Abstract
Background:
Expanded RAS/BRAF mutations have not been assessed as predictive for single-agent cetuximab in metastatic colorectal cancer (mCRC) and low mutant allele frequency (MAF) mutations are of unclear significance. We aimed to establish cetuximab efficacy in optimally selected patients using highly sensitive BEAMing, capable of detecting alterations below standard clinical assays.
Methods:
CO.17 compared cetuximab versus best supportive care (BSC) in RAS/BRAF unselected mCRC. We performed RAS/BRAF analysis on micro-dissected tissue of 242 patients in CO.17 using BEAMing for KRAS/NRAS (codons 12/13/59/61/117/146) and BRAF V600E. Patients without BEAMing but with previous Sanger sequencing detected mutations were included.
Results:
KRAS, NRAS, and BRAF mutations were present in 53%, 4%, and 3% of tumors, respectively. Cetuximab improved overall survival (OS) (HR 0.51, 95% CI 0.32–0.81, P=0.004) and progression free survival (PFS) (HR 0.25, 95% CI 0.15–0.41, P<0.0001) compared to BSC in RAS/BRAF wild type patients. Cetuximab did not improve OS/PFS for KRAS, NRAS, or BRAF mutated tumors and tests of interaction confirmed expanded KRAS (P=0.0002) and NRAS (P=0.006) as predictive, while BRAF mutations were not (P=0.089). BEAMing identified 14% more tumors as RAS mutant than Sanger sequencing and cetuximab lacked activity in these patients. Mutations at MAF<5% were noted in 6/242 patients (2%). One patient with a KRAS A59T mutation (MAF=2%) responded to cetuximab. More NRAS than KRAS mutations were low MAF (OR 20.50, 95% CI 3.88–96.85, P=0.0038).
Conclusions:
We establish single-agent cetuximab efficacy in optimally selected patients and show that subclonal RAS/BRAF alterations are uncommon and remain of indeterminate significance.
Keywords: colon, rectal, metastatic, mutation, KRAS, NRAS, subclonal
Introduction:
The anti-epidermal growth factor receptor (anti-EGFR) antibodies cetuximab and panitumumab are important treatment options for patients with metastatic colorectal cancer (mCRC). KRAS/NRAS (RAS) mutation status and primary tumor location guide treatment selection, with left sided RAS wild type tumors showing greatest benefit from anti-EGFR antibodies.1–7 Patients with BRAF V600E mutations may also have reduced benefit from anti-EGFR therapy.8 However, it is unclear whether BRAF mutations obviate all benefit, and a test of interaction for the predictive utility of BRAF V600E mutations has not been established.
Though RAS and BRAF V600E sequencing helps identify the optimal population to treat with anti-EGFR antibodies, eventually patients develop resistance through acquired RAS mutations, which appear to be expanded from rare clones pre-existing in the tumor.9 Longitudinal assessment of circulating tumor DNA (ctDNA) has provided evidence that resistant clones decay with time, creating an opportunity for anti-EGFR re-challenge.10 In the CRICKET phase 2 trial, patients previously treated with anti-EGFR antibodies who subsequently progressed were re-challenged with cetuximab + irinotecan after intervening therapy. Among patients who were RAS wild type by ctDNA preceding re-challenge, progression free survival (PFS) was 4.0 months.11 Numerous other re-challenge protocols are underway, many with drug combinations, and it is essential to understand the magnitude of benefit of single-agent anti-EGFR therapy to establish a bench mark for re-challenge.9 Expanded RAS and BRAF V600E have not been previously assessed in a randomized single-agent cetuximab trial to establish predictive capacity and their utility are extrapolated from multi-agent or panitumumab trials.
It is also unclear whether mutations below the 5% mutant allele frequency (MAF) limit of detection of standard assays are of importance.12 Although historic PCR and Sanger sequencing methods identified mutations occurring at MAFs above 10–20%, newer techniques have sensitivities down to 0.1% and may lead to improved outcomes.13 In the CAPRI-GOIM trial evaluating FOLFIRI + cetuximab in mCRC, next generation sequencing revealed an additional 15.9% of patients with KRAS exon 2 mutations beyond Sanger sequencing. These patients had inferior outcomes compared to patients with RAS wild-type tumors and similar prognosis to high allele frequency RAS mutations.14 These findings have been replicated by many retrospective studies, however it remains unclear whether low allele frequency mutations obviate all benefit.15–17 In the CRYSTAL trial evaluating cetuximab and FOLFIRI in the first line, the use of high sensitivity BEAMing demonstrated a relationship between the RAS MAF and anti-EGFR efficacy and it was unclear whether low MAF mutations prevent all benefit.18
Given the current gaps in knowledge, we undertook a retrospective analysis of the CO.17 trial comparing cetuximab with best supportive care (BSC) to establish 1) the efficacy of single-agent cetuximab in optimally selected RAS/BRAF wild type patients relative to BSC, and 2) the frequency and clinical relevance of low allele frequency RAS mutations detected by an ultra-sensitive assay.
Methods:
Patient population:
CO.17 was a phase III clinical trial that randomized patients 1:1 to receive either cetuximab or BSC after institutional review board approval (NCT00079066).1 The study was IRB approved, with written consent for all subjects, and conducted in accordance with the Declaration of Helsinki and International Ethical Guidelines for Biomedical Research Involving Human Subjects. Patients consented to enrollment and correlative studies and had either progressed on or were intolerant of a fluoropyrimidine, oxaliplatin, and irinotecan. No prior anti-EGFR therapy was allowed. Enrollment was unselected for RAS/BRAF.
This correlative analysis assessed all patients with remaining evaluable tissue (N=242). Median time from tissue collection to randomization was 2.2 years for patients who underwent analysis with BEAMing and did not differ between arms of the study (P=0.22). Of 211 samples with known site of origin, 207 arose from primary tumors (98.1%), while 4 were from metastases (1.9%). There were 84 patients without remaining tissue that were historically identified to have a KRAS exon 2 (N=76) or BRAF V600E (N=8) mutation using Sanger sequencing from previously published analyses that were included.1,19 Two additional patients were not analyzable for KRAS in the BEAMing assay but historically had a KRAS exon 2 which was used to fill in the missing result. Patients with a prior mutation were included, however patients with no remaining tissue and a prior result that did not identify a mutation were excluded, as the previous assessments lacked coverage of all KRAS/NRAS codons.
Treatment:
Cetuximab treatment consisted of an intravenous loading dose of 400 mg/m2 followed by 250 mg/m2 given weekly until progression.
RAS and BRAF Testing:
Archival formalin fixed and paraffin embedded (FFPE) blocks were evaluated for sample quality prior to sectioning five slides for DNA extraction. Areas of highest tumor content were selected and micro-dissected. DNA was extracted using QIAMP DNA FFPE tissue kits with barcoding to maintain sample continuity.
Prior to sequencing, samples underwent a repair step using the New England Biolabs PreCR repair mix. DNA isolated from FFPE was subjected to LINE-1 qPCR for quantification and quality control.20 Only PCR-accessible, inhibition-free and amplifiable target regions qualified for subsequent analysis. For any sample with amplicons exhibiting insufficient amplification, PCR products underwent additional analysis on an agarose gel to confirm successful and target-specific amplification before BEAMing analysis was performed. 2/242 samples had one or more amplicons in RAS that was not analyzable due to unamplifiable DNA. These samples were still analyzed for BRAF mutations.
A previously described highly sensitive beads, emulsion, amplification, and magnetics (BEAMing) analysis was utilized to detect mutations in KRAS/NRAS (codons 12, 13, 59, 61, 117, & 146) and BRAF V600E with coverage outlined in Supplemental table 1 and a 1% MAF limit of detection. Sequencing was carried out by Sysmex Inostics (Baltimore, MD).15,18,21
Statistical methods:
Survival was summarized with Kaplan-Meier curves and compared using stratified log-rank tests adjusted for performance status at randomization. Hazard ratios and 95% confidence intervals (95% CI) were calculated from stratified Cox-regression models with treatment group as the single factor. Overall survival (OS) was defined as the time from randomization until death from any cause. Progression free survival (PFS) was defined as the time from randomization until progression or death from any cause. To determine whether expanded RAS and BRAF V600E mutations were predictive, we used a Cox model with treatment, mutation status, and their interaction term as covariates. Objective response rate (ORR) was defined according to modified Response Evaluation Criteria in Solid Tumors.22 Between group comparisons used Kruskal-Wallis tests for continuous variables or a χ2/Fisher’s exact test as appropriate.
Results:
Patient Population:
Of 572 patients, 242 (42%) underwent analysis with BEAMing. BEAMing was successful in all samples for BRAF, but 3 had inconclusive RAS analysis. Baseline characteristics are summarized in Table 1. Prevalence in the BEAMing population was 97 (41%) RAS/BRAF V600E wild type, 126 (53%) KRAS, 9 (4%) NRAS, and 7 (3%) BRAF V600E mutated, with specific mutations noted in Supplemental Table 2. There were 5 patients with 2 concurrent KRAS mutations, while 1 patient had 3 concurrent KRAS mutations. Patients with multiple alterations frequently had second mutations of low allele frequency. These cases were excluded from analysis of low allele frequency variants as they had both high and low allele frequency alterations (Supplemental Figure 1).
Table 1.
Baseline patient characteristics
| Characteristic | |||||
|---|---|---|---|---|---|
| RAS and BRAF V600E Wild Type (N=97) | KRAS Mutated (N=204) | NRAS Mutated (N=9) | BRAF V600E Mutated (N=15) | P | |
| Median Age (range) | 64 (29–88) | 63 (37– 86) | 69 (47–75) | 64 (39– 77) | 0.79 |
| Gender | |||||
| Female | 28 (29) | 72 (35) | 2 (22) | 5 (33) | 0.63 |
| Male | 69 (71) | 132 (65) | 7 (78) | 10 (67) | |
| ECOG | |||||
| 0 | 26 (27) | 46 (23) | 1 (11) | 1 (7) | 0.29 |
| 1 | 58 (60) | 117 (57) | 5 (56) | 9 (60) | |
| 2 | 13 (13) | 41 (20) | 3 (33) | 5 (33) | |
| Side of tumor | |||||
| Right | 18 (19) | 71 (37) | 3 (38) | 10 (67) | 0.0005 |
| Left | 78 (81) | 123 (63) | 5 (63) | 5 (33) | |
| Prior Treatment | |||||
| 5-FU | 97 (100) | 204 (100) | 9 (100) | 15 (100) | 1 |
| Irinotecan | 92 (95) | 199 (98) | 8 (89) | 14 (93) | 0.36 |
| Oxaliplatin | 96 (99) | 201 (99) | 9 (100) | 15 (100) | 0.93 |
| Site of Disease | |||||
| Liver | 87 (90) | 159 (78) | 8 (89) | 10 (67) | 0.038 |
| Lung | 56 (58) | 133 (65) | 5 (56) | 10 (67) | 0.60 |
| Nodes | 51 (53) | 80 (39) | 3 (33) | 7 (47) | 0.16 |
| Treatment | |||||
| Cetuximab | 54 (56) | 101 (50) | 3 (33) | 7 (47) | 0.45 |
| BSC | 43 (44) | 103 (51) | 6 (67) | 8 (53) | |
Percentages represent the % of known, N=number, BSC = best supportive care
Overall Survival:
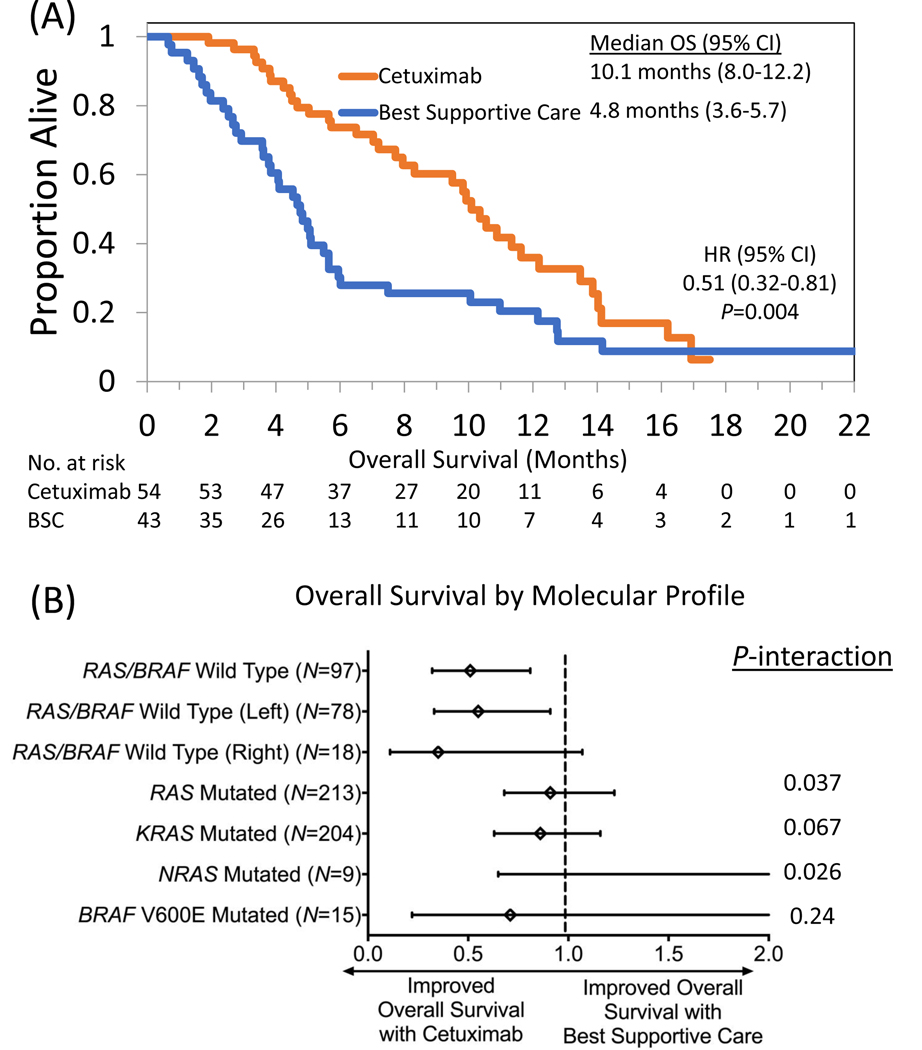
OS was significantly improved with cetuximab compared to BSC (median 10.1 vs 4.8 months, HR 0.51, 95% CI 0.32–0.81, P=0.004) in patients with RAS/BRAF V600E wild type tumors (Figure 1A). No improvement in OS was noted following cetuximab in patients with KRAS (HR 0.86, 95% CI 0.63–1.16, P=0.32), NRAS (HR 3.93, 95% CI 0.65–23.89, P=0.11), combined RAS (HR 0.91, 95% CI 0.68–1.23, P=0.55) or BRAF V600E mutated tumors (HR 0.71, 95% CI 0.22–2.27, P=0.56) compared to BSC (Figure 1B). A test of interaction was positive for combined RAS (P=0.037) and NRAS (P=0.026) but not KRAS alone (P=0.067) or BRAF V600E mutations (P=0.24) as predictive biomarkers for OS following cetuximab. Among RAS/BRAF V600E wild-type patients, left sided tumors (median 10.4 vs 4.8 months, HR 0.55, 95% CI 0.33–0.91, P=0.019) had improved OS with cetuximab relative to BSC but this was not significant for right sided tumors (median 5.7 vs 3.7 months, HR 0.35, 95% CI 0.11–1.07, P=0.055). We repeated our analysis but included patients with mutations <5% MAF as wild type (N=6) and noted no differences in results (Supplemental Table 3).
Figure 1.
Impact of cetuximab on overall survival in patients with (A) RAS/BRAF V600E wild type metastatic colorectal cancer compared to best supportive care in the CO.17 trial and stratified by (B) molecular subgroup.
Progression Free Survival:
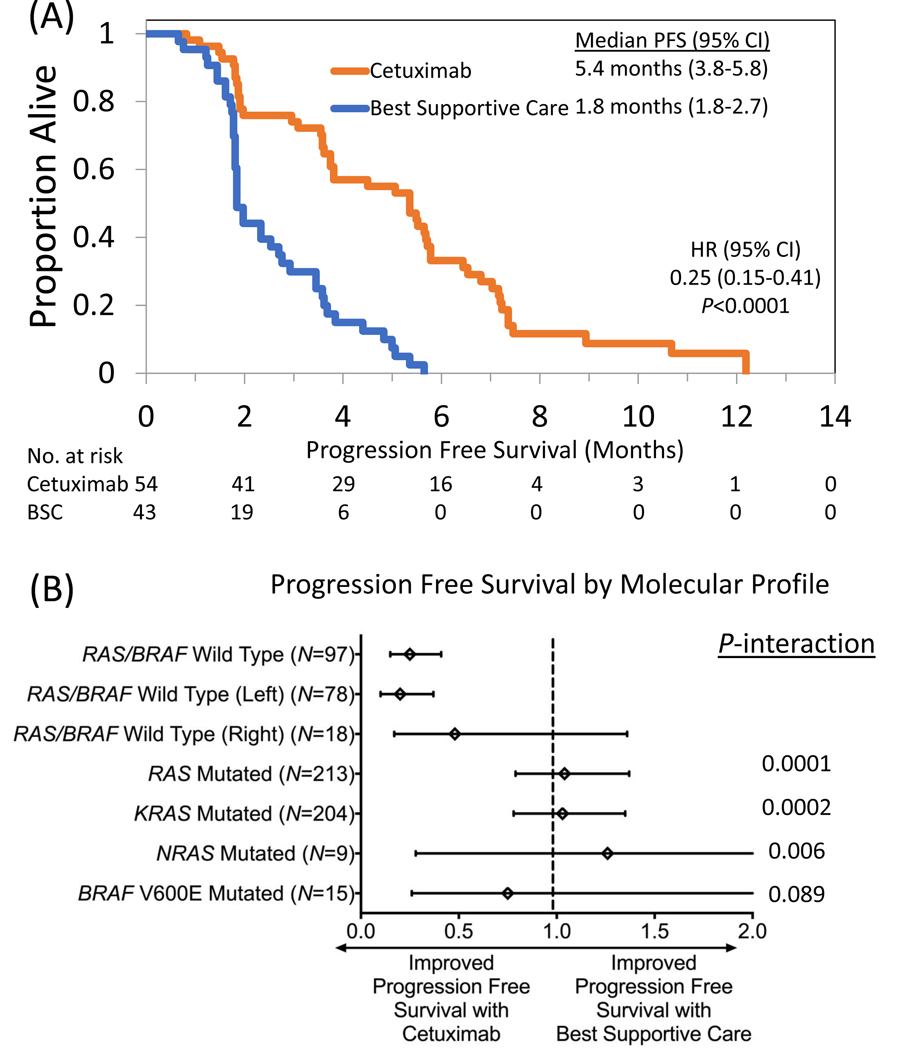
Among patients with RAS/BRAF V600E wild type tumors, PFS improved following cetuximab relative to BSC (median 5.4 vs 1.8 months, HR 0.25, 95% CI 0.15–0.41, P<0.0001) (Figure 2A). There did not appear to be any prolongation of PFS with the use of cetuximab for patients with KRAS (HR 1.03, 95% CI 0.78–1.35, P=0.86), NRAS (HR 1.26, 95% CI 0.28–5.74, P=0.76), combined RAS (HR 1.04, 95% CI 0.79–1.37, P=0.76) or BRAF V600E mutations (HR 0.75, 95% CI 0.26–2.19, P=0.60) (Figure 2B). KRAS (P=0.0002), NRAS (P=0.006), and combined RAS mutations (P=0.0001) were predictive of lack of benefit from cetuximab for PFS using a test of interaction, while BRAF V600E mutations neared significance for predictive utility (P=0.089). Left sided RAS/BRAF V600E wild type tumors had prolonged PFS following cetuximab (median 5.5 vs 2.0, HR 0.20, 95% CI 0.10–0.37, P<0.0001), while right sided tumors did not meet significance (median 3.6 vs 1.8 months, HR 0.48, 95% CI 0.17–1.36, P=0.16). Similar to OS, when we categorized the 6 patients with mutations occurring at MAF<5% as wild type and repeated the analysis, we noted no change in the PFS end point (Supplemental Table 3).
Figure 2.
Impact of cetuximab on progression free survival in patients with (A) RAS/BRAF V600E wild type metastatic colorectal cancer compared to best supportive care in the CO.17 trial and stratified by (B) molecular subgroup.
Response Rate:
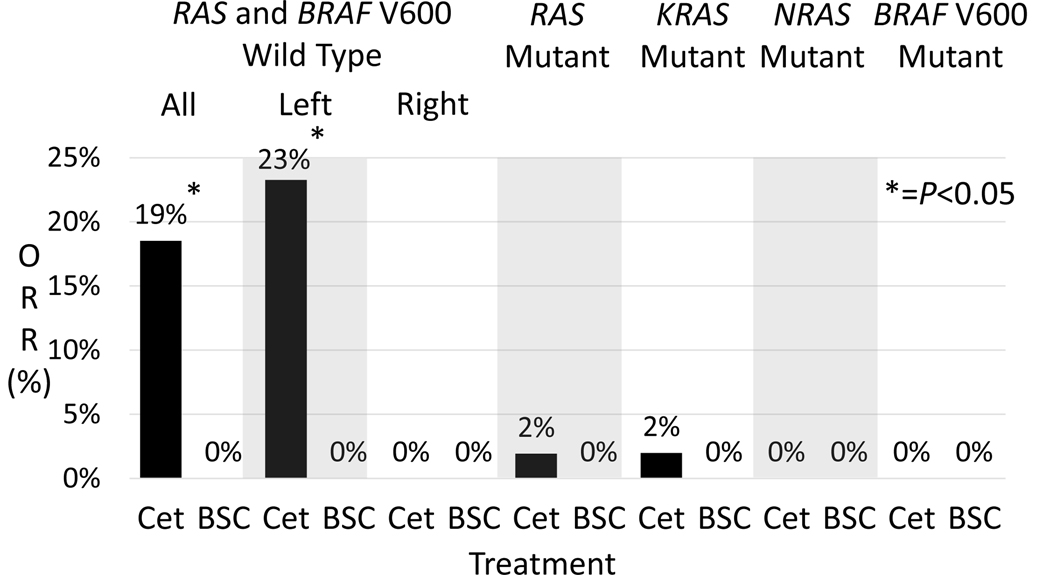
ORR (19% vs 0%, P=0.002) was significantly improved with cetuximab compared to BSC in RAS/BRAF V600E wild type CRC (Figure 4). Among patients with KRAS (ORR 2%), NRAS (ORR 0%), combined RAS (ORR 2%), and BRAF V600 mutations (ORR 0%) there was no difference in ORR relative to BSC where ORR was 0% in all molecular groups. In left sided RAS/BRAF V600E wild type tumors ORR was higher than right sided tumors (23% vs 0%, P=0.18) but not significantly different. Categorizing mutations <5% MAF as wild type did not change the ORR for patients with RAS/BRAF V600E wild type tumors but did decrease KRAS and combined RAS ORR to 1%.
Figure 4.
Objective response rate of patients in CO.17 receiving cetuximab or best supportive care (BSC).
Low Mutant Allele Frequency (MAF) Mutations:
Low allele frequency mutations (MAF<5%) occurred in 6/242 patients (2%). In these six tumors, 3 KRAS (G12V, A59T, A59T) and 3 NRAS mutations (G13R, A146T, A59T) were identified. Mutations in NRAS were more likely to occur at low allele frequency than KRAS (OR 20.5, 95% CI 3.9–96.9, P=0.0038). A59T RAS mutations were present in 3/6 patients (2 with KRAS and 1 NRAS) with low MAF alterations compared with 0/136 patients with mutations occurring at MAF>5% (OR ∞, 95% CI 26.28−∞, P<0.0001).
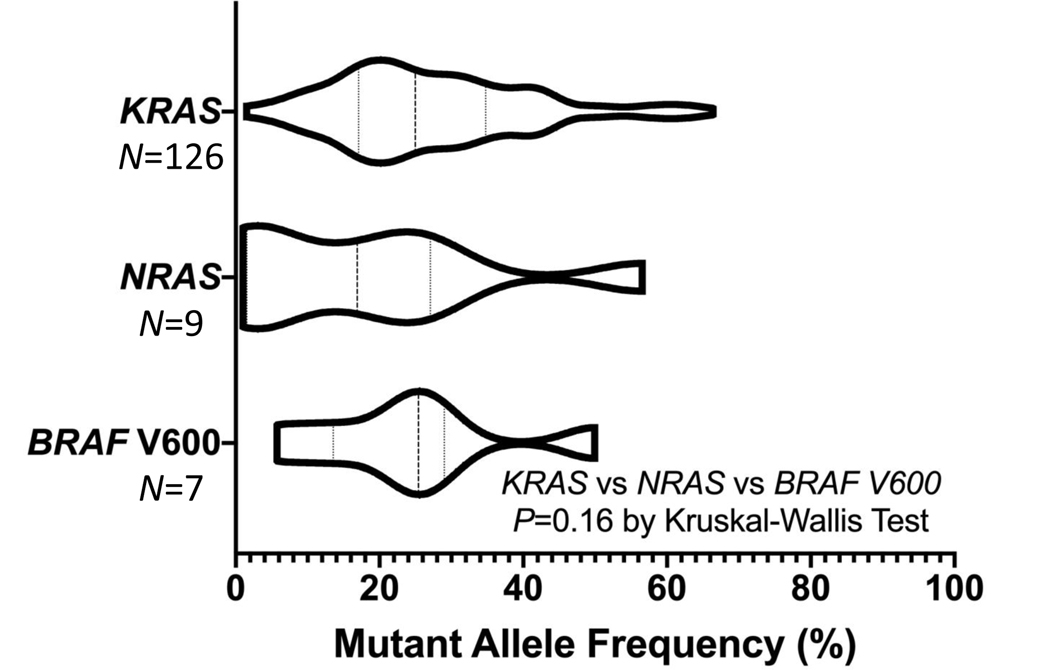
Most mutations occurred at high allele frequencies consistent with a clonal mutation (Figure 3). KRAS variants trended towards higher MAF than NRAS (P=0.058), but did not differ from BRAF V600E (P=0.69). NRAS and BRAF V600 allele frequencies did not differ (P=0.32). There were 34 (14%) patients who had results for KRAS exon 2 available from Sanger sequencing who were previously wild-type but now had a mutation in KRAS exon 2 detected with BEAMing. The median MAF for these 34 patients was 20% (range 2%−60%) and treatment with cetuximab did not improve OS (median 6.8 vs 5.4 months, HR 0.60, 95% CI 0.27–1.35, P=0.21), PFS (median 1.9 vs 1.8 months, HR 0.72, 95% CI 0.36–1.46, P=0.36) or response rate (7% vs 0% with BSC, P=0.41) among these patients, suggesting they were clinically relevant. Seven patients (2.9%) previously had Sanger detected KRAS mutations but were re-classified as wild type. All discordant cases had high quality assay results with BEAMing and were reviewed.
Figure 3.
Violin plot displaying the mutant allele frequency distribution density of detected mutations in KRAS, NRAS and BRAF.
Of the 3 patients with RAS mutations at MAF <5% who received cetuximab, two progressed after 2.7 and 3.7 months with only one patient having a partial response that lasted 11.2 months, while those with low MAF RAS mutations in the BSC arm progressed after 1.9 and 3.6 months with one patient withdrawing and none having a response. The one response to cetuximab occurred in a male patient with a KRAS A59T mutation (MAF=2%) occurring in a left sided tumor. The patient had received prior fluoropyrimidine, oxaliplatin, and irinotecan and had liver limited metastatic disease. OS for patients with low MAF RAS mutations was 11.6, 18.2, and 12.4 months following cetuximab and 2.7, 10.7, and 12.0 months following BSC.
Discussion:
This updated analysis of CO.17 refines our understanding of the magnitude of benefit from single-agent cetuximab in optimally selected patients. Compared to the previous assessment of only KRAS exon 2 mutations (mPFS of 3.7 months with cetuximab), mPFS increased to 5.4 months with improved molecular profiling.1 Using highly sensitive BEAMing we identified an additional 14% of patients who were wild type by Sanger sequencing and lacked benefit from cetuximab, highlighting the utility of more sensitive assays. This work also enhances our knowledge of predictive biomarkers in mCRC. Previously, a test of interaction for anti-EGFR interacting with expanded RAS mutations was only available for panitumumab, not cetuximab3. Additionally, despite the non-significant (P=0.089) test of interaction for BRAF V600E mutations being a predictive biomarker, this work highlights reduced benefit from anti-EGFR therapy in this population. These findings support early incorporation of combination BRAF directed treatment rather than single agent anti-EGFR therapy.23
Our prevalence estimate of expanded RAS mutations (56%) agrees with other series, where pooled estimates suggest RAS mutations occur in 55.9% of mCRC.24 By combining RAS and BRAF V600E alterations, the population expected to benefit from single-agent anti-EGFR therapy drops to only 41% in our study. Interestingly, we only detected an additional 6 (2%) patients with low allele frequency mutations. This is lower than others have reported, and may reflect the impact of tumor micro-dissection or utilization of a threshold for the assay associated with low rates of false-positive results (1% instead of 0.1%). Improved methodologies for high-depth sequencing have been developed, although the clinical relevance of such higher sensitivity approaches remain unclear given the low prevalence of this population and difficulty confirming lack of benefit.
In our study, 3 individuals had tumors harboring mutations at MAF<5% who received anti-EGFR therapy. One of these patients had a response to cetuximab, suggesting a potential gradient of efficacy based on the MAF of mutant RAS in a tumor. This is supported by the CRYSTAL trial, where a gradient of activity was noted among patients based on allele frequency of RAS mutations.18 By using BEAMing technology, we were able to provide better stratification of patients. Not only were there 6 patients with mutations occurring between 1–5% MAF, but we also identified 34 patients that were KRAS wild type by Sanger sequencing. This suggests there were “intermediate” allele frequency mutations not detected with Sanger sequencing (threshold for detection between MAF 10–20%), however current next generation sequencing assays may have identified them.1,13 Indeed, the median MAF of these 34 discordant cases was 20%. Although many of these samples should have had variants detected by Sanger’s threshold, an important distinction between the original assessment of KRAS for CO.17 and our current analysis is that microdissection was performed in our updated analysis. Therefore, the detected allele frequencies are likely higher than would have been noted in the original analysis that used whole slides. Taken together, our results lend further support to the need for high sensitivity assays in the clinic.
Current guidelines suggest assays need a 5% MAF limit of detection for RAS mutations and our work suggests the number of additional patients identified with more sensitive assays is relatively small.12 While only 3 patients with low MAF RAS mutations were treated with cetuximab, 1 of these patients had a response and a PFS of 11.2 months, while 2 others with low MAF mutations had PFS of 2.7 and 3.7 months. In the CRYSTAL trial of FOLFIRI +/− cetuximab, high sensitivity BEAMing identified 23/430 (5.3%) patients with RAS mutations outside of codon 12/13 occurring at allele frequencies of 0.1–5%. In this group, the addition of anti-EGFR agents provided a signal towards benefit (HR 0.57, 95% CI 0.33–1.01).25 While the low allele frequency of the responding patient’s mutation in CO.17 may explain the activity of cetuximab, the mutation was KRAS A59T which has previous case reports of response and is one of these least well studied RAS mutations, with only 7 patients harboring this alteration in the PRIME trial that defined expanded RAS as a biomarker.3,26 Taken together, both low allele frequency mutations and certain expanded RAS mutations are sufficiently uncommon that it is unlikely we will ever conclusively establish their role as predictive biomarkers. Hopefully increasing use of ctDNA will provide further insights into subclonal RAS dynamics.
ctDNA provides great promise for detecting acquired resistance to targeted therapies and evaluating evolutionary changes in cancers. Previous work has demonstrated that RAS mutant clones develop during anti-EGFR therapy27,28 These variants tend to be lower allele frequency than mutations present at baseline, and in Morelli et al’s report, 35% of them were found in primary tissue when assessed with high sensitivity BEAMing with sensitivity beyond standard clinical tests. It remains unclear whether the utility of ctDNA may better select baseline RAS status compared to tissue, however it does allow dynamic surveillance of resistance which is unique. In mCRC, many acquired resistance mechanisms have been shown to decay over time, allowing anti-EGFR re-challenge as a treatment consideration.10,29 The improvement in median PFS from 1.8 to 5.4 months in RAS/BRAF V600 wild type patients in CO.17 sets a target for these re-challenge efforts. Given that many anti-EGFR re-challenge concepts include additional agents and in the context of the ever rising costs of oncology drugs, it is essential that combinatorial strategies demonstrate clear superiority to single agent re-challenge.9
This study also further supports the combinatorial treatment strategy for BRAF V600E mCRC as single agent anti-EGFR does not improve PFS in BRAF V600E mutated mCRC.23 Unfortunately, only 15 patients with BRAF V600 mutations were evaluable in CO.17 for a test of interaction, which neared significance (P=0.089) despite small numbers. As CO.17 accrued in the treatment refractory population, it is not surprising that we saw a low prevalence of BRAF V600 mutations given their poor prognosis. Given the lack of benefit to date with anti-EGFR therapy, patients with BRAF V600E mutations should be prioritized for combinatorial strategies which have shown significant activity in this population.23
Despite the important findings of our study, it must be interpreted in the context of several limitations. As CO.17 completed enrollment over a decade ago, previous correlative analyses have exhausted much of the tissue and we could only analyze a subset of patients. Bias may be introduced into some analyses by the fact that certain patients had remaining tissue while others did not. However, we noted no differences in OS, PFS, or RR between the historic analysis and the updated analysis when assessing the best supportive care arm for prognosis in either the RAS/BRAF mutant group or the wild type group. The small number of patients who had BRAF or low allele frequency mutation means that findings among these groups must be interpreted in the context of the wide confidence interval surrounding treatment effect. When the trial was planned, the importance of RAS was not understood and as such our analyses are post-hoc and were not part of the original statistical plan. This is often the case with biomarker discovery, and all current evidence supporting RAS mutations as predictive are post-hoc.
In conclusion, we provide updated evidence that patients with mCRC harboring expanded RAS or BRAF V600E mutations lack benefit following single agent cetuximab. Our work demonstrates improved patient selection with the use of a high sensitivity assay that re-classified 14% of tumors as RAS mutated compared to Sanger sequencing. Subclonal mutations <5% MAF were uncommon, occurring in 2% of patients and remain of unclear significance. We hope this updated work informs future anti-EGFR combinatorial strategies by setting a benchmark for the activity of single agent cetuximab using a modern high sensitivity assay.
Supplementary Material
Translational Relevance:
The predictive utility of expanded RAS and BRAF for anti-EGFR therapy in colorectal cancer arises from panitumumab trials and it is unclear whether low mutant allele frequency mutations in these genes impact efficacy. We evaluated tissue from the CO.17 trial that randomized patients to cetuximab or best supportive care with a high sensitivity assay (BEAMing) and micro-dissection for expanded RAS/BRAF mutations (MAF>1%). Cetuximab improved overall and progression free survival in patients with RAS/BRAF V600E wild type tumors relative to supportive care and a test of interaction confirmed RAS (P<0.01) but not BRAF (P=0.089) mutations as predictive for cetuximab benefit. BEAMing showed increased sensitivity and identified 14% more KRAS mutations than historic Sanger sequencing. Mutations in RAS with allele frequency <5% were noted in 2% of patients, one of whom responded to cetuximab.
Acknowledgements:
JML was the recipient of an ASCO Young Investigator Award and a Michael Smith Health Professional Investigator Award that supported this work. SK is the recipient of NIH R01 grants that supported this research (CA172670 & CA187238).
Funding: NIH R01 grants (CA 172670 & CA 187238) to Scott Kopetz.
Footnotes
Conflicts of Interest: Daniel Edelstein, Hannah Quinn and Frank Holtrup disclose potential conflicts of interest as employees of Sysmex-Inostics.
Clinical Trial Identifier: NCT00079066
References:
- 1.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385 [DOI] [PubMed] [Google Scholar]
- 2.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized controlled trials. Ann Oncol. 2014;(August 2014):1–27. doi: 10.1093/annonc/mdu378 [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Siena S, Cassidy J, et al. Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25(7):1346–1355. doi: 10.1093/annonc/mdu141 [DOI] [PubMed] [Google Scholar]
- 4.Venook A, Niedzwiecki D, Ou F, et al. Impact of primary tumor location on Overall Survival and Progression Free Survival in patients with metastatic colorectal cancer: Analysis of all RAS wt subgroup on CALGB/SWOG 80405 (Alliance). JCO. 2016;34:Supplemental Abstract 3504. [Google Scholar]
- 5.Arnold D, Lueza B, Douillard J-YY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28(8):1713–1729. doi: 10.1093/annonc/mdx175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 7.Boeckx N, Koukakis R, Beeck K, et al. Primary tumor sidedness impacts on prognosis and treatment outcome: results from three randomized studies of panitumumab plus chemotherapy versus chemotherapy or chemotherapy plus bevacizumab in 1st and 2nd line RAS/BRAF WT mCRC. In: Annals of Oncology. ; 2016:Supplement 6, 89P. [Google Scholar]
- 8.Rowland A, Dias MM, Wiese MD, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112(12):1888–1894. doi: 10.1038/bjc.2015.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parseghian CM, Napolitano S, Loree JM, Kopetz S. Mechanisms of Innate and Acquired Resistance to Anti-EGFR therapy: A Review of Current Knowledge with a Focus on Rechallenge Therapies. Clin Cancer Res. July 2019. doi: 10.1158/1078-0432.CCR-19-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parseghian CM, Loree JM, Morris VK, et al. Anti-EGFR Resistant Clones Decay Exponentially After Progression: Implications for Anti-EGFR Re-challenge. Ann Oncol. November 2018. doi: 10.1093/annonc/mdy509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cremolini C, Rossini D, Dell’Aquila E, et al. Rechallenge for Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer With Acquired Resistance to First-line Cetuximab and Irinotecan. JAMA Oncol. 2018:1–8. doi: 10.1001/jamaoncol.2018.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: Guideline from The American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35(13):1453–1496. doi: 10.1200/JCO.2016.71.9807 [DOI] [PubMed] [Google Scholar]
- 13.Loree J, Kopetz S, Raghav K. Current companion diagnostics in advanced colorectal cancer; getting a bigger and better piece of the pie. J Gastrointest Oncol. 2017;8(1):199–212. doi: 10.21037/jgo.2017.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciardiello F, Normanno N, Maiello E, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next generation sequencing: findings from the CAPRI-GOIM trial. Ann Oncol. 2014;(June):1756–1761. doi: 10.1093/annonc/mdu230 [DOI] [PubMed] [Google Scholar]
- 15.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551–559. doi: 10.1038/nmeth898 [DOI] [PubMed] [Google Scholar]
- 16.Tougeron D, Lecomte T, Pagès JC, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2013;24(5):1267–1273. doi: 10.1093/annonc/mds620 [DOI] [PubMed] [Google Scholar]
- 17.Dono M, Massucco C, Chiara S, et al. Low Percentage of KRASMutations Revealed by Locked Nucleic Acid Polymerase Chain Reaction: Implications for Treatment of Metastatic Colorectal Cancer. Mol Med. 2012;18:1519–1526. doi: 10.2119/molmed.2012.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Cutsem E, Lenz H-J, Köhne C- H, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Ctations in Colorectal Cancer.etuximab Treatment and RAS Mu. J Clin Oncol. 2015;33(7):33(7):692–700. doi: 10.1200/JCO.2014.59.4812 [DOI] [PubMed] [Google Scholar]
- 19.Karapetis CS, Jonker D, Daneshmand M, et al. PIK3CA, BRAF, and PTEN status and benefit from cetuximab in the treatment of advanced colorectal cancer-results from NCIC CTG/AGITG CO.17. Clin Cancer Res. 2014;20(3):744–753. doi: 10.1158/1078-0432.CCR-13-0606 [DOI] [PubMed] [Google Scholar]
- 20.Rago C, Huso DL, Diehl F, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67(19):9364–9370. doi: 10.1158/0008-5472.CAN-07-0605 [DOI] [PubMed] [Google Scholar]
- 21.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci U S A. 2003;100(15):8817–8822. doi: 10.1073/pnas.1133470100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisenhauer E a, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 23.Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N Engl J Med. 2019;381(17):1632–1643. doi: 10.1056/nejmoa1908075 [DOI] [PubMed] [Google Scholar]
- 24.Peeters M, Kafatos G, Taylor A, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer. 2015;51(13):1704–1713. doi: 10.1016/j.ejca.2015.05.017 [DOI] [PubMed] [Google Scholar]
- 25.Van Cutsem E, Lenz H-J, Kohne C-H, et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J Clin Oncol. 2015;33(7). doi: 10.1200/JCO.2014.59.4812 [DOI] [PubMed] [Google Scholar]
- 26.Loree JM, Kopetz S. Why a one size fits all approach to RAS might not fit colorectal cancer. JNCCN J Natl Compr Cancer Netw. 2017;15(4):545–547. http://europepmc.org/abstract/med/28404763. [DOI] [PubMed] [Google Scholar]
- 27.Kim TW, Peeters M, Thomas A, et al. Impact of emergent circulating tumor DNA Ras mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin Cancer Res. 2018;24(22):5602–5609. doi: 10.1158/1078-0432.CCR-17-3377 [DOI] [PubMed] [Google Scholar]
- 28.Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26(4):731–736. doi: 10.1093/annonc/mdv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.