Abstract
Fueled by key technological innovations during the last several decades, chromatin-based research has greatly advanced our mechanistic understanding of how genes are regulated by epigenetic factors and their associated histone-modifying activities. Most notably, the landmark finding that linked histone acetylation by Gcn5 of the Spt–Ada–Gcn5–acetyltransferase (SAGA) complex to gene activation ushered in a new area of chromatin research and a realization that histone-modifying activities have integral genome functions. This review will discuss past and recent studies that have shaped our understanding of how the histone-modifying activities of SAGA are regulated by, and modulate the outcomes of, other histone modifications during gene transcription. Because much of our understanding of SAGA was established with budding yeast, we will focus on yeast as a model. We discuss the actions of cis- and trans-histone crosstalk pathways that involve the histone acetyltransferase, deubiquitylase, and reader domains of SAGA. We conclude by considering unanswered questions about SAGA and related complexes.
Keywords: SAGA, gene transcription, histone modifications, histone acetylation, histone methylation, histone deubiquitylation, Gcn5, Ubp8, crosstalk pathways, bromodomain, Tudor domains
1. Introduction
1.1. A brief timeline leading to SAGA-associated histone-modifying activities
In 1951, Stedman and Stedman [1] proposed the notion that histones act as an inhibitory barrier for genes, and, in 1961, Allfrey confirmed that histones inhibit RNA synthesis [2] (Figure 1). Later, biochemical and cell biology approaches led Phillips [3] and Murray [4] to discover histone modifications such as acetylation and methylation, respectively. On the basis of these post-translational modifications (PTMs) and their association with RNA synthesis, Allfrey hypothesized that histone acetylation/deacetylation act as a switch to alter RNA synthesis [5]. Since Allfrey’s 1964 hypothesis [5], the scientific community has been on a continuous “SAGA” that details how transcription is initiated, maintained, and regulated.
Figure 1. Histone acetylation and SAGA timeline.
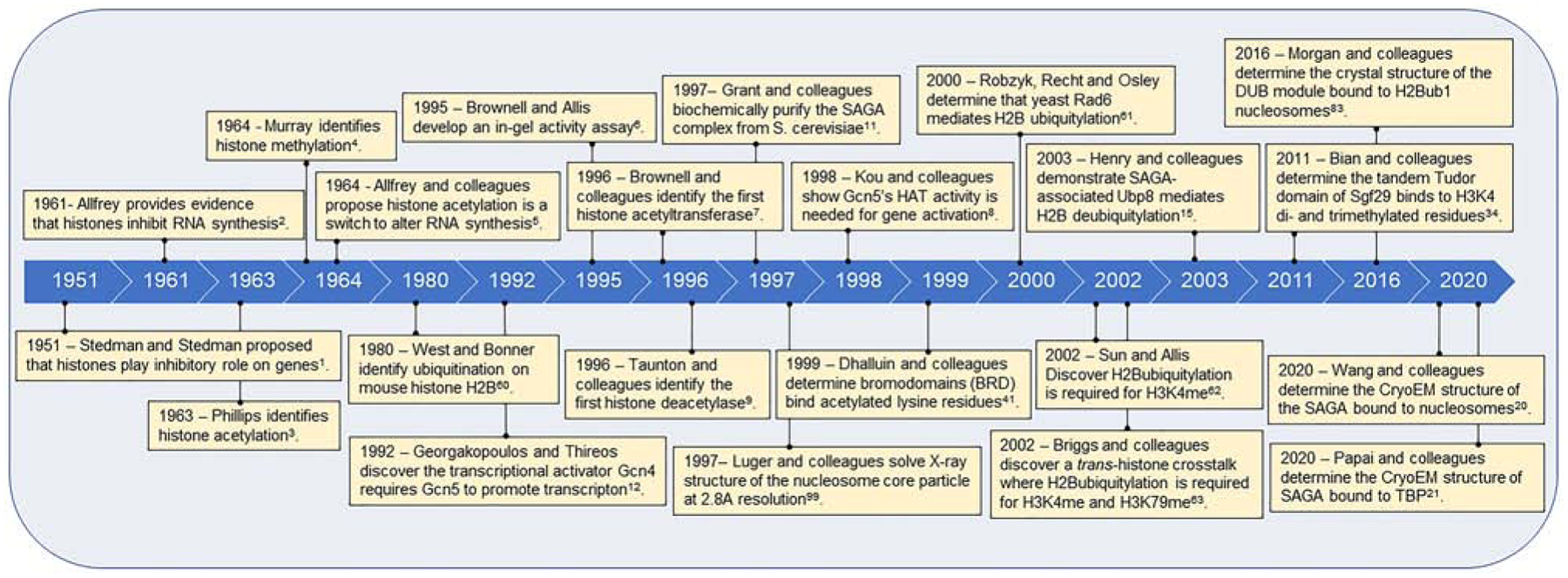
Notable discoveries contributing to the current understanding of SAGA’s function.
Allfrey’s intriguing proposal notwithstanding, the importance of histone modifications acting as a switch wasn’t realized until the mid-1990’s when Allis and colleagues biochemically and genetically defined yeast Gcn5 (General control nonderepressible 5) as the first transcription-linked histone acetyltransferase (HAT) [6–8]. Similarly, Schrieber and colleagues also identified the first mammalian histone deacetylase, HDAC1, which was homologous to yeast Rpd3, a known transcriptional repressor [9]. These pivotal discoveries of enzymes that add or remove acetylation from histones completed our genetic understanding of a known activator (Gcn5) and repressor (Rpd3) in transcriptional regulation.
After Gcn5 was recognized as a specific HAT for histone H3 lysine 14 (H3K14) [10], Workman and colleagues showed that Gcn5 was part of a large multi-subunit complex coined SAGA that showed catalytic activity towards nucleosomes and an expanded set of lysine residues on histone H3 and other histones [11]. The significance of the SAGA complex in transcriptional regulation was initially established in classical genetic screens by various groups including the Thireos, Guarente, and Winston lab’s, which provided an early view of how SAGA subunits contributed to gene regulation [12–14]. Interestingly, the HAT activity SAGA was not its sole activity. The discovery that SAGA contained histone H2B deubiquitylase activity expanded the toolkit by which this complex is able to contribute to chromatin organization and gene transcription [15]. From these two SAGA activities, we learned new twists and plots in how these activities are regulated by other histones and PTMs and how SAGA-associated activities modulate the outcomes of other histone PTMs.
In this review, we will focus on discoveries from mainly, but not limited to, investigations with the model organism Saccharomyces cerevisiae (sc). These discoveries have promoted the “rise” of cis- and trans-histone crosstalk pathways centered around the SAGA complex and its associated histone modifying activities. We will conclude by describing additional avenues of research and pertinent questions whose answers are necessary to fully understand how SAGA and chromatin merge to control gene expression and other DNA-templated processes.
2. SAGA (Spt-Ada-Gcn5 acetyltransferase)
SAGA is a multi-subunit protein complex that is conserved in many species such as Saccharomyces cerevisiae (scSAGA), Drosophila melanogaster (dmSAGA), Homo sapiens (hsSAGA) and Arabidopsis thaliana (atSAGA) [16–18]. Yeast SAGA (Spt-Ada-Gcn5 acetyltransferase) is a 2.0 MDa multi-subunit protein complex that is composed of 19 distinct proteins [16,19]. These proteins are separated into four distinct modules: the HAT module (Gcn5, Ada2, Ada3, and Sgf29), a deubiquitylase (DUB) module (Ubp8, Sgf11, Sgf73 (hSca7), and Sus1), the SPT (Suppressor of Ty) module (Tra1, Ada1, Spt3, Spt7, Spt8, and Spt20), and TAF (TATA-binding protein (TBP)-associated factors) module (Taf5, Taf6, Taf9, Taf10, and Taf12) (Figure 2) [17,19]. However, new Cryo-EM studies showed most of the proteins from the SPT and TAF module form a structural “core” complex involving Spt7, Spt3, Spt20, sgf73, Ada1, Taf5, Taf6, Taf9, Taf10, and Taf12 (Figures 2 and 3) [20,21].
Figure 2. SAGA subunits and their domains.
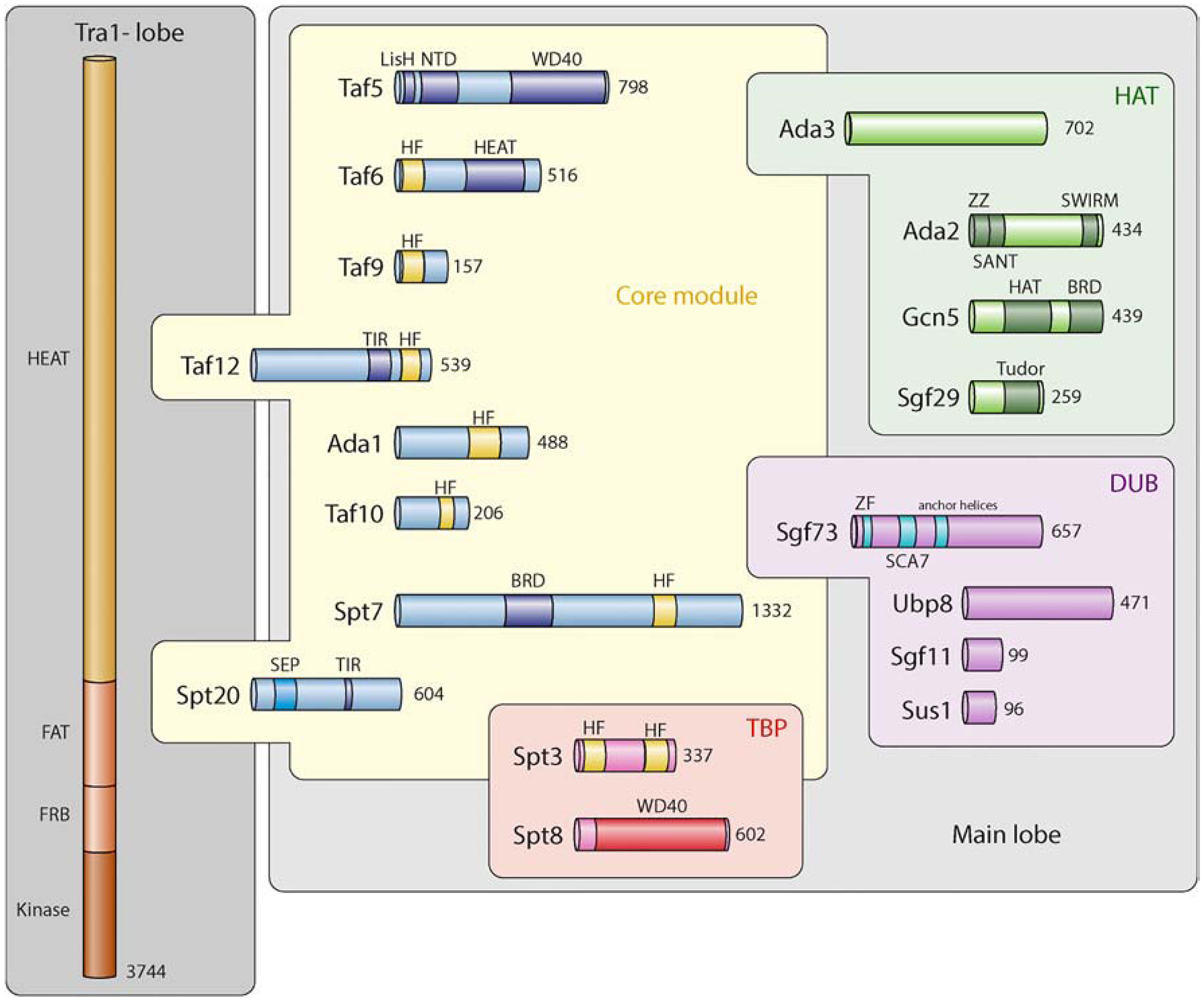
Cartoon description showing the individual subunits of the SAGA complex with their functional domains highlighted. The distinct lobes containing various SAGA subunits are grouped together with subunits that connect the various lobes together intersecting. HEAT = Huntington, Elongation Factor 3, PR65/A, TOR; FAT = FRAP, ATM and TRRAP; FRB = FKBP12-rapamycin binding domain; TIR = Tra1-interacting; SEP = SEP domain; HF = histone fold domain; LisH = lis homology domain; BRD = bromodomain; WD40 = WD domain, G-beta repeat; ZF = Zinc Finger; SWIRM = SWIRM domain; HAT = histone acetyltransferase; Tudor = Tudor domain; ZZ = Zinc finger, ZZ type. Figure design is adapted from Papai et al. [21].
Figure 3. HAT and DUB subunits within SAGA that contribute to chromatin engagement.
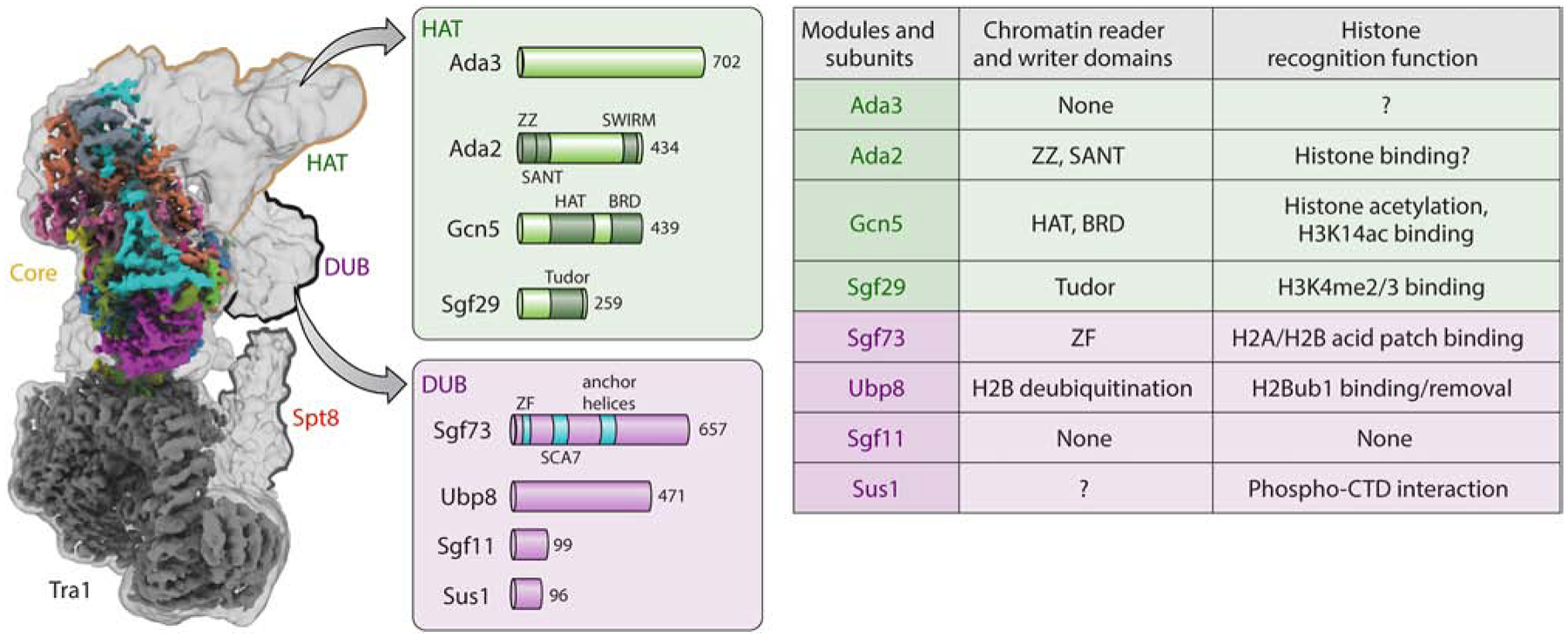
Left, Cryo-EM structure of the SAGA complex with HAT and DUB modules represented (from Wang et al. [20]. Arrows extending from the SAGA structure show the individual subunits of the HAT and DUB modules with their functional domains shown (see Figure 2). Right, Table highlighting the functional domains found in each DUB or HAT subunit with the documented histone or RNA polymerase II interactions made.
SAGA from all species contains two distinct catalytic activities, HAT and DUB that are mediated by scGcn5 and scUbp8 (human ortholog hsUsp22), respectively. In contrast, there are also specific metazoan subunits which include two components of the SF3B spliceosomal complex, SF3B3 and SF3B5. These subunits have been found associated with dmSAGA and hsSAGA (STAGA) but not scSAGA complexes suggesting a specialized function of these subunits in metazoan gene regulation [22,23]. In addition, there are key structural components such as Tra1, the largest subunit, that interacts with transcriptional activators; similarly, Spt3 and Spt8 interact with TBP for its promoter recruitment to TATA-containing promoters of stress responsive genes [24–26].
Though initially thought to only target stress responsive genes, SAGA appears to acts as a general coactivator, similar to Mediator, for pre-initiation complex (PIC) formation and all RNA Polymerase II (RNAPII) transcription [27,28]. In addition, several proteins in SAGA have distinct protein-binding or “reader” domains (Tudor or Bromodomains) that enable the SAGA complex to associate with chromatin or other proteins via PTMs. Moreover, several SAGA proteins are also associated with other protein complexes (reviewed in [16]). For example, Gcn5 is found in human ATAC and yeast SALSA/SILK complexes, Tra1 is part of the Nua4 acetyltransferase complex, and five TAF proteins (Taf5, Taf6, Taf9, Taf10, Taf12) are components in the TFIID complex [16,19,29]. Many of the aforesaid aspects of SAGA will be discussed further in other excellent reviews in this issue. In the following, we will specifically discuss how the HAT and DUB modules and their associated reader modules affect cis- and trans-histone crosstalk pathways.
2.1. HAT module and cis-histone crosstalk pathways
Gcn5 was originally discovered in a genetic screen for the inability of the yeast mutants to grow under amino acid-starved conditions [30]. However, Gcn5 was first cloned in 1992 by Georgakopoulos and Thireos, who also showed that Gcn5 was required for the transcriptional activator function of Gcn4 [12]. Once Gcn5 was identified to have enzymatic activity [7], Gcn5’s HAT activity was initially shown to have an in vitro preference for histone H3 at lysine 14 (H3K14) [10]. However, Gcn5 could also acetylate additional histone lysine residues, such as H3K9, H3K18, H3K23, H3K27, H3K36 and other histones such as H4 and H2B [10,11,31,32]. Nonetheless, other proteins were needed for Gcn5 to acetylate nucleosomal substrates [11,33]. These additional proteins where found in the highly conserved HAT module of SAGA in which the association of Gcn5 with Ada2 and Ada3 subunits dictates Gcn5 specificity for nucleosomal substrates, whereas Sgf29 recognizes Set1-mediated H3K4 trimethylation [11,33–35]. Within the HAT module, there are two distinct reader domains - a single bromodomain within Gcn5 and a tandem Tudor domain located in the C-terminus of Sgf29 (Figures 2 and 3). Both reader domains have significant functions in cis-histone crosstalk pathways, as discussed below.
One of the earliest examples of a cis-histone crosstalk pathway involving Gcn5 was the connection between histone H3 phosphorylation and H3 acetylation in which Gcn5 prefers to acetylate histone H3K14 when H3 serine 10 (H3S10) is phosphorylated [36]. This type of crosstalk was further supported by in vivo analysis of the yeast INO1 promoter where histone H3S10 phosphorylation by the Snf1 kinase is required for Gcn5 to acetylate H3K14 [37]. More recently, Shimada et al. reported that histone H3 threonine 11 (H3T11) phosphorylation by Chk1 facilitates Gcn5-mediated H3K14 acetylation and transcription of cell-cycle genes [38]. However, upon DNA damage, this cis-histone crosstalk pathway is reversed such that Chk1 is released from chromatin, thereby reducing H3T11 phosphorylation and H3K14 acetylation and resulting in transcriptional repression [38]. Intriguingly, histone H3 phosphorylation may also impact chromatin remodeling because H3S10 phosphorylation can antagonize binding of the Rsc4 bromodomain to H3K14 acetylation; thus, phosphorylation in this situation may act as a switch to inhibit and/or release chromatin remodeling factors from chromatin [39].
Gcn5 also contains a single bromodomain (BRD) that is adjacent to its catalytic HAT domain. The BRD of Gcn5 not only increases the retention of the SAGA complex on chromatin but it also facilitates cis-histone crosstalk [40,41]. For example, the BRD of Gcn5 binds to Gcn5-mediated H3K14 acetylated resides to facilitate acetylation at other histone lysine residues (H3K9, H3K18, K27, K36) neighboring H3K14, with H3K18 acetylation being the most dependent on the BRD [42]. Beyond facilitating acetylation sites on the same histone, the BRD can also enhance acetylation of the adjacent histone H3 within the same nucleosome and possibly within nearby nucleosomes [43]. In addition, the bromodomain of Gcn5 and H3K14 acetylation can promote chromatin remodeling by Switch/Sucrose non fermenting (SWI/SNF) and Remodels the Structure of Chromatin (RSC) complexes in which local hyperacetylation by SAGA marks promoters leading to subsequent recruitment of chromatin remodeling factors [39,44,45]. Interestingly, protein subunits that contain BRDs are common to most ATP-dependent chromatin remodeling complexes. SWI/SNF and RSC complexes contain subunits Swi2/Snf2 and Rsc4, respectively that harbor BRDs. Swi2/Snf2 has a single BRD and Rsc4 contains a tandem bromodomain (BRD1 and BRD2). The BRD of Swi2/Snf2 is needed for efficient remodeling of nucleosomes and displacement of SAGA from chromatin [46]. For Rsc4, Gcn5 HAT activity promotes its binding to H3K14 acetylated peptides via its BRD2 and inhibits Rsc4 binding to acetylated substrates via direct acetylation of Rsc4 at K25. Subsequently, BRD1 of Rsc4 binds to K25 acetylation, and this intramolecular interaction prevents BRD2 from binding acetylated histone substrates [39]. In a similar but distinct manner, Swi2/Snf2, the catalytic subunit of SWI/SNF complex, is also acetylated by Gcn5, which enables an intermolecular interaction between Swi2/Snf2’s bromodomain and its acetylated lysine residues that prevents or releases the bromodomain of Swi2/Snf2 from interacting with acetylated histone H3 [47]. Together, these regulatory intramolecular interactions would provide a temporal switch to release RSC and/or SWI/SNF complexes from the already remodeled chromatin so that chromatin remodeling complexes can be recycled for the next round of transcription.
Additional cis-histone crosstalk pathways were discovered using genome-wide chromatin immunoprecipitation (ChIP) and mass-spectrometry analyses. In these studies, histone H3K4 methylation and H3K14 acetylation were colocalized on the same genomic loci or on the same peptide when isolated from endogenous histones [48–50]. The importance and mechanism of the suggested cis-crosstalk between these two co-associated histone modifications was not realized until the discovery of histone H3K4 methyl readers that include chromodomain, plant homeodomain (PHD) fingers, and Tudor domains. In yeast, these readers include, Chd1 (chromodomain), Yng1 (PhD finger), and Sgf29 (tandem Tudor domain) [34,35,51–53]. Furthermore, each of these methyl-binding proteins is associated with a histone acetyltransferase complex where Yng1 is a subunit in the NuA3 complex, whereas Chd1 and Sgf29 are found associated with the SAGA complex [51,54]. Interestingly, both of these complexes utilize the same general mechanism in which the reader domains (PhD finger vs Tudor domain) recruit the HAT complexes to their histone methylated target genes to acetylate the histone H3 N-terminus [34,35,52,53]. In context of the HAT module, the tandem Tudor domains of Sgf29 also are required to stimulate processive multisite acetylation on H3K4 methylated nucleosomes [55]. Conversely, SAGA can also associate with the 5’ region of the ORF where histone acetylation appears to promote H3K4 methylation [56]. In this case, histone acetylation may directly stimulate Set1-mediated H3K4 methyltransferase activity, a function that has been observed for mammalian H3K4 methyltransferases MLL1 and MLL4 that prefer to methylate pre-acetylated substrates [57,58]. Alternatively, because Jhd2-mediated H3K4 demethylation is inhibited by histone H3 acetylation and prevents binding of Jhd2 to acetylated substrates ([59] and Briggs unpublished observations), acetylation by Gcn5 may also indirectly increase the steady state amount of H3K4 methylation on ORFs by preventing Jhd2 to recognize and demethylate its substrate. Overall, the HAT module is designed so that two distinct cis-histone crosstalk pathways enable local hyperacetylation of histone H3. In addition, these methyl-acetyl cis-histone crosstalk pathways appear to be bidirectional and may act as a feedback or feedforward loop to drive and control the histone PTM steps required for transcriptional initiation and elongation (see section 2.3).
2.2. DUB module and trans-histone crosstalk pathways
In 1980, West and Bonner first identified H2B ubiquitination in mouse histones. However, the function of H2B ubiquitination remained unknown until work performed by the Osley group in 2000 [60,61]. This ubiquitin-conjugating enzyme) in gene regulation and DNA repair to the mono-ubiquitylation of H2B at lysine 123 (H2Bub1). H2Bub1 is a highly conserved and dynamic modification that is localized to the promoters and bodies of actively transcribed genes. In addition to being required for proper tunable gene induction, H2Bub1 is necessary for the study critically linked the previously known functions of Rad6 (an E2 trans-histone crosstalk pathway of Set1/COMPASS-mediated H3K4 and Dot1-mediated H3K79 methylation [62–64].
Although the mechanistic basis of this histone crosstalk eluded discovery for nearly two decades, recent cryo-EM studies have defined the structural basis for how H2Bub1 directs the nucleosome binding and allosteric regulation of COMPASS and Dot1 methylation [65–68]. In the case of COMPASS, H2Bub1 prevents Set1 from making autoinhibitory contacts with deubiquitinated nucleosomes, thereby enabling Set1 to make key contacts with other COMPASS subunits so that histone H3K4 methylation can occur. In the case of Dot1, H2Bub1 binds and sequesters Dot1 in a more favorable orientation on the nucleosome for H3K79 methylation [65–68]. One striking difference between H2Bub1 and histone H3 methylation at K4 and K79 is their dynamics. H2Bub1 has a short half-life of ~1 min [69]. The H2B ubiquitin machinery (Rad6, Bre1 and Lge1) is associated with the Polymerase-Associated Factor 1 Complex (PAF1C) and the phosphorylated forms (serine 2 and 5) of the C-terminal domain (CTD) of RNAPII, which suggests that H2Bub1 is written and erased in a co-transcriptional manner during transcription elongation [70–75]. Unlike H2Bub1, however, H3K4 and H3K79 methylation are significantly more stable [69], and, in the case of H3K79me3, the modification is removed only passively via cell division [76]. These intriguing findings have helped to formulate the notion that, whereas H2Bub1 functions directly in transcription elongation, histone H3K4 and H3K79 methylation may function to generate transcriptional memory – perhaps to recruit or repel chromatin-associated proteins (e.g., ATP-dependent remodelers) that contain reader domains which maintain a chromatin state more amenable to further rounds of transcription [75,77].
Consistent with H2Bub1 functioning in transcription elongation, Chandrasekharan et al. and Batta et al. showed that deposition and removal of this chromatin mark correlated with the stability of nucleosomes genome-wide and at individual genes [78,79]. Although it was initially reasoned that the ubiquitin moiety on H2B serves as a “wedge” that “opens” chromatin domains during RNAPII elongation [80], high H2Bub1 levels are, in fact, associated with nucleosome assembly and/or stability, whereas low H2Bub1 levels lead to nucleosome loss. This finding is still poorly understood but may be due to the function of H2Bub1 in regulating the recruitment and/or activity of histone chaperones and/or ATP-dependent chromatin remodelers that contribute to the transient disruption of nucleosomes during transcription [81]. The precise mechanism for how H2Bub1 contributes to nucleosome stability is not well understood. However, the concept that H2Bub1 removal promotes nucleosome destabilization is consistent with the fact that SAGA, which stimulates transcriptional initiation and elongation, contains one of two major DUBs that remove this mark (i.e., Ubp8). The other major DUB involved in H2Bub1 removal is Ubp10, which functions independent of SAGA but in association with the FACT complex in gene bodies [82]. Although Ubp8 removes H2Bub1 more prominently at the 5’ ends of genes compared with 3’ ends, absence of Ubp8 greatly increases nucleosome assembly across the entire lengths of transcribed genes [79]. This finding is not only consistent with studies that showed SAGA within gene bodies, but it also implies that both DUBs contribute to the complete removal of H2Bub1 across genes undergoing transcription.
Within the SAGA complex, Ubp8, Sgf11, Sus1 and Sgf73 form a distinct DUB submodule responsible for H2Bub1 removal (Figure 2). Although the HAT and DUB modules are distinct structurally, they are in close proximity in cryo-EM analyses (Figure 3) [20,21]. Prior to binding the nucleosome, the DUB and HAT modules contact each another. However, the modules become displaced upon nucleosome binding, when the DUB module binds the nucleosomal H2A/H2B disk and the HAT module is more flexible and presumably free or accessible to acetylate histone H3. Detailed structural studies of the Ubp8 DUB module showed that an arginine cluster in the zinc finger of Sgf11 interacts directly with the H2A/H2B acidic patch, thereby stabilizing the DUB module on the nucleosomal disk [83]. In an earlier study, this zinc finger was proposed to also interact with DNA [84]. Additionally, Ubp8 makes contacts on H2B near the site of ubiquitin at K123 [83]. Although the cryo-EM structure of the Upb8 DUB module in isolation suggests that Ubp8 is bound to both sides of the nucleosomal disk, recent cryo-EM analyses of SAGA revealed that only one SAGA complex is likely to be bound to a single nucleosome [20].
An obvious expectation of having the HAT module in close proximity to the Ubp8-containing DUB module is that these two distinct activities are coupled events that enable transient passage of RNAPII during transcription. In this scenario, SAGA-dependent histone acetylation releases histone tails wrapped around DNA in the nucleosome to enable transient release of DNA and RNAPII passage. In parallel, Upb8 deubiquitylation may contribute to RNAPII passage by further facilitating nucleosome destabilization (likely in coordination with histone chaperones such as FACT). The combined activities would provide rapid and transient nucleosome destabilization needed for efficient RNAPII elongation through the nucleosome (Figure 4). Although it is widely assumed that nucleosomes are disassembled during transcription elongation, more recent cryo-EM studies that examined the passage of RNAPII through the nucleosome suggest that nucleosomes may be held together throughout elongation [85]. Additionally, in promoting transcription elongation, it may be that the key functions for histone chaperones and elongation factors (e.g., FACT, Spt6 and PAF) are to employ their histone-interacting acidic regions to maintain nucleosome integrity and decrease pausing during elongation [86]. In this model (see Figure 4), Ubp8-directed removal of H2Bub1, along with histone H3 acetylation by the HAT module in SAGA, may couple histone-DNA tail release activity with nucleosome destabilization to transiently facilitate RNAPII passage through the nucleosome. Following RNAPII passage, the PAF complex, Rad6/Bre1, Spt6 and Set2 (note that many of these factors are biochemically and genetically in the same pathway) may create nucleosome stabilization and re-association of histone tails with DNA and perhaps neighboring nucleosomes [77,87]. Although H2Bub1 is transient and may be removed rapidly, the deposition of histone H3K36 methylation by Set2 and the deacetylation “memory” that this pathway has would ensure long-term nucleosome integrity during cell growth.
Figure 4. Transcription model for SAGA.
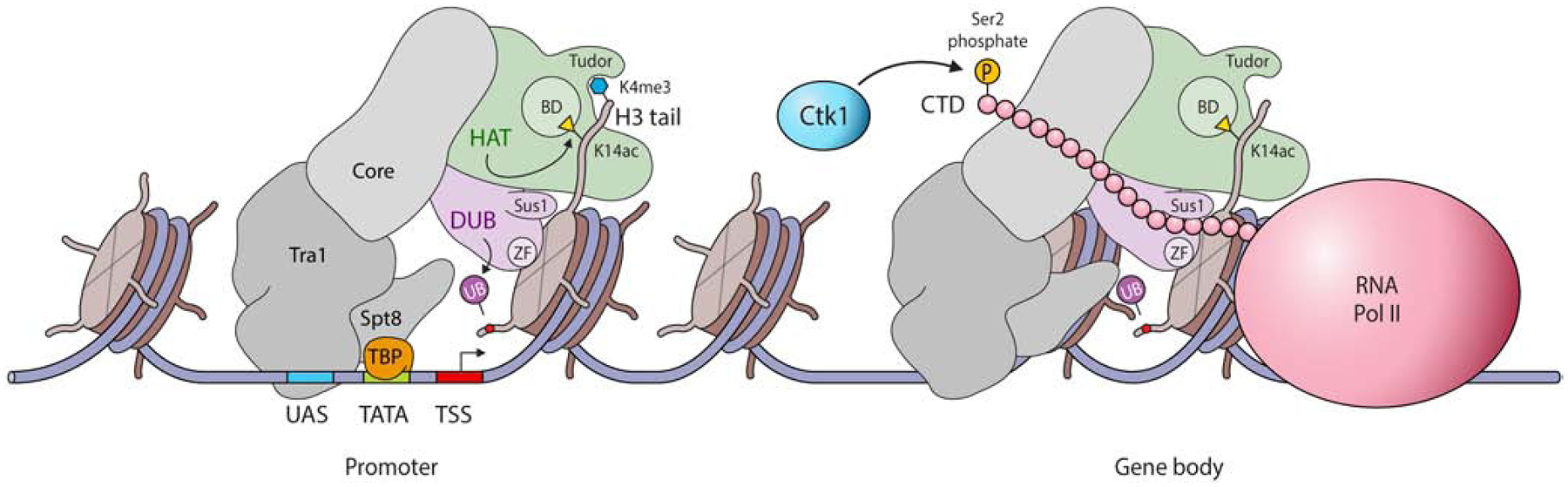
A cartoon model of the function of SAGA and its DUB and HAT modules in gene transcription. The left side shows the role of various subunits in mediating histone-binding and crosstalk at promoters, while the right-side highlights various activities of the SAGA complex during transcription elongation.
Although the function of SAGA-mediated Ubp8 deubiquitylation during RNAPII elongation is still in need of investigation, there is a more defined activity of Ubp8 deubiquitylation at the 5’ ends of genes. Specifically, and during the transition from serine 5 phosphorylation to serine 2 phosphorylation of the CTD of RNAPII, H2Bub1 removal by Ubp8 is required for proper recruitment and function of the major serine 2 CTD kinase, Ctk1 [88]. This work not only defined a key activity of SAGA and deubiquitylation of H2Bub1 in the transition step from transcription initiation to elongation but revealed how deubiquitylation at 5’ ends is essential for establishing downstream activities in chromatin and transcription (Figure 4). It is likely that the function of Ubp8 at the 5’ end to control Ctk1 activity is different from its nucleosome destabilizing activity that is performed across the transcribed regions of genes in coordination with RNAPII.
2.3. SAGA reader modules facilitate stable feedback loops that promote and create transcriptional memory
In addition to the function of SAGA-associated HAT and DUB modules in the control of nucleosome stability and histone PTMs during RNAPII elongation and nucleosome passage, multiple subunits of these two SAGA catalytic modules also “read” histones or specific histone PTMs (see Figure 3). This so called “read-write” capacity of chromatin-associated enzyme complexes like SAGA to read their own products and/or use embedded reader domains to engage the nucleosome in the correct context has become a common theme in chromatin biology. Below, we describe how SAGA histone reading, and writing, regulates its cellular functions and histone crosstalk abilities.
The BRD of Gcn5 binds to histone H3K14ac, and this association is critical for SAGA recruitment, chromatin retention, and HAT activity [40]. Additionally, and in the central SAGA module, Spt7 contains a BRD domain that, at least in vitro, can interact with histone H3K9ac [89]. These findings have reinforced the concept that SAGA reads its own product, thereby stabilizing itself on the nucleosome in a feed-forward loop that maintains robust HAT activity on that nucleosome or perhaps neighboring nucleosomes. This feed-forward circuit may be a critical rheostat in gene expression control, whereby high activator recruitment of SAGA promotes greater histone acetylation at the promoters to reinforce promoter escape and enhance transcription elongation by RNAPII.
Similar to histone H3 N-terminal acetylation that stabilizes or recruits SAGA by way of its BRDs, Sgf29 binding of histone H3K4 trimethylation (H3K4me3) via its tandem Tudor domain may further enhance and stabilize SAGA binding and activity on nucleosomes, specifically the +1 nucleosomes at gene promoters that are largely marked with H3K4me3 [34,55] (Figure 4). Perhaps, because Gcn5 exists at both promoters and in the transcribed regions of genes, H3K4me3 marked +1 nucleosomes may have a unique function on SAGA HAT activity at gene promoters that is not necessarily carried over to the gene bodies that lack high levels of H3K4me3. Thus, H3K4me3 may provide a robust feed-forward loop at promoters to ensure significant SAGA recruitment and histone acetylation that enables either RNAPII initiation or release into gene bodies. Finally, it is intriguing to consider how the crosstalk between H3K4me3 and H3 N-terminal acetylation may contribute to SAGA’s feed-forward circuit and chromatin association. Specifically, the BRDs within SAGA promote further levels of histone H3 tail acetylation that should repel the H3K4me3-specific demethylase Jhd2 ([59], Briggs unpublished observation), thereby increasing H3K4me3 levels that increase still more SAGA binding by Sgf29. Thus, SAGA’s read-write (and repel) capabilities likely have a critical function in SAGA’s association with chromatin and likely help to direct histone modifications that control gene transcription. In addition, it appears that a feedback loop may exist wherein H3K4 methylation is stimulated by Gcn5-mediated H3 acetylation [56]. This feedback loop may be a conserved function because histone acetylation appears to stimulate H3K4 methylation via scSet1, MLL1, and MLL4 H3K4 methyltransferases as mentioned in section 2.1 [56–58].
Another potential feed-forward loop may exist to control histone H3 methylation by SAGA/Ubp8-mediated removal of H2Bub1. Because H2Bub1 regulates the trans-histone crosstalk methylation pathway, one could envision that SAGA-directed removal of H2Bub1 also acts to decrease the levels of H3K4 and H3K79 methylation. Although the potential for such regulation is possible, even low levels of H2Bub1 are sufficient to promote at least some or normal levels of H3K4 and H3K79 methylation [90]. Thus, although some H2Bub1 is needed for trans-histone methylation, the normal cycle of its removal by SAGA does not appear to contribute to reducing these histone methylation events.
Lastly, SAGA may have yet another putative histone interacting domain. Within the HAT module, the ADA2 subunit contains a conserved ZZ-SANT domain (Figure 3) that, in several human proteins was shown to interact with unmodified histone H3 tails [91]. Thus, this domain may read histone tails to further enhance SAGA recruitment or function on chromatin. The importance of this domain in SAGA biology awaits confirmation.
3. Future perspectives
Although we have learned about how SAGA contributes to transcription, there remain many unanswered questions and unsolved mysteries concerning how SAGA functions. Some of these issues include the functions of the different SAGA-like complexes, how histone acidic and basic patches contribute to SAGA activity, and how SAGA modification of non-histone targets contributes to transcriptional regulation.
The discovery and purification of multiple Gcn5-containing SAGA-like complexes has prompted many unanswered questions about their functions. In most cases, these noncanonical SAGA-like complexes share similarities to SAGA, but they also differ in subunit composition and function. For example, SALSA/SLIK possesses two major differences compared with SAGA. First, SALSA/SLIK contains an N-terminal truncated version of Spt7 [92,93]. Second, the Spt8 subunit in the SAGA complex is replaced by Rtg2 in the SALSA/SLIK complex, thereby linking SALSA/SLIK to the retrograde pathway [94]. Clarification is needed as to whether this altered complex is associated with gene bodies and is governed by the same crosstalk and regulation described for SAGA. The Berger group showed that SAGA and SALSA/SLIK differentially regulate gene transcription [95]; thus, the mechanisms by which SAGA and SALSA/SLIK contribute to this differential regulation may be due to the differences these complexes have in regulating the multiple crosstalk mechanisms discussed above. Additionally, Gcn5 was observed in yet even smaller subcomplexes called ADA and HAT-A2 [16]. These subcomplexes still possess the histone-reading subunit Sgf29; thus, it is likely that their functions and localizations across genes are also influenced by histone crosstalk and the histone PTM landscape.
Acidic and basic (charged) patches on histones appear to exert distinct but important activities in SAGA regulation. For example, we recently revealed the ability of the basic patch in the H4 tail to restrict SAGA histone acetylation and H2B deubiquitylation [96]. Yet, how the basic patch contributes to this regulation is unknown, but it appears to be independent of the ability of the basic patch to control ATP-dependent remodelers or Dot1 [96,97]. An attractive hypothesis is that the basic patch contributes to chromatin accessibility by its interaction with the H4 tail, thereby limiting the DUB and HAT modules from interacting with nucleosomes. Another charged patch, the acidic patch of H2A/H2B, is critical for the interaction and function of the Upb8-containing DUB module of SAGA. An obvious question is how other chromatin regulators that require this same acidic patch (e.g., RSC) and that presumably work in coordination with SAGA can simultaneously use this same region of the nucleosome. Furthermore, it is notable that SAGA, and many of the other acidic-patch utilizing complexes [98], operates at the +1 nucleosome that is defined as having H2A.Z-containing nucleosomes that create a larger acidic patch [99,100]. The interesting question is how this unique +1 acidic patch regulates the activities of SAGA and other complexes.
An important regulatory mechanism for gene expression involves SAGA-mediated HAT activity via direct acetylation of non-histone subunits of RSC and SWI/SNF ATP-dependent chromatin remodeling complexes. As mentioned in section 2.1, Rsc4 and Snf2 are acetylated by Gcn5, which results in an intramolecular interaction with their respective bromodomains [39,47]. Could the BRD of the SAGA-associated subunits Spt7 or Gcn5 interact with a yet to be identified PTM on SAGA? Furthermore, this type of intramolecular interaction may exist for other chromatin-associated factors that have reader domains. For example, several proteins contain tandem Tudor domains, such as Sgf29, JMJD2A, FMR1, SND1, Rad9, and 53BP1 [34]. Although each of these proteins contains tandem Tudor domains classified in distinct subfamilies, it is possible that non-histone methylation provides mechanism distinct from, but analogous to the mechanism of non-histone acetylation-BRD intramolecular interactions. Interestingly, many readers also have single or double PhD domains that may be regulated not only by non-histone methylation but by non-histone acetylation because PhD fingers also bind to acetylated histone substrates [101]. Additionally, histone deacetylases and demethylases may target non-histone substrates to reverse the above-mentioned intramolecular interactions to recycle the “active” state for recognizing their chromatin substrates. The discovery of other key intramolecular interactions will explain how chromatin-associated factors are released or prevented from interacting with their substrates. These types of intramolecular switches could also contribute to feedforward and/or feedback loops that control gene expression and transcriptional memory. Overall, the questions raised here will likely be addressed in the coming years, thereby ensuring that the “SAGA” of understanding how gene expression is fundamentally regulated will continue to unfold and yield new insights.
Highlights:
Discusses advances in understanding SAGA regulation of transcription via histone post-translational modifications (PTMs).
Discusses the functions of the HAT and DUB modules of SAGA in establishing, and regulating, histone PTMs.
Discusses the activities of cis- and trans-histone crosstalk pathways and how reader domains promote feedforward and feedback loops.
Acknowledgements
This work was supported by grants from to National Institutes of Health to S.D.B (AI136995) and B.D.S (GM126900). Additional funding support was provided by the National Institute of Food and Agriculture to S.D.B (1007570). We like to thank Drs. Patrick Cramer and Haibo Wang for sharing the PDB files for the SAGA Cryo-EM structure and Patrick Lane for help in generating SAGA and chromatin figures. In addition, we thank Kortany Baker, Raghuvar Dronamraju, and Howard Fried or editorial suggestions. Due to space limitations, we apologize to our colleagues for not being able to cite all of the important papers contributing to SAGA’s discovery, structure and function. Finally, we dedicate our review in memory of Dr. Susan Abmayr and Dr. Craig Mizzen for their seminal contributions to the field.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Stedman E, Stedman E, The Basic Proteins of Cell Nuclei, Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 235 (1951) 565–595. 10.1098/rstb.1951.0008. [DOI] [Google Scholar]
- [2].Allfrey V, The Functional Biochemistry of Cell Structures (Symp. Intern. Congr. Biochem, 5th, Lindberg Moscow, O., ed.), Pergamon Press, Oxford, 1961, pp. 127. [Google Scholar]
- [3].Phillips DMP, The presence of acetyl groups in histones, Biochemical Journal, 87 (1963) 258–263. 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Murray K, The Occurrence of iε-N-Methyl Lysine in Histones, Biochemistry, 3 (1964) 10–15. 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- [5].Allfrey VG, Faulkner R, Mirsky AE, Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis, Proc Natl Acad Sci U S A, 51 (1964) 786–794. 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Brownell JE, Allis CD, An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei, Proc Natl Acad Sci U S A, 92 (1995) 6364–6368. 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD, Tetrahymena Histone Acetyltransferase A: A Homolog to Yeast Gcn5p Linking Histone Acetylation to Gene Activation, Cell, 84 (1996) 843–851. 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- [8].Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD, Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo, Genes & development, 12 (1998) 627–639. 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Taunton J, Hassig CA, Schreiber SL, A Mammalian Histone Deacetylase Related to the Yeast Transcriptional Regulator Rpd3p, Science, 272 (1996) 408 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- [10].Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD, Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines, Nature, 383 (1996) 269–272. 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- [11].Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL, Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex, Genes & development, 11 (1997) 1640–1650. 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- [12].Georgakopoulos T, Thireos G, Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription, EMBO J, 11 (1992) 4145–4152. 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Berger SL, Piña B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L, Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains, Cell, 70 (1992) 251–265. 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- [14].Roberts SM, Winston F, SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae, Molecular and cellular biology, 16 (1996) 3206–3213. 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Henry KW, Wyce A, Lo W-S, Duggan LJ, Emre NCT, Kao C-F, Pillus L, Shilatifard A, Osley MA, Berger SL, Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8, Genes & development, 17 (2003) 2648–2663. 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee KK, Workman JL, Histone acetyltransferase complexes: one size doesn’t fit all, Nature Reviews Molecular Cell Biology, 8 (2007) 284–295. 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- [17].Wang L, Dent SY, Functions of SAGA in development and disease, Epigenomics, 6 (2014) 329–339. 10.2217/epi.14.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moraga F, Aquea F, Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses, Frontiers in Plant Science, 6 (2015) 10.3389/fpls.2015.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Helmlinger D, Tora L, Sharing the SAGA, Trends Biochem Sci, 42 (2017) 850–861. 10.1016/j.tibs.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang H, Dienemann C, Stützer A, Urlaub H, Cheung ACM, Cramer P, Structure of the transcription coactivator SAGA, Nature, 577 (2020) 717–720. 10.1038/s41586-020-1933-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, Ben-Shem A, Structure of SAGA and mechanism of TBP deposition on gene promoters, Nature, 577 (2020) 711–716. 10.1038/s41586-020-1944-2. [DOI] [PubMed] [Google Scholar]
- [22].Stegeman R, Spreacker PJ, Swanson SK, Stephenson R, Florens L, Washburn MP, Weake VM, The Spliceosomal Protein SF3B5 is a Novel Component of Drosophila SAGA that Functions in Gene Expression Independent of Splicing, Journal of molecular biology, 428 (2016) 3632–3649. 10.1016/j.jmb.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG, Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo, Molecular and cellular biology, 21 (2001) 6782–6795. 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL, Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit, Science, 292 (2001) 2333–2337. 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- [25].Sermwittayawong D, Tan S, SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment, EMBO J, 25 (2006) 3791–3800. 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laprade L, Rose D, Winston F, Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8, Genetics, 177 (2007) 2007–2017. 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S, Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID, Molecular cell, 68 (2017) 118–129.e115. 10.1016/j.molcel.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, Tora L, SAGA Is a General Cofactor for RNA Polymerase II Transcription, Molecular cell, 70 (2018) 1163–1164. 10.1016/j.molcel.2018.06.007. [DOI] [PubMed] [Google Scholar]
- [29].Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA, Redundant roles for the TFIID and SAGA complexes in global transcription, Nature, 405 (2000) 701–704. 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- [30].Penn MD, Galgoci B, Greer H, Identification of AAS genes and their regulatory role in general control of amino acid biosynthesis in yeast, Proc Natl Acad Sci U S A, 80 (1983) 2704–2708. 10.1073/pnas.80.9.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL, Expanded lysine acetylation specificity of Gcn5 in native complexes, The Journal of biological chemistry, 274 (1999) 5895–5900. 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- [32].Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD, Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification, The Journal of biological chemistry, 282 (2007) 7632–7640. 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S, Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation, The Journal of biological chemistry, 277 (2002) 7989–7995. 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- [34].Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, Fleharty BE, Paulson A, Allali-Hassani A, Zhou JQ, Mer G, Grant PA, Workman JL, Zang J, Min J, Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation, EMBO J, 30 (2011) 2829–2842. 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M, Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers, Cell, 142 (2010) 967–980. 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- [36].Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD, Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation, Molecular cell, 5 (2000) 905–915. 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- [37].Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL, Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription, Science, 293 (2001) 1142–1146. 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- [38].Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M, Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression, Cell, 132 (2008) 221–232. 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- [39].VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR, Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation, Molecular cell, 27 (2007) 817–828. 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL, Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes, Cell, 111 (2002) 369–379. 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- [41].Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM, Structure and ligand of a histone acetyltransferase bromodomain, Nature, 399 (1999) 491–496. 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- [42].Cieniewicz AM, Moreland L, Ringel AE, Mackintosh SG, Raman A, Gilbert TM, Wolberger C, Tackett AJ, Taverna SD, The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3, Molecular & cellular proteomics : MCP, 13 (2014) 2896–2910. 10.1074/mcp.M114.038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li S, Shogren-Knaak MA, The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes, The Journal of biological chemistry, 284 (2009) 9411–9417. 10.1074/jbc.M809617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Syntichaki P, Topalidou I, Thireos G, The Gcn5 bromodomain co-ordinates nucleosome remodelling, Nature, 404 (2000) 414–417. 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- [45].Barbaric S, Walker J, Schmid A, Svejstrup JQ, Hörz W, Increasing the rate of chromatin remodeling and gene activation--a novel role for the histone acetyltransferase Gcn5, EMBO J, 20 (2001) 4944–4951. 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hassan AH, Awad S, Prochasson P, The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes, The Journal of biological chemistry, 281 (2006) 18126–18134. 10.1074/jbc.M602851200. [DOI] [PubMed] [Google Scholar]
- [47].Kim JH, Saraf A, Florens L, Washburn M, Workman JL, Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2, Genes & development, 24 (2010) 2766–2771. 10.1101/gad.1979710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA, Genome-wide map of nucleosome acetylation and methylation in yeast, Cell, 122 (2005) 517–527. 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- [49].Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ, Single-nucleosome mapping of histone modifications in S. cerevisiae, PLoS biology, 3 (2005) e328 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jiang L, Smith JN, Anderson SL, Ma P, Mizzen CA, Kelleher NL, Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. LYS4 trimethylation correlates with degree of acetylation on the same H3 tail, The Journal of biological chemistry, 282 (2007) 27923–27934. 10.1074/jbc.M704194200. [DOI] [PubMed] [Google Scholar]
- [51].Pray-Grant MG, Daniel JA, Schieltz D, Yates JR 3rd, Grant PA, Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation, Nature, 433 (2005) 434–438. 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- [52].Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD, Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs, Molecular cell, 24 (2006) 785–796. 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L, The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3, Molecular and cellular biology, 26 (2006) 7871–7879. 10.1128/mcb.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA, Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry, Molecular and cellular biology, 22 (2002) 4723–4738. 10.1128/mcb.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ringel AE, Cieniewicz AM, Taverna SD, Wolberger C, Nucleosome competition reveals processive acetylation by the SAGA HAT module, Proc Natl Acad Sci U S A, 112 (2015) E5461–5470. 10.1073/pnas.1508449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG, Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions, Molecular cell, 25 (2007) 31–42. 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- [57].Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL, MLL targets SET domain methyltransferase activity to Hox gene promoters, Molecular cell, 10 (2002) 1107–1117. 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- [58].Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM, Crosstalk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation, The Journal of biological chemistry, 282 (2007) 4408–4416. 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- [59].Maltby VE, Martin BJ, Brind’Amour J, Chruscicki AT, McBurney KL, Schulze JM, Johnson IJ, Hills M, Hentrich T, Kobor MS, Lorincz MC, Howe LJ, Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae, Proc Natl Acad Sci U S A, 109 (2012) 18505–18510. 10.1073/pnas.1202070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].West MH, Bonner WM, Histone 2B can be modified by the attachment of ubiquitin, Nucleic acids research, 8 (1980) 4671–4680. 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Robzyk K, Recht J, Osley MA, Rad6-dependent ubiquitination of histone H2B in yeast, Science, 287 (2000) 501–504. 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- [62].Sun ZW, Allis CD, Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast, Nature, 418 (2002) 104–108. 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- [63].Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD, Gene silencing: trans-histone regulatory pathway in chromatin, Nature, 418 (2002) 498 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- [64].Fingerman IM, Du HN, Briggs SD, Controlling histone methylation via trans-histone pathways, Epigenetics, 3 (2008) 237–242. 10.4161/epi.3.5.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hsu PL, Shi H, Leonen C, Kang J, Chatterjee C, Zheng N, Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS, Molecular cell, 76 (2019) 712–723.e714. 10.1016/j.molcel.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Worden EJ, Hoffmann NA, Hicks CW, Wolberger C, Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L, Cell, 176 (2019) 1490–1501.e1412. 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Anderson CJ, Baird MR, Hsu A, Barbour EH, Koyama Y, Borgnia MJ, McGinty RK, Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase, Cell reports, 26 (2019) 1681–1690.e1685. 10.1016/j.celrep.2019.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Valencia-Sánchez MI, De Ioannes P, Wang M, Vasilyev N, Chen R, Nudler E, Armache J-P, Armache K-J, Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination, Molecular cell, 74 (2019) 1010–1019.e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yumerefendi H, Lerner AM, Zimmerman SP, Hahn K, Bear JE, Strahl BD, Kuhlman B, Light-induced nuclear export reveals rapid dynamics of epigenetic modifications, Nat Chem Biol, 12 (2016) 399–401. 10.1038/nchembio.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xiao T, Kao C, Krogan N, Sun Z, Greenblatt J, Osley M, Strahl B, Histone H2B ubiquitylation is associated with elongating RNA polymerase II, Molecular and cellular biology, 25 (2005) 637–651. 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dronamraju R, Strahl BD, A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation, Nucleic acids research, 42 (2014) 870–881. 10.1093/nar/gkt1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Van Oss SB, Shirra MK, Bataille AR, Wier AD, Yen K, Vinayachandran V, Byeon IL, Cucinotta CE, Heroux A, Jeon J, Kim J, VanDemark AP, Pugh BF, Arndt KM, The Histone Modification Domain of Paf1 Complex Subunit Rtf1 Directly Stimulates H2B Ubiquitylation through an Interaction with Rad6, Molecular cell, 64 (2016) 815–825. 10.1016/j.molcel.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF, RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach, Molecular and cellular biology, 22 (2002) 6979–6992. 10.1128/mcb.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kim J, Roeder RG, Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast, The Journal of biological chemistry, 284 (2009) 20582–20592. 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wood A, Schneider J, Dover J, Johnston M, Shilatifard A, The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p, The Journal of biological chemistry, 278 (2003) 34739–34742. 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- [76].Katan-Khaykovich Y, Struhl K, Heterochromatin formation involves changes in histone modifications over multiple cell generations, EMBO J, 24 (2005) 2138–2149. 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wozniak GG, Strahl BD, Hitting the ‘mark’: interpreting lysine methylation in the context of active transcription, Biochim Biophys Acta, 1839 (2014) 1353–1361. 10.1016/j.bbagrm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- [78].Chandrasekharan MB, Huang F, Sun ZW, Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability, Proc Natl Acad Sci U S A, 106 (2009) 16686–16691. 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF, Genome-wide function of H2B ubiquitylation in promoter and genic regions, Genes & development, 25 (2011) 2254–2265. 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Henry KW, Berger SL, Trans-tail histone modifications: wedge or bridge?, Nat Struct Biol, 9 (2002) 565–566. 10.1038/nsb0802-565. [DOI] [PubMed] [Google Scholar]
- [81].Shema-Yaacoby E, Nikolov M, Haj-Yahya M, Siman P, Allemand E, Yamaguchi Y, Muchardt C, Urlaub H, Brik A, Oren M, Fischle W, Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription, Cell reports, 4 (2013) 601–608. 10.1016/j.celrep.2013.07.014. [DOI] [PubMed] [Google Scholar]
- [82].Nune M, Morgan MT, Connell Z, McCullough L, Jbara M, Sun H, Brik A, Formosa T, Wolberger C, FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics, Elife, 8 (2019) 10.7554/eLife.40988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C, Structural basis for histone H2B deubiquitination by the SAGA DUB module, Science, 351 (2016) 725–728. 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Koehler C, Bonnet J, Stierle M, Romier C, Devys D, Kieffer B, DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt-Ada-Gcn5-acetyltransferase (SAGA), The Journal of biological chemistry, 289 (2014) 8989–8999. 10.1074/jbc.M113.500868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine SI, Kurumizaka H, Structural basis of the nucleosome transition during RNA polymerase II passage, Science, 362 (2018) 595–598. 10.1126/science.aau9904. [DOI] [PubMed] [Google Scholar]
- [86].Kujirai T, Kurumizaka H, Transcription through the nucleosome, Curr Opin Struct Biol, 61 (2020) 42–49. 10.1016/j.sbi.2019.10.007. [DOI] [PubMed] [Google Scholar]
- [87].McDaniel SL, Strahl BD, Shaping the cellular landscape with Set2/SETD2 methylation, Cell Mol Life Sci, 74 (2017) 3317–3334. 10.1007/s00018-017-2517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL, H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex, Molecular cell, 27 (2007) 275–288. 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- [89].Hassan AH, Awad S, Al-Natour Z, Othman S, Mustafa F, Rizvi TA, Selective recognition of acetylated histones by bromodomains in transcriptional co-activators, The Biochemical journal, 402 (2007) 125–133. 10.1042/bj20060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD, BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex, Curr Biol, 15 (2005) 1487–1493. 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- [91].Zhang Y, Mi W, Xue Y, Shi X, Kutateladze TG, The ZZ domain as a new epigenetic reader and a degradation signal sensor, Crit Rev Biochem Mol Biol, 54 (2019) 1–10. 10.1080/10409238.2018.1564730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Spedale G, Mischerikow N, Heck AJ, Timmers HT, Pijnappel WW, Identification of Pep4p as the protease responsible for formation of the SAGA-related SLIK protein complex, The Journal of biological chemistry, 285 (2010) 22793–22799. 10.1074/jbc.M110.108787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mischerikow N, Spedale G, Altelaar AF, Timmers HT, Pijnappel WW, Heck AJ, In-depth profiling of post-translational modifications on the related transcription factor complexes TFIID and SAGA, J Proteome Res, 8 (2009) 5020–5030. 10.1021/pr900449e. [DOI] [PubMed] [Google Scholar]
- [94].Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR 3rd, Grant PA, The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway, Molecular and cellular biology, 22 (2002) 8774–8786. 10.1128/mcb.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sterner DE, Belotserkovskaya R, Berger SL, SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription, Proc Natl Acad Sci U S A, 99 (2002) 11622–11627. 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Meriesh HA, Lerner AM, Chandrasekharan MB, Strahl BD, The histone H4 basic patch regulates SAGA-mediated H2B deubiquitination and histone acetylation, The Journal of biological chemistry, 295 (2020) 6561–6569. 10.1074/jbc.RA120.013196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Fingerman IM, Li HC, Briggs SD, A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway, Genes & development, 21 (2007) 2018–2029. 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].McGinty RK, Tan S, Recognition of the nucleosome by chromatin factors and enzymes, Curr Opin Struct Biol, 37 (2016) 54–61. 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature, 389 (1997) 251–260. 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [100].Suto RK, Clarkson MJ, Tremethick DJ, Luger K, Crystal structure of a nucleosome core particle containing the variant histone H2A.Z, Nat Struct Biol, 7 (2000) 1121–1124. 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- [101].Sanchez R, Zhou MM, The PHD finger: a versatile epigenome reader, Trends Biochem Sci, 36 (2011) 364–372. 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


