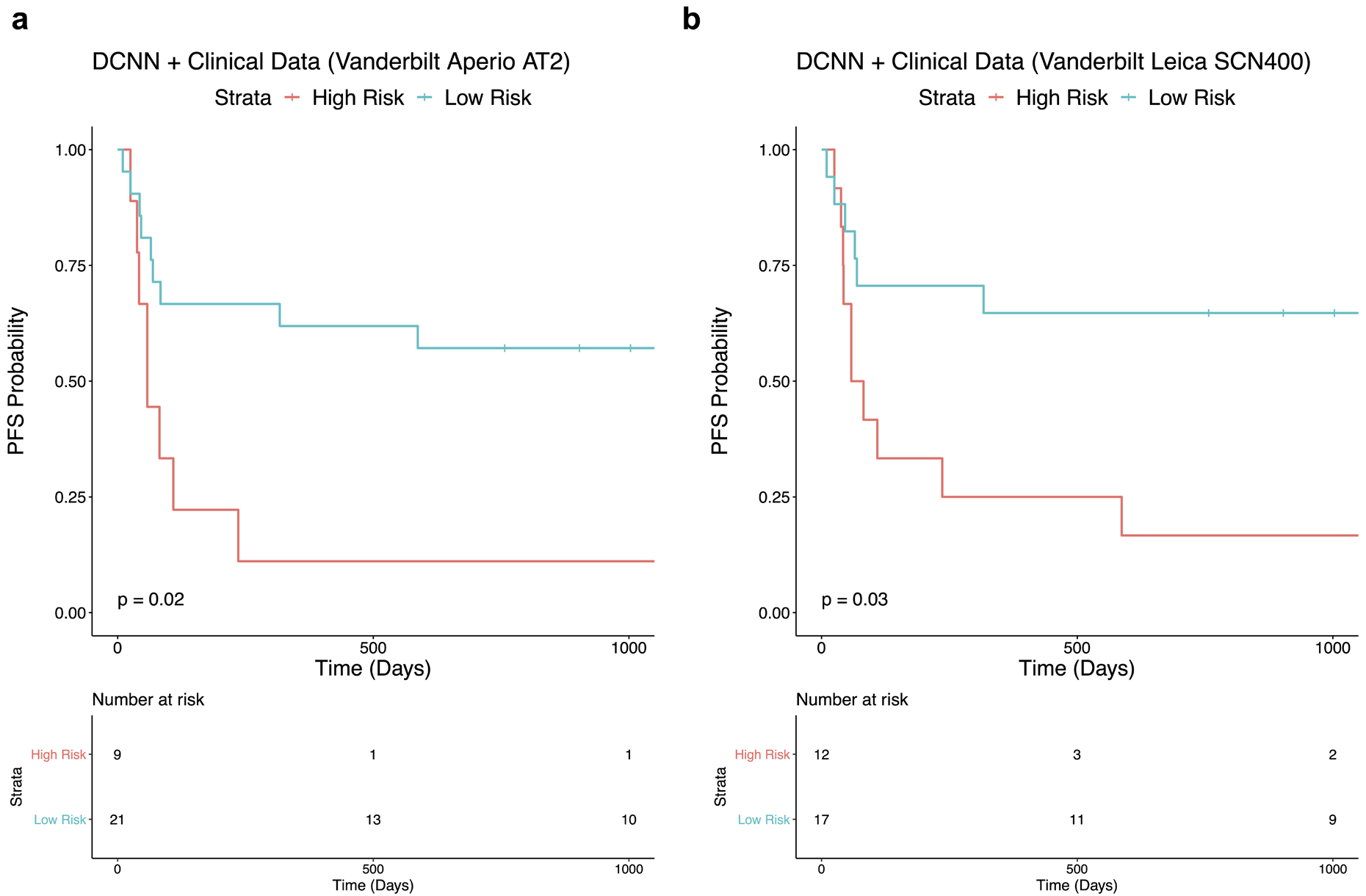
Prognostic potential of the multivariable classifier is demonstrated on slides from the independent test cohort that were scanned with an (a) Aperio AT2 scanner and (b) Leica SCN400 scanner.
Discussion
Immune checkpoint blockade has fundamentally changed the treatment landscape for advanced melanoma, but many individuals do not achieve long-term clinical benefit. Oncologists urgently need predictors of response to immunotherapy, but the models proposed to date have myriad limitations. Although PD-L1 expression is a widely implemented assay, its expression is inducible and can change after treatment initiation, which precludes its utility as a predictor of long-term response [26]. Chen et al (2018) showed that changes in exosomal PD-L1 expression predict immunotherapy response with AUC 0.9184, but this approach requires purification of exosomes, which limits its generalizability [9]. Several other robust prediction models were recently constructed using transcriptome expression profiles of immune checkpoint or T-cell activity. These perform with AUC ~0.8, but utilize RNA-sequencing and thus are not yet scalable to clinics outside of academic centers [7, 8]. Recent evidence supports tumor mutation burden as another potential predictor for ICI efficacy. Samstein et al. (2019) found that higher somatic mutation load was associated with better OS (HR 0.52; p=1.6 ×10−6) among 1662 advanced cancer patients who received immunotherapy. However, for the subset of melanoma patients in their study, the association between higher TMB and better OS was not statistically significant (p=0.067) [10]. Tumor mutation load was not associated with treatment response in our analysis either, which adds to the ongoing debate regarding whether and to what degree TMB has prognostic value in melanoma.
In two recent studies, neural network-based analyses of WSIs proved to be an effective tool for prognosticating survival outcomes in melanoma patients [27, 28]. Given the success of these and other computer vision models, there is growing interest in whether neural networks can be used to predict response to treatments. In 2019, Harder et al. proposed a workflow for predicting response to ipilimumab that relies on DCNNs to robustly segment cell nuclei and classify CD3+, CD8+, and melanin objects of interest [29]. Here, we present an approach to predicting treatment response that similarly draws upon the automated assessment of digital histology images. The neural network component to our model offers several capabilities with immediate translational relevance. It assesses routine H&E slides and thus utilizes data collected as a part of standard clinical care, which would ultimately facilitate swift clinical decision-making. Moreover, we don’t restrain the neural network’s prediction to specific geometric features from select cells. Instead, our DCNN independently analyzes entire tumor regions to generate its predictions. As such, our method only requires H&E-stained tissue as opposed to also needing CD3+ and CD8+ staining data; thus, it is less time and resource intensive. Additionally, there was minimal relationship between the accuracy of our classifier and the amount of time between tissue resection and treatment initiation. As applied to the clinical setting, this means that patients who underwent remote biopsy could potentially be spared repeat procedural intervention without compromising the utility of the assay, which would in turn mitigate delays in starting treatment. We also found that performance accuracy was minimally associated with the number of tiles used to generate the prediction. In practice, this means that a lower number of tiles from smaller tissue samples would not preclude accurate predictions, which would similarly obviate the need for repeat procedures. This also suggests that tissue from excisional, incisional, and core biopsies can be used since all provide an adequate amount of tissue for our model. Finally, our neural network also performs consistently when applied to WSIs from different slide scanners. This has important implications for clinical practice as it would allow smaller facilities to send digital pathology data to centers with the computing ability to run DCNN classifiers. Given the size of our validation dataset and concomitant possibility of underpowering, these findings should be verified in additional institutional datasets in order to confirm that our approach is consistent regardless of biopsy date, amount of tumor tissue, and slide scanner.
Using multiple logistic regression to merge our Response Classifier output with known clinical predictors was crucial to generate a model with enhanced accuracy. In prior studies, ECOG performance status predicted survival and response to immunotherapy in patients with melanoma [30, 31]. We too found that the incorporation of performance status augmented prediction accuracy, likely because it accounts for critical patient information not necessarily reflected in tissue histology. Prediction accuracy further improved by incorporating the patients’ treatment regimen to account for the heterogeneity of patients’ responses to various choices of ICI. However, there was no significant interaction effect between the Response Classifier and ECOG score or treatment regimen, which suggested that the prediction from the Response Classifier was significant, independent of ECOG score and choice of treatment. Ultimately, the final model accurately stratified patients into groups with significantly different PFS, which, when applied to clinical practice, could help optimize patient selection for treatment with immunotherapy. Importantly, the sensitivity and specificity of the final model’s prediction of response versus POD were comparable to those for PD-L1 immunohistochemistry. For determining objective response rate, which is the proportion of patients with CR or PR, the Dako 22C3 bioassay for pembrolizumab is reported to have a sensitivity of 80% and specificity of 60% at a stain cutoff of 1%. Dako 28–8 has a stain cutoff of 5%; at this point, the sensitivity and specificity are 58% and 49% for nivolumab monotherapy, and 57% and 54% for combination ipilimumab and nivolumab therapy [32]. In contrast, the sensitivity and specificity of our multivariable classifier is 64% and 84%, respectively. However, the comparison of these two assays is limited by the fact that the majority of our training cohort received ipilimumab monotherapy.
It is noteworthy that our model performed consistently despite being trained on a cohort who primarily received anti-CTLA-4 therapy and tested on a cohort who mostly received anti-PD-1 therapy. One recent study showed that biomarkers derived from anti-CTLA-4 response datasets have limited applicability to patients treated with anti-PD-1 agents [33]. However, several other studies introduce predictive models that perform equally well when applied to patients who receive anti-CTLA-4 or anti-PD-1 therapy [7, 8]. Taken together, these mixed data suggest that some biomarkers are not necessarily specific to checkpoint target. Based on our CAM analyses, the DCNN appears to predict disease progression based on regions where nuclei are larger and more numerous. This could reflect an appreciation for greater ploidy, which has been shown to be associated with immunotherapy response [34]. Genome instability and higher cancer neoantigen load can inform likelihood of an immune response irrespective of checkpoint target, which could explain why our algorithm performs well when applied to different treatments. Notwithstanding the above, CTLA-4 and PD-1 blockade have different response rates and patients who don’t respond to one regimen might have responded to another. Although we controlled for different therapies in our multivariable analyses, larger studies would enable the construction of classifiers specific to each treatment modality.
This proof-of-principle study has several other limitations. First, as mentioned above, we were constrained by the limited amount of available data, which comes from 151 patients in total. This created a ceiling for peak neural network accuracy because the weights and biases within a DCNN are fine-tuned through back propagation, so more data naturally allows for more training epochs. In fact, recent research suggests that larger datasets with thousands of WSIs are necessary to achieve a level of tissue classification performance acceptable for clinical implementation [35]. Second, we found that both the neural network and multivariable classifiers generated more accurate predictions on lymph node than on soft tissue. Performance on soft tissue may be worse because there were fewer soft tissue slides in the training set, or because segmentation of soft tissue is worse, or because of other unspecified inherent features of soft tissue samples that make it difficult to generate predictions of response. Future studies with larger datasets should train and test the efficacy of models on selectively lymph node or soft tissue. Third, we found that, despite color normalization, the Response Classifier was sensitive to differences in staining, which can be a consequence of both different staining procedures as well as variability in the age of the slides. In this study, we tested the algorithm against 2 different staining protocols that use variable amounts of hematoxylin. Knowing that hematoxylin stains nucleic acids, it follows that protocols which use more hematoxylin could compromise the neural network’s ability to differentiate the density of nuclei. For this study, we therefore concentrated on using a single staining protocol for the training and test sets, but in the future, training the algorithm to adapt for differences in slide appearance will be requisite to improve the generalizability of the assay. To make it work on a broad range of stains will require training on larger datasets that include slides stained with different protocols.
In conclusion, we demonstrate the feasibility of predicting immunotherapy response by combining neural network classification on histology slides with clinicodemographic information. Our proposed model overcomes the limitations of the temporal and spatial heterogeneity that impede the performance of PD-L1 as well as the resource scarcity that precludes using RNA-sequencing, while simultaneously maintaining its validity across multiple slide scanners. With further optimization of the model using larger datasets, and following prospective validation in the clinical trial setting, we believe this computational approach has the potential for integration into clinical practice. This could help oncologists identify patients who are at high versus low risk for progression through immunotherapy. Going forward, it will be interesting to test the model’s efficacy when applied to primary melanoma tissue as well as to tissue from other cancers. Ultimately, we suspect that a combination of methods will be used to predict immunotherapy response. In this setting, our fast and readily available approach could provide rapid first assessments to pre-select candidates for treatment or identify those who require further analysis using complementary predictive models.
Supplementary Material
Statement of translational relevance.
We present a computational method that integrates deep learning on histology specimens with clinicodemographic variables to predict treatment outcomes in advanced melanoma. Using hematoxylin and eosin stained slides of metastatic lymph node and subcutaneous tissue, we trained a neural network classifier to identify whether individuals responded to checkpoint blockade or suffered disease progression. We then developed a logistic regression classifier that combines neural network output with clinicodemographic variables to generate predictions with enhanced accuracy. Our approach is time-efficient, reproducible, and requires minimal resource allocation, thus overcoming multiple common barriers to generalizability for contemporary biomarkers.
Acknowledgements
This work was supported by the NYU Melanoma SPORE (P50CA225450 to I. Osman and J. Weber), Melanoma Research Alliance (622668 to I. Osman), American Cancer Society (RSG-15-189-01-RMC to A. Tsirigos), Laura and Isaac Perlmutter Cancer Center (P30CA016087 to I. Osman), and Onassis Foundation (F ZP 036-1/2019-2020 to S. Noumikou). For this work, we used computing resources at the High-Performance Computing Facility at NYU Langone Medical Center. We thank the Center for Biospecimen Research and Development (CBRD) and Experimental Pathology Research Laboratory (EPRL) at NYU Langone Medical Center for scanning the H&E-slides. We thank the Applied Bioinformatics Laboratories for providing computational support and helping with the analysis and interpretation of the data. We thank Dr. Randie Kim and Zarmeena Dawood for their assistance in the preliminary steps of this project. We thank Runyu Hong for his assistance with the Class Activation Mapping. We thank Dr. Narges Razavian and Dr. Theodoros Sakellaropoulos for their advice on the deep-learning section of this work.
Footnotes
Conflicts of interest
Author J.W. is a paid consultant for Merck, Genentech, Astra Zeneca, GSK, Novartis, Nektar, Medivation, Celldex, Incyte, EMD Serono, and BMS. J.W. holds equity in CytoMx, Biond and Altor. J.W. is on advisory boards for Celldex, CytoMx, Incyte, Biond, Protean, CV6 and Sellas. J.W. is on a patent filed by Moffitt Cancer Center for an Ipilimumab biomarker. J.W. is on a PD-1 patent filed by Biodesix. Author D.B.J. serves on advisory boards for Array Biopharma, BMS, Merck, and Novartis, and receives research funding from BMS and Incyte. All remaining authors have declared no conflicts of interest.
Data availability
Data is available from the authors upon request but may require data transfer agreements. No personalized health information will be shared.
Code availability
The code for the neural network and logistic regression classifiers is available on GitHub at the following location: https://github.com/ncoudray/DeepPATH/tree/master/DeepPATH_code. The CellProfiler pipeline is available at: https://github.com/sofnom/HistoPathNCA_pipeline
References
- 1.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Eng. J. Med 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Robert C, Ribas A, Schachter J, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 20, 1239–1251 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 19, 1480–1492 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Michot JM, Bigenwald C, Champiat S, Collins M, Carbonnel F, Postel-Vinay S, et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer 54, 139–148 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Jacquelot N, Roberti MP, Enot DP, Rusakiewicz S, Ternes N, Jegou S, et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun 8, 592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auslander N, Zhang G, Sang Lee J, Frederick D, Miao B, Moll T, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med 24, 1545–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Huang A, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian Y, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrera C, Gual A, Diaz A, Puig-Butille JA, Nogues S, Vilalta A, et al. Prognostic role of the histological subtype of melanoma on the hands and feet in Caucasians. Melanoma Res. 27, 315–320 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Lattanzi M, Lee Y, Simpson D, Moran U, Darvishian F, Kim R, et al. Primary melanoma histologic subtype: impact on survival and response to therapy. J. Natl. Cancer. Inst 111, 180–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pizzichetta MA, Massi D, Mandala M, Queirolo P, Stanganelli I, De Giorgi V, et al. Clinicopathological predictors of recurrence in nodular and superficial spreading cutaneous melanoma: a multivariate analysis of 214 cases. Journal of Trans. Med 15, 227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li CX, Shen CB, Xue K, Shen X, Jing Y, Wang ZY, et al. Artificial intelligence in dermatology: past, present, and future. Chin. Med. J 132, 2017–2020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haenssle HA, Fink C, Schneiderbauer R, Toberer F, Buhl T, Blum A, et al. “Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists.” Ann. Oncol 29, 1836–1842 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Esteva A, Kuprel B, Novoa R, Ko J, Swetter S, Blau H, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). Brit. J. Cancer 93, 387–391 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szegedy C, Vanhoucke V, Ioffe S, Shlens J & Wojna Z. Rethinking the Inception Architecture for Computer Vision In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2818–2826 (Boston, MA, USA, 2015). [Google Scholar]
- 20.Reinhard E, Ashikhimin M, Gooch B, and Shirley P. Color transfer between images. IEEE Comput. Graph. Appl 21, 34–41 (2001). [Google Scholar]
- 21.Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 35, 1962–1971 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kim R, Nomikou S, Coudray N, Jour G, Dawood Z, Hong R, et al. 2020. A deep learning approach for rapid mutational screening in melanoma. bioRxiv doi: 10.1101/610311 [DOI] [Google Scholar]
- 23.Zhou B, Khosla A, Lapedriza A, Oliva A, & Torralba A. Learning deep features for discriminative localization 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, pp. 2921–2929 (2016). [Google Scholar]
- 24.Hong R, Liu W, DeLair D, Razavian N, Fenyo D. 2020. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. bioRxiv doi: 10.1101/2020.02.25.965038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilm A, Poh Kim Aw P, Bertrand D, Hui Ting Yeo G, Hoe Ong S, Hua Wong C, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tray N, Weber JS, & Adams S. Predictive biomarkers for checkpoint immunotherapy: current status and challenges for clinical application. Cancer Immunol. Res 6, 1122–1128 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Acs B, Ahmed FS, Gupta S, Fai Wong P, Gartrell RD, Pradhan JS, et al. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat. Commun 10, 5440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni PM et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin. Cancer Res Epub ahead of print doi: 10.1158/1078-0432.CCR-19-1495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harder N, Schonmeyer R, Nekolla K, Meier A, Brieu N, Vanegas C, et al. Automated discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Scientific Reports. 9, 7449, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diem S, Kasenda B, Martin-Liberal J, Lee A, Chauhan D, Gore M, Larkin J. Prognostic score for patients with advanced melanoma treated with ipilimumab. Eur. J. Cancer 51, 2785–2791 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Wong A, Williams M, Milne D, Morris K, Lau P, Spruyt O, et al. Clinical and palliative care outcomes for patients of poor performance status treated with antiprogrammed death-1 monoclonal antibodies for advanced melanoma. Asia Pac. J. Clin. Oncol 13, 385–390 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Diggs LP & Hsueh EC. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark Res. 5, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davoli T, Uno H, Wooten EC, & Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 355, eaaf8399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck V, Silva K, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


