Abstract
Biomolecular condensation partitions cellular contents and has important roles in stress responses, maintaining homeostasis, development and disease. Many nuclear and cytoplasmic condensates form from RNA and RNA-binding proteins (RBPs), which undergo liquid-liquid phase separation (LLPS). Whereas the role of RBPs in condensates has been well studied, less attention has been paid to the contribution of RNA to LLPS. In this Review, we discuss the role of RNA in biomolecular condensation, and highlight considerations for designing condensate reconstitution experiments. We focus on RNA properties such as composition, length, structure, modifications and expression level. These properties can modulate the biophysical features of native condensates, including their size, shape, viscosity, liquidity, surface tension and composition. We also discuss the role of RNA–protein condensates in development, disease and homeostasis, emphasizing how their properties and function can be determined by RNA. Finally, we discuss the multifaceted functions of biomolecular condensates, including cell compartmentalization through RNA transport and localization, supporting catalytic processes, storage and inheritance of specific molecules, and buffering noise and responding to stress.
Introduction:
The term liquid-liquid phase separation (LLPS) describes the formation of two immiscible fluids from a single homogenous mixture. Many, but not all phase-separated biological condensates arise from RNA and protein. Although proteins, lipids and DNA can undergo phase separation independently of RNA, in this Review we discuss only RNA-containing condensates. Many phase-separated condensates studied to date are enriched in and depend on RNA, and proteins involved in the formation and function of these condensates often have canonical RNA-binding domains. Many properties of RNA can promote its phase separation with RNA-binding proteins (RBPs). The most basic property is the negative charge of RNA, which promotes complex coacervation — de-mixing of polymers owing to opposing charges and likely promotes non-specific RNA interactions that contribute to condensation1. However, sequence-specific features are also known or likely to be relevant to RNA-driven phase separation. For example, the number and spacing of RBP binding sites are analogous to the ‘stickers and spacers’ models describing peptides undergoing phase separation2–4. Other features encoded in RNA sequences that have been shown to, or likely influence the LLPS include RNA modifications and RNA–RNA interactions, which can be considered a source of multivalency5, 6. Finally, it is well appreciated that peptides in condensates contain low complexity sequences or intrinsically disordered regions [G] (IDRs) that enable the weak, multivalent interactions to promote liquid-like properties; a similar role for unstructured sequences in RNAs likely contributes to LLPS, but the role of disordered domains in RNA has not yet been shown. Thus, although various physiochemical properties of RNA support their widespread association with condensates, the molecular understanding of these properties is still limited.
In cases where LLPS has been reconstituted using RNA and RBPs, RNA had potent effects on phase behaviour, both controlling the phase boundaries and material properties of the condensed state. For example, proteins of P granules [G] in Drosophila melanogaster and Caenorhabditis elegans, and cytoplasmic condensates of the RBP Whi3 in Ashbya gossypii will undergo phase separation at lower protein concentrations and in more physiological buffers in the presence of RNA7, 8. This is thought to be owing to a combination of charge effects and the ability of a single RNA molecule to recruit multiple RBPs, bringing proteins into close proximity to promote phase separation. Interestingly, whereas low concentrations of RNA support the formation of condensates, RNA at a sufficiently high concentration can dissolve reconstituted condensates; indeed, the high concentration of RNA in the nucleus is thought to prevent certain proteins from spontaneously demixing9. This dissolving capacity potentially causes re-entrant phase behaviour [G] of components in nuclear condensates, which can be predicted from polymer chemistry for particular combinations of concentrations of components10. Furthermore, depending on the condensate, the presence of RNA can promote distinct material properties of the whole or a part of a condensate5, 8, 11–13. Thus, RNA can regulate the formation, material properties, composition and permanence of condensates. Akin to proteins, RNAs are likely to function as ‘scaffolds’, which are essential structural components of condensates, as well as ‘clients’, which are non-essential molecules that are recruited depending on the composition and function of a given condensate14.
In this Review, we discuss the current knowledge of, and key open questions related to how RNA regulates the assembly, properties and functions of condensates. We first focus on how RNA affects the formation and biophysical properties of condensates, discuss considerations for using RNA in condensate-reconstitution experiments and finally highlight some of the functions of biomolecular condensates in vivo. In vitro reconstitution of condensates from purified RNA and proteins is an essential step in demonstrating their mechanism of formation (LLPS or otherwise) and minimal biochemical complexity. Reconstitution of RNA–protein condensates can be achieved using recombinant protein and in vitro-transcribed RNA at appropriate ratios and buffer conditions. Considerations for the protein component in these reconstitution experiments have been extensively reviewed elsewhere15, 16, so here we highlight RNA contributions for the formation of condensates. The composition, length, structure, modifications and expression level of RNA can all potentially contribute to the features of native condensates such as size, shape, viscosity, liquidity, surface tension, and composition, and thus should be considered in reconstitution experiments.
In the second part of the Review, we discuss key functional roles of RNA-based condensates. The physiological functions of some condensates have been inferred from loss of function studies of an essential component of the condensate at the cellular level and/or in whole organisms17–23. The functions of condensates can be classified into four categories, namely cell compartmentalization through RNA transport and localization, supporting catalytic processes, storage and inheritance of specific molecules, and buffering noise and responding to stress. Particular condensates may act by performing one or more of these functions and, in disease states; mutations may disrupt or create new, pathological functions. In section two, we will provide examples of the 4 roles for RNA-protein condensates in cell and organismal biology.
The formation of condensates
In vitro reconstitution is a powerful tool in the study of phase separation, as this simplification of the process facilitates the assessment of mechanisms and comparisons with computational simulations and physical modeling. In this section we discuss RNA features that have been demonstrated to have important roles in condensate formation. Although no in vitro experiment can perfectly recapitulate, measure or address all in vivo RNA features, we encourage the reader to apply the considerations presented in this section in their research, think critically about what RNA feature or features are most relevant to their studies and design their experiments in a way that minimizes artifacts and maximizes usefulness and physiological relevance.
Challenges in studying the role of RNA in condensates
RNA-rich condensates are often complex mixtures of many different RNA molecules, making it difficult to recapitulate their compositional complexity outside of the cell. It is a common practice in in reconstitution assays to use homotypic RNA polymers — molecules that consist of a single type of nucleoside such as poly(A), and which are shorter than native target RNAs (RNAs that are enriched in a particular granule), and tend to be single stranded and poorly structured. Importantly, different single-nucleoside polymers (poly(A), poly(G), etc.) and different polymer lengths can yield considerably different condensate properties 24 (Figure 1A). Another important consideration is the sequence binding preference of the RNA-binding domain of the target protein (protein that is enriched in a particular granule), as different RNA-binding domains may favour different sequences, for example purine-rich vs. pyrimidine-rich or single stranded vs. double stranded14, 25. By using non-native, homopolymeric RNAs, assays fail to capture the complex contribution of the diverse native cellular RNA population, which can dramatically alter all aspects of phase diagrams [G], material properties and molecular composition of condensates.
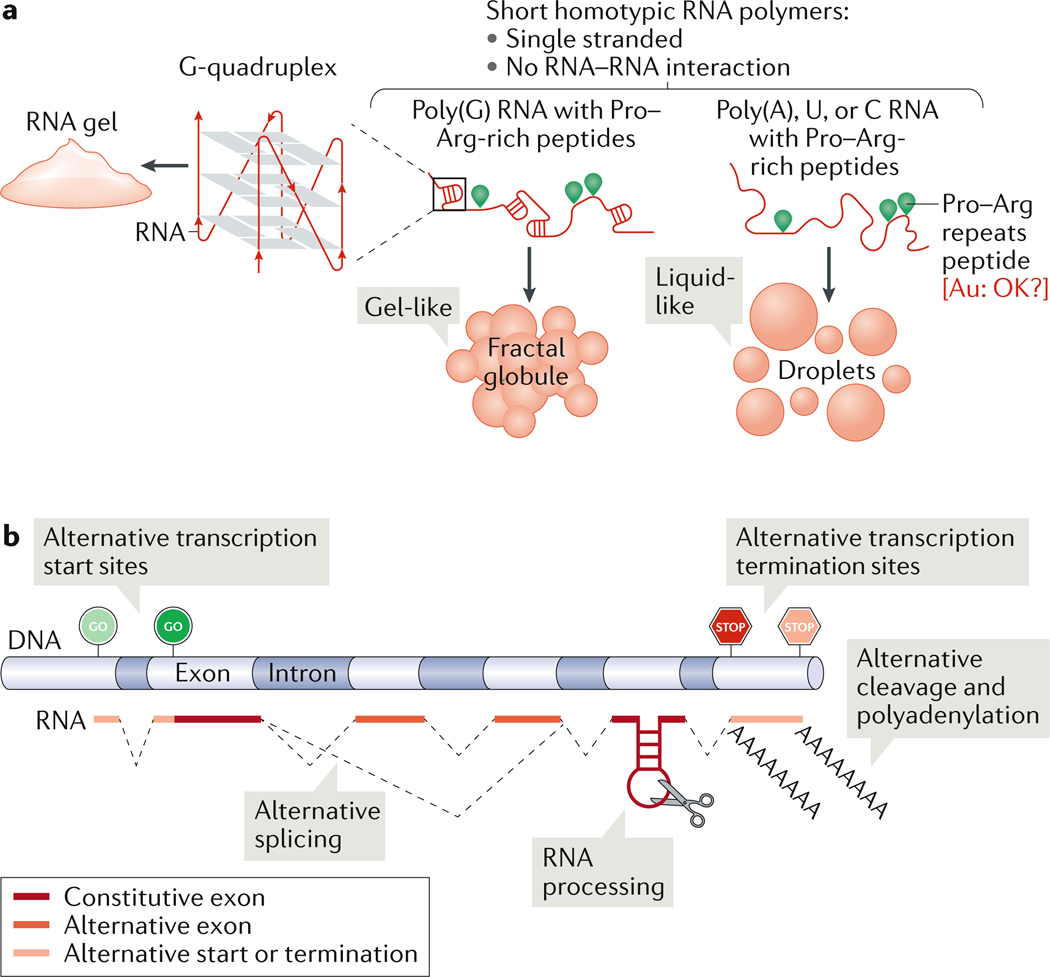
Figure 1: Regulation of condensate properties through RNA sequence and length.
(A) Homotypic RNA polymers mixed with proline–arginine-repeats peptides yield either liquid-like (poly(A), U or C) or gel-like (poly(G)) condensates. At high-enough concentrations, poly(G) RNAs can form G-quadruplexes and RNA gels. (B) Generating RNA sequence (chemical) complexity. RNA sequence length can be regulated by choice of transcription start or termination sites, alternative splicing, RNA processing or alternate cleavage and polyadenylation.
Thus, caution must be applied when interpreting the results of in vitro studies of non-native RNAs. Such experiments probe only a limited dimension of RNA molecules — their function as an anionic polymer — while neglecting the rich array of native RNA properties. Ideally, condensate reconstitution would employ native target RNA sequences, which can confer distinct biophysical properties to the condensates8, but these might be difficult or expensive to synthesize in vitro in sufficient quantities for phase separation assays. Furthermore, RNAs of higher eukaryotes are often too long for in vitro transcription, particularly in un-spliced form. Finally, another complication is that most RNA–protein condensates likely arise from a collection of many hundreds or thousands of RNA species present in a single condensate (for example, in stress granules [G] 26 and P granules 27). P granule RNAs appear to be arranged in specific spatial patterns13, 28, and the protein component of stress granules (and likely also the RNA component) is assembled in a specific temporal order29. Reconstitution of the full cohort of recruited RNAs can only be realistically achieved using whole cell extracts and/or through isolation from condensates, which requires large amounts of biomass that can be obtained from Xenopus laevis oocytes30, budding yeast31 or cultured mammalian cells32, but may be more difficult to obtain from condensates observed difficult to culture cell types such as neurons. It is important to characterize the RNAs present and enriched in condensates, and a number of techniques such as crosslinking immunoprecipitation followed by RNA-seq, physical condensate isolation, and cell free induction of condensate formation have been employed for recapitulating complex condensates such as stress granules33, P bodies32 and neuronal transport granules34. Profiling the variety of RNA targets of condensates may provide hints to condensate function, and is useful for identifying target RNAs for reconstitution experiments.
Despite the limited complexity of composition, there is tremendous value in employing reductionist approaches to reconstitute RNA-based LLPS. Because reconstitution is based on tailored experiments with either native sequences or carefully engineered RNAs that have particular sequence features, it is the most feasible way to begin to dissect the mechanisms by which RNA affects condensates. Next, we consider the distinct features of RNA that are functionally relevant for determining and informing about condensate properties, including nucleotide composition and sequence, polymer length, structure, modifications and higher-order assemblies.
Regulation through RNA sequence and length
RNA is a polymer consisting of purines (A and G) and pyrimidines (C, U). Analogous to the contribution of amino acid sequence to protein biochemistry, purines and pyrimidines confer different properties to the RNA molecule and in turn to the condensate, which is composed of various RNAs. It was first noted already in 1910 that guanosine in high concentration forms a gel 36. In studies with homopolymers of RNA, condensate properties were conferred by RNA sequence, RNA–RNA interactions and RNA–protein interactions2. For example, condensates formed of poly(G) RNA form fractal-like and gel-like networks with proline–arginine-repeats peptides, in contrast to other homotypic polymers (A, U or C), which form more liquid-like condensates (Figure 1A). Condensates consisting of poly(A) RNA are more viscous then those comprising poly(U) or poly(C) RNA, whereas mixtures of poly(A) and poly(U) RNA form more solid-like gels24. Thus, even artificial homopolymers can have considerably different effects on condensate properties depending on the nucleotide composition.
RNA polymers in cells are typically complex mixtures of nucleotides. Sequence (chemical) complexity in RNA is generated during transcription and processing. Variable sequences can be made from the same gene through use of alternative transcription start and termination sites, polyadenylation sites, alternative splicing (especially in higher eukaryotes)37, or through degradation by RNAses38 (Figure 1B). These changes could deliver an RNA to a particular condensate in one cell type but not in another cell type through as simple a mechanism as extending or shortening the length of the RNA. For example, in mammalian cells, relatively long and less translated mRNAs are thought to be favoured over short RNAs for phase separation in stress granules and possibly other condensates 33. Another example of regulation of phase separation by RNA length is observed in mammalian cells with the long non-coding RNA NEAT1 (nuclear enriched abundant transcript 1)39. NEAT1 has two splice isoforms: the longer isoform is recruited to paraspeckles [G] whereas the shorter isoform is recruited into microspeckles [G] 40. A particular sequence or structure in NEAT1 that varies between isoforms may change specificity for particular compartments through its interactions with a specific RBP, or through changing physiochemical features of the RNA such as length or structure that could influence localization. Therefore, it is important to verify that the correct RNA isoform is present in a condensate of interest using techniques such as fluorescence in situ hybridization41. In vitro production of the particular condensate-relevant splice isoform could be ideal. Pure, full-length RNA must be used for reconstitution as contamination with shorter RNA fragments can alter phase separation in multiple ways, making it impossible to quantify the concentration of full length RNA and altering or abolishing phase separation by forming spurious RNA–protein or RNA–RNA interactions.
RNA sequence and length is tightly controlled; multiple condensate-associated diseases are caused by alteration of RNA primary sequences, independently of, or in addition to the alteration of the encoded proteins — the best example being alterations in C9ORF72 in familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD)42. Alterations of the target RNA sequence can be divided into RNA mutations that modify the levels of transcript (phase separation is less likely to occur if RNA levels are reduced) or of translation (active translation shields RNA from phase separation). Alteration of the RNA sequence can cause RNA-dependent and protein-independent phase separation, for example in the disease-causing G4C2 repeat expansion in the C90orf72 gene, where phase separation of RNA can occur independently of protein, likely through formation of G-quadruplex structures42, 43.
Alteration of the RNA sequence can also remove or alter the protein component of condensates in disease and in normal physiology. Alternative splicing of mRNAs encoding RBPs that undergo phase separation can regulate phase separation of the corresponding condensates, potentially through changing both RNA and protein length. This could occur through alternative splicing (inclusion or exclusion) of RNA-binding domains (although these exons tend to be constitutive) or, more likely, through alternative splicing of IDR exons. An example of regulation of phase separation through IDR sequences and their alternative splicing is found in fragile X mental retardation syndrome-related protein 1 (FXR1), the mammalian autosomal homologue of fragile X mental retardation protein 1 (FMR1). A mutation in the IDR of the muscle-specific exon 15 of FXR1 results in congenital minicore myopathy44. Multiple FXR1 splice isoforms are developmentally regulated and thus alter the IDR composition of FXR1 and modulate material properties of FXR1 condensates45. Overall, particular care should be taken to study physiologically-relevant RNA and protein sequences, as these are more likely to be representative of in vivo condensates.
Tuning condensates through RNA modifications
Analogous to post-translational modifications of proteins, RNA post-transcriptional modifications help regulate important aspects of RNA function. All four RNA nucleotides are chemically modified46, 47. Among the most abundant and well-studied RNA modifications are N6-methyladenosine (m6A), N6,2’-O-dimethyladenosine (m6Am), deamination of adenine to inosine (I), 5-methylcytidine (m5C) and 5-hydroxylmethylcytidine (hm5C), N1-methyladenosine (m1A) and pseudouridine (Ψ). These modifications could influence phase separation in at least three ways: by altering the protein interactions of modified RNA, by altering the RNA secondary structure48, and/or by inhibiting or enhancing RNA–RNA interactions. Although even for the most abundant modification (m6A), the levels of modified RNA are much lower in cells then the levels of unmodified RNA, modifications can be enriched on particular transcripts and locations in the mRNA, for example m6A modifications are enriched near the translation stop codon 49, 50. This differential modification of RNAs suggests their role in condensate formation is restricted to a subset of target RNAs.
RNA modifications are deposited in a highly regulated manner by ‘writer’ enzymes, removed by ‘eraser’ enzymes and recognized by ‘reader’ proteins38, 51. Binding of m6A by the reader YTHDF2 has been reported to induce LLPS in mammalian cells6 and the m6A binding protein YTHDF which is found in clusters at the periphery of stress granules, potentially promoting their formation by reducing the activation energy barrier and critical size52.
It is likely that other RNA modifications and their regulatory proteins can undergo LLPS or potentially block RNAs from undergoing LLPS (Figure 2A). RNA modifications may be primarily found within specific condensates. Mutations in the proteins that regulate RNA modifications, such as adenosine deaminase acting on RNA 1 (ADAR1; also known as DRADA) and APOBEC proteins, can cause human diseases 53–56. Furthermore, RNA modifications are commonly mis-regulated in disease57 and have important roles in the response to viral infection58. The presence or absence of a modification can alter RNA structure and sequence, as shown for ADAR159 and APOBEC proteins53. APOBEC proteins deaminate cytosine to uridine and ADAR1 deaminates adenine to inosine. Inosine is chemically similar to guanine (and is read as guanosine during translation), and is capable of pairing with cytosine in forming RNA structures. A major unanswered question is how RNA modifications that tune RNA structure (Figure 2B) and RNA–RNA interactions (Figure 2C) can regulate the formation of condensates.
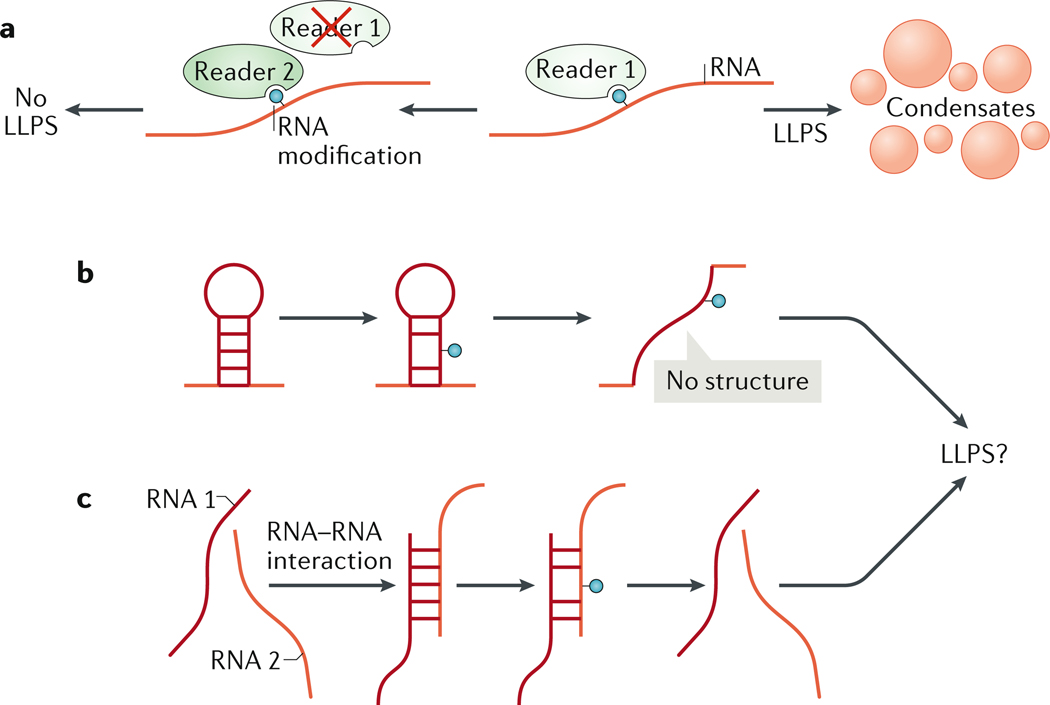
Figure 2: Tuning condensate properties through RNA modifications.
(A) RNA modifications are recognized by ‘reader’ proteins. Interaction with a reader can enhance or inhibit RNA undergoing liquid–liquid phase separation (LLPS). (B and C) RNA modifications can tune RNA structure by blocking or promoting RNA–RNA interactions in cis, within the same RNA molecule (B) or in trans between different RNA molecules(C). Modification of RNA sequence and structure could alter phase separation of target RNA.
The difficulty of studying RNA modifications in vivo is that many thousands of RNAs may be modified in a particular cell type, and mutation in a regulatory protein (writer, reader or eraser) can affect most or all of them. How then does one assign a particular function to a particular modification of a particular RNA? Localization of modified RNA to, or absence from a condensate of reader proteins may be an interesting avenue for future functional studies. Another experimental option would be to change or remove the modified nucleotide or modification consensus site and assess the effect on RNA localization in condensates and their function.
In vitro transcription reactions typically include a small amount of a randomly incorporated fluorescently labeled nucleotide (usually uridine). The problem of studying RNA modifications in a cell-free system is thus fourfold: producing the modified nucleotide required for in vitro transcription reactions, efficiently incorporating the modified nucleotide into the nascent transcript, incorporating it in the correct location, and not incorporating it ubiquitously. Addressing all these issues can be difficult. To minimally demonstrate protein binding to modified RNA and their phase separation together, random replacement of modified bases could be a first step to analyze potential consequences of modified bases. Although this does not often faithfully recapitulate native modifications, it could show whether RNAs modified with particular moieties modulate phase separation of a target protein.
The main unanswered questions regarding the roles of RNA modifications in condensates are: do RNA modifications other than m6A induce phase separation? Do modifications serve as a sorting mechanism for protein delivery into, or exclusion from condensates (through an RNA-structure dependent or independent manner)? And do condensates facilitate the addition or removal of RNA modifications?
Probing the roles of RNA structure
RNA structure can have a vital regulatory role in the formation and identity of condensates, both with and without proteins5, 42. Thus, any in vitro reconstitution experiment ideally should consider the structure of target RNAs. Whereas DNA favours the formation of Watson–Crick base pairings, RNA can form non-Watson–Crick base pairs, which are necessary for the formation of triplex and G-quadruplex [G] structures and give rise to other complex secondary and tertiary RNA structures60. Functional consequences of RNA structures on condensates in cells are discussed below.
Techniques to measure RNA structure (particularly of long, complex RNAs) are in their infancy relative to methods that determine protein structure, generally cannot capture 3D shapes and provide static reflections of structure ensembles (that is, they average a continuum of multiple RNA structures)61. Regardless of these limitations, methods such as SHAPE/DMS and mutational profiling (SHAPE, or DMS)62, 63 and psoralen analysis of RNA interactions and structures (PARIS)64 are available and in some cases have been used to study how RNA structure contributes to condensate identity. The basis of structure-probing methods such as selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE), is that single-stranded RNA is more reactive to chemical modification than double-stranded RNA and that the modified nucleotides are mis-read during reverse transcription and produce detectable mutations. Mutation profiling data are fed into RNA structure-prediction computational pipelines to predict RNA folding patterns. Computational prediction of RNA structure can be undertaken independently of RNA structure probing; however, this approach may be less accurate65–68. When examining multiple RNA sequences, or highly conserved, simple RNA structures, computational prediction remains a viable alternative to RNA structure probing when the latter is unfeasible.
How should the experimental design take RNA structure into consideration? RNA structures are sensitive to many factors, so the buffer should be carefully selected with regards to salt concentration, pH, ion composition and inclusion of a crowding agent (Figure 3A). It is important to select a buffer that yields optimal behavior of both condensate components (RNA and RBPs) and, if possible, is physiologically relevant (considerations for proteins have been extensively reviewed elsewhere69). The species that the reconstituted condensate originates from is an important determinant in the selection of ion concentrations and no universal buffer exists for phase separation assays, because ion composition varies between different cells of different species. For example, the concentration of potassium ions (K+) in budding yeast can reach 300mM70, whereas most mammalian cells have half that concentration of K+ (150mM). The concentration of K+ and of sodium ions (Na+), which are the most physiologically relevant monovalent cations, can stabilize G-quadruplexes 71, 72. However, by far the most important criterion for salt selection when working with RNA is the inclusion or exclusion of magnesium ions (Mg2+). Addition of the chelating agents like EDTA, will remove magnesium from the solution. Magnesium ions stabilize RNA structures and facilitate folding73. If Mg2+ is included in the buffer (in addition to K+ and NA+), it is important to consider in what form (buffered or unbuffered) and in what concentration. In the cell, magnesium is mostly bound to proteins or other biological molecules; therefore, for a buffer to be physiologically relevant, Mg2+ must be either present in very low concentration or buffered by another biological molecule such as glutamate74. As magnesium and manganese (Mn2+) are chemically similar, manganese can effectively replace magnesium in many contexts, and so a similar caution should be applied when using this ion in phase separation buffers. Thus, the choice of salt is critical in reconstitution experiments, in which RNA structure is an important feature.
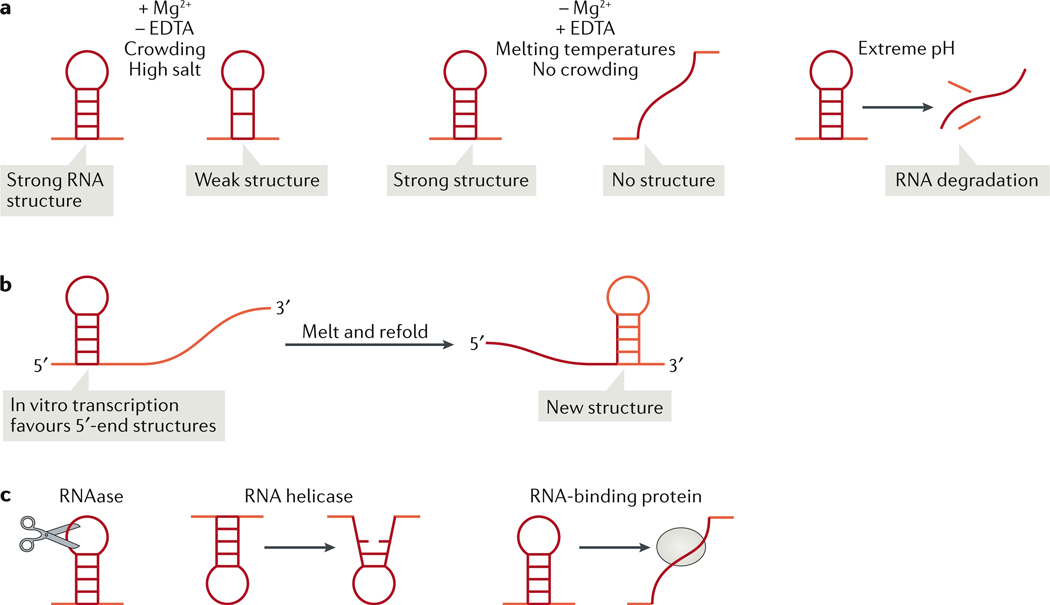
Figure 3: Modifying RNA structure in vitro and in vivo.
(A) In vitro, buffers that include magnesium ions (Mg2+) and a crowding agent and exclude the chelating agent EDTA, will support the formation of both strong RNA structures (structures with many base pairs) and weak RNA structures (fewer base pairs) at lower temperatures (left), whereas in the opposite buffer conditions or in higher temperature, the weaker RNA structures will unfold (middle). Extreme pH will denature and cause the degradation of even strong RNA structures (right). (B) In vitro transcription favours interactions at the 5’-end region, as this region is transcribed first. Melting and refolding of the in vitro transcribed RNA will allow and the formation of interactions between the 5’ and 3’ regions of the molecule. (C) In vivo, RNA structures are modulated by cellular RNAses, which actively remove particular RNA sequences and structures; by RNA helicases, which unwind double-stranded RNA; and by RNA-binding proteins, which can shield RNA sequences from RNA–RNA interaction or block structure formation. The cellular environment helps to explain the discrepancy between RNA-structure probing data obtained in vitro or from cells. This discrepancy may be explained also by RNA modifications (Figure 2).
Another consideration for buffer selection is the inclusion and choice of crowding agent. Crowding agents mimic the dense intracellular environment and facilitate phase separation, but their use requires caution, as addition of crowding agent in sufficient quantities induces phase separation of any protein or RNA. Commonly used inert crowding agents include PEG, ficoll, dextrans and spermine; however, these compounds are unlikely to be truly inert. Addition of crowding agents stabilizes RNA structure in particular confirmations75–77. This effect has been observed in ribozymes (RNAs that catalyze biochemical reactions). Ribozyme activity is facilitated by addition of molecular crowding 78 and addition of crowding agents dampens the impact of structure-disrupting mutations on ribozyme catalysis79. Crowding agents likely have a similar effect on structured RNAs that undergo phase separation; in agreement with this possibility, RNA structures can be different in the condensed phase [G] compared with the dilute phase [G] 5. Thus, some condensates might stabilize or favour particular RNA structures, or promote specific inter-molecular interactions.
A final consideration for buffer selection is the pH and the buffering agent. Not only will the pH alter the phase separation of a protein depending on its isoelectric point, RNA stability and structure is also affected by pH. RNA is most stable at pH 4–5 and unstable at more alkaline pH80. Extreme pH levels can help melt RNA structures 81. Many important downstream RNA experiments such as reverse transcription and RNA structure probing are optimized for pH 8 and the use of TRIS buffer82. Certain stress conditions can alter the pH of the cytosol83 and thus pH can be an especially important component in studying RNA structure in stress-induced condensates.
The interplay between transcription kinetics and RNA folding can influence which secondary RNA structures will form out of a range of theoretically possible structures. This is evident in in vitro transcription reactions, which can give rise to a different RNA structure than that formed when RNA undergoes denaturation and refolding (Figure 3B). This is due to fewer sites being sampled for RNA–RNA interaction (in trans and in cis) at the 5’-end region of the RNA than at the 3’-end region, because sites at the 5’-end are transcribed first. Melting the RNA by heating it to 95°C and refolding it can allow the formation of RNA–RNA interactions between the 5’ and 3’ end regions, which would not be observed in vitro but may occur in a cell owing to the activity of RBPs. This effect is the most readily observed in the reconstitution of in vitro transcribed long, highly structured RNAs such as the CLN3 mRNA, melting and refolding of which alters multiple condensate properties5. Thus, ideally the decision of whether or not to melt and refold the RNA would depend on which RNA production method provides RNA that most closely mimics the cellular RNA structure profiles. RNA structures can be measured in vivo using chemical structure probing as described above. RNA structure can vary between particular subcellular compartments. For example, in SHAPE data of RNA derived from the chromatin, nucleus and cytoplasm, the same RNA may have different reactivity, suggesting it is adopting different structures in different compartments86. Thus, it may be necessary to fractionate cells to probe structures at different cellular compartments.
How do different RNA structures fold from the same sequence? Just as chaperones regulate proteins folding, RBPs and helicases regulate RNA structure and function87–89 (Figure 3C). RBPs can inhibit or promote RNA–RNA interactions in cis and in trans, and RNA helicases can rearrange RNA structures in vivo and in turn regulate condensate properties90. The extensive regulation of RNA structures by RBPs is thought to be a major cause of the discordance between cellular and cell-free RNA-structure profiling85. Therefore, reconstitution of native or native-like RNA structures in a cell-free or protein-free environment can be tricky. The longer RNA targets may sample more secondary structures than shorter RNAs, and have different structures in vitro and in vivo. Furthermore, more than one structural conformation could have a regulatory function (for example, an alternative RNA structure can regulate alternative splicing)91. It is also possible that a particular structure may be favoured for inclusion in condensates. Thus, the cellular environment carefully regulates RNA structures, like protein structures, and this regulation may have a role in determining the formation and properties of condensates in vivo.
Although RNA structure remains one of the most difficult–to-measure features of RNA, it has the capacity to profoundly alter condensate properties. RNA structures can license specific RNA–RNA interactions that influence the molecular composition of condensates5; however, RNA structure could influence condensates also at biochemical and biophysical levels. RNA structures can change the affinity and valency of the RNA molecule to RBPs, which would change the stoichiometry of condensate components (see below). Structures can potentially change the flexibility of the RNA molecule, will influence the volume occupied by RNAs and direct the formation of higher-order RNA structures and long-range interactions92.
RNA–RNA interactions
RNA–RNA interactions are likely an important driver of condensate formation and could control their composition5, 92. RNA features that can promote RNA–RNA interactions include length; high GC content; lack of RNA structure, binding by RBPs or RNA modifications; and poor translation. Thus, enrichment of these features is likely to promote RNA undergoing phase separation. A single RNA molecule can associate within itself (cis RNA interactions) or with another RNA molecule (trans interactions). Trans interactions can occur between two RNAs of the same type or between two or more different RNA types, and can be promiscuous (as in the case of stress granules) or specific, as in the case of the cytoplasmic Whi3 condensates of the multinucleate filamentous fungus A. gossypii, 5, 92,5, 92,5,90,5,90,5, 78,5, 78,5, 78,5, 78,5, 91,5, 90,5, 89,5, 88 where the BNI1 and SPA2 mRNAs assemble together5, 92. RNAs can (self-) associate on their own, or proteins can facilitate RNA–RNA interactions by bringing RNAs together, as in the case of Argonaute proteins that bring siRNA, shRNAs and microRNAs together with their target mRNAs. The organization of mRNAs in germ granules of D. melanogaster highlights roles of yet another type of RNA interaction. A recent study using super-resolution imaging in vivo showed that after being targeted to granules in a sequence-dependent manner, mRNAs of the same sequence cluster within the granule separately of other mRNAs93. This indicates the existence of a mechanism of spatial organization inside granules, which is dependent on the identity of the RNA. Interestingly, the study also revealed that such self-sorting is based on a sequence-independent RNA–RNA interaction; thus, such like-RNAs self-assemble, but in a manner that does not require specific sequence elements.93 This study suggests that RNA organization in germ granules, and perhaps in different RNA-rich condensates, is the result of an ensemble of properties including bound proteins and length, modifications and structures of RNAs, which results in homotypic condensation of RNAs within a larger condensate.
Crosslinking of RNAs inside or outside cells (using psoralen64, AMT94, 95 or dimethyl sulfide96) can be used to identify RNA–RNA interactions without a priori knowledge. RNA-RNA interactions likely have a specialized role in condensate formation in at least four ways: lowering the threshold for phase separation (nucleation), controlling condensate growth rates, sorting particular RNAs to a particular condensate, and creating a meshwork of interconnected RNAs that scaffold condensates and confer important material properties92. With regard to facilitating nucleation, RNA dimerization or multimerization could promote phase separation by concentrating RBPs. Independently of nucleation, RNA–RNA interactions can promote condensate growth by providing more sources of multivalency. Interestingly, the RNA–RNA interaction can regulate sorting of particular RNAs into either the same or different condensates, with RNA structure masking or revealing sites of interaction5.
Like RNA structure, measuring RNA–RNA interactions accurately is challenging. RNA–RNA interaction is easiest to prove between RNAs of two different types, as most crosslinking approaches rely on the sequencing of chimeric reads to identify interactions between two RNAs. RNA–RNA interactions can be estimated using RNAhybrid97 or similar programs. The role of RNA–RNA interactions in phase separation has also been discussed elsewhere92, but this understudied feature of RNA is an important emergent property that is informed by RNA sequence and structure.
Balancing the stoichiometry of RNA and protein in condensates
RNA expression levels are tightly regulated through transcription, stabilization and degradation, and RNA levels are important for the formation and maintenance of condensates. For example, transcription of rRNA can induce nucleoli formation98, and transcription of NEAT1 leads to paraspeckle formation in mammalian cells. Additional RNA regulation occurs at the level of cellular localization and interactions with RBPs. For example, concentrations of soluble RNA are typically higher in the nucleus than in the cytoplasm and consequently multiple RNA–protein condensates (paraspeckles, speckles [G], Cajal bodies [G], nucleoli, etc.) can be found in the nucleus. However, RNA in the nucleus has also been proposed to solubilize some phase-separating proteins and thus prevent pathological aggregation in mammalian cells 9. Many diseases are associated with a failure of phase-separating RBPs to localize to the nucleus9, because in the cytosol they assemble aggregates at least in part due to the lower levels of RNA99. Cytoplasmic RNA–protein condensates may be less common than nuclear condensates, because cytoplasmic RNA is protected from phase separation by translation and free RNA is destroyed by antiviral sensing mechanisms. Thus, RNA and RBP components of condensates must be tightly regulated in abundance and location.
When reconstituting phase separation in vitro, it is therefore important to consider physiological RNA and protein concentrations and ratios, as inferred from cellular measurements, to ensure that the cell-free observations are relevant. Particular care must be taken when considering the required concentrations of RNA and protein: if cellular concentrations are insufficient to promote phase separation, it is likely that additional regulation of LLPS is at play in cells. Altering RNA and protein ratios can result in more liquid-like or more gel-like condensates, which is similar to disease condensate states resulting from imbalance of RNA or protein components that lead to condensate ‘hardening’100–103. Therefore, in vitro reconstitutions must be informed by in vivo observations of RNA and RBP concentrations measured using methods such as mass spectrometry, quantitative western and northern blotting or fluorescence correlation spectroscopy.
Another factor to consider when choosing RNA and protein concentrations is valency, or the number of protein contact sites for a given RNA. If the RNA-binding domain binds highly specific RNA motifs, valency can be estimated by counting the number of times the sequence appears in the RNA (Figure 4A and B), although interactions of RNA with disordered, low complexity domains of RBPs are more difficult to assess. For example, disordered RNA-binding domains such as RGG (Arg–Gly-rich) regions may bind a variety of different RNA sequences —RGG regions were shown to bind to poly(G), poly(A), poly(U) and weakly to poly(C) sequences104. Altering the number of interactions between RNA and RBPs may shift the phase diagram with increasing valency, thereby reducing the saturation concentration (Figure 4C and D) of RNA for phase separation and vice versa (Figure 4E, F and G), but specific situations might deviate from this simple prediction. Ideally, the binding of RNA to an RBP will be assessed first in vivo using crosslinking to identify genuine sites of contact, and if possible by removing the RNA-binding sites (through mutation or deletion) to abolish phase separation105. Although the valency of a specific RNA is unlikely to be variable in a given species (unless the RNA undergoes modification), it may be subjected to evolutionary selection, thereby enabling interesting analysis of condensates across species.
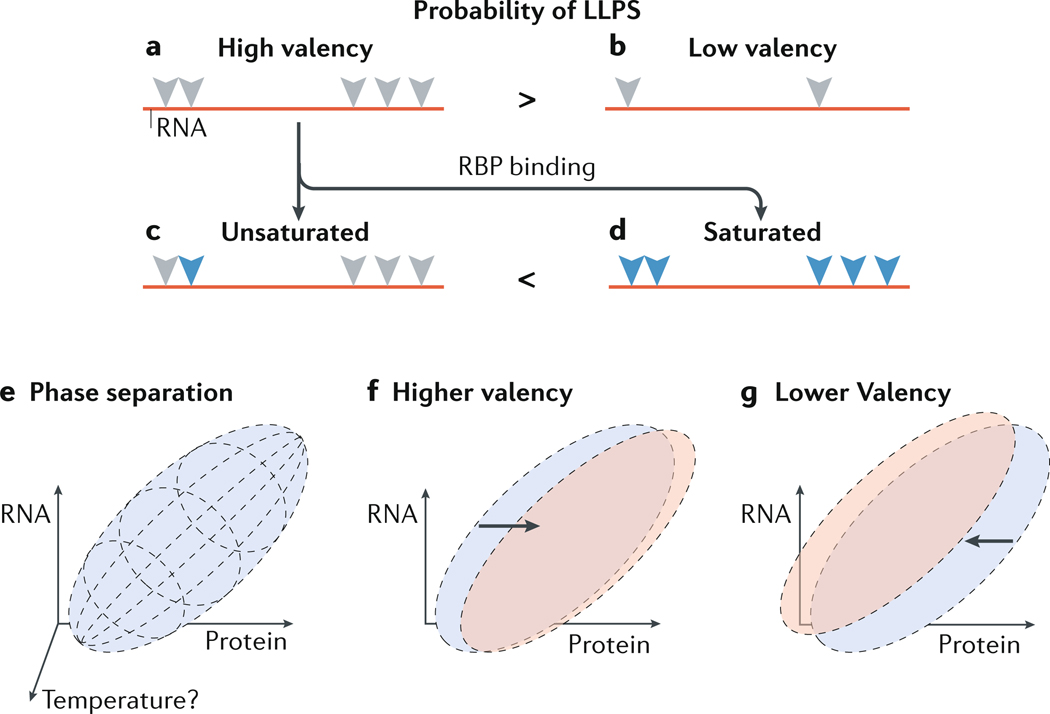
Figure 4: Balancing RNA and protein ratios in condensates.
(A and B) RNA with high valency (A) refers to an RNA molecule that contains multiple sequences and/or structures (grey triangles) that can be recognized by an RNA-binding protein (RBP). Low valency (B) refers to an RNA with few such sequences or structures. High valency may favor liquid–liquid phase separation (LLPS) over low valency. A fraction, or all of the potential binding sites for RBPs could be actually bound by RBPs (blue triangles), resulting in unsaturated (C) or saturated (D) RNA to protein ratios, respectively. Saturation may favor LLPS. (E) A possible phase diagram of a mixture of RNA and protein at a particular temperature. Phase separation between the RNA and protein occurs within the parameters of the purple ellipsoid and is a factor of the concentrations of RNA, of protein, and of buffer (not shown). (F) Increasing the valency of the RNA target may shift the phase diagram to the right (red ellipse), indicating that a lower RNA concentration is required to induce LLPS. (G) Reducing RNA valency may have an opposite effect.
In summary, the physiochemical role of RNA in condensate formation remains an important understudied topic. Many challenges remain in studying the role of native and native-like RNA targets in a cell-free context, including chemical complexity (RNA sequence, length, variability, modifications) and sequence-dependent properties such as RNA structure, RNA–RNA interactions and RNA–protein interactions. Incorporation of these considerations into experimental design will result in reconstitution of more native-like condensates and improve our understanding of how the biophysical properties of condensates inform in vivo functions.
Physiological roles of RNA condensates
Condensates can regulate the localization and function of cellular molecules in space and time. In this section, we provide examples of RNA-rich condensates and their cellular functions.
Cell compartmentalization through RNA transport and localization
Cell size varies between and within organisms. Regardless of size, all cells must compartmentalize their contents to optimize their function, and one mechanism for achieving this is by compartmentalizing processes in different condensates106, 107 (Figure 5). Cells of many sizes form condensates; however, diseases with condensate involvement seem to be associated with large cells such as neurons (Figure 5A), oocytes (Figure 5B) and syncytial cells such as muscle cells and filamentous fungi (Figure 5C). This association likely exists because large cells must expend more effort to actively compartmentalize their cytoplasm than smaller cells. In large cells, it is crucial to enclose mRNAs in condensates for long-range transport and for the often ensuing localized, on-demand translation. RNA compartmentalization is conserved across multiple cell types and species. For example, the Staufen family of RBPs compartmentalize both oocytes and neurons in fruit fly and mouse108,109. With regard to active RNA transport, Staufen proteins (Staufen in the fruit fly and STAU1 and STAU2 in humans) bind both double-stranded RNA and microtubules, and thus provide both RNA-target specificity (through the preference of their RNA-binding domain) and a mechanism of transport (microtubules), which is conserved from fruit flies to humans108, 110. Condensates can also ‘hitchhike’ on other cytoplasmic components. For example, annexin A11 helps transport RNA granules in neurons by tethering them to moving lysosome111.
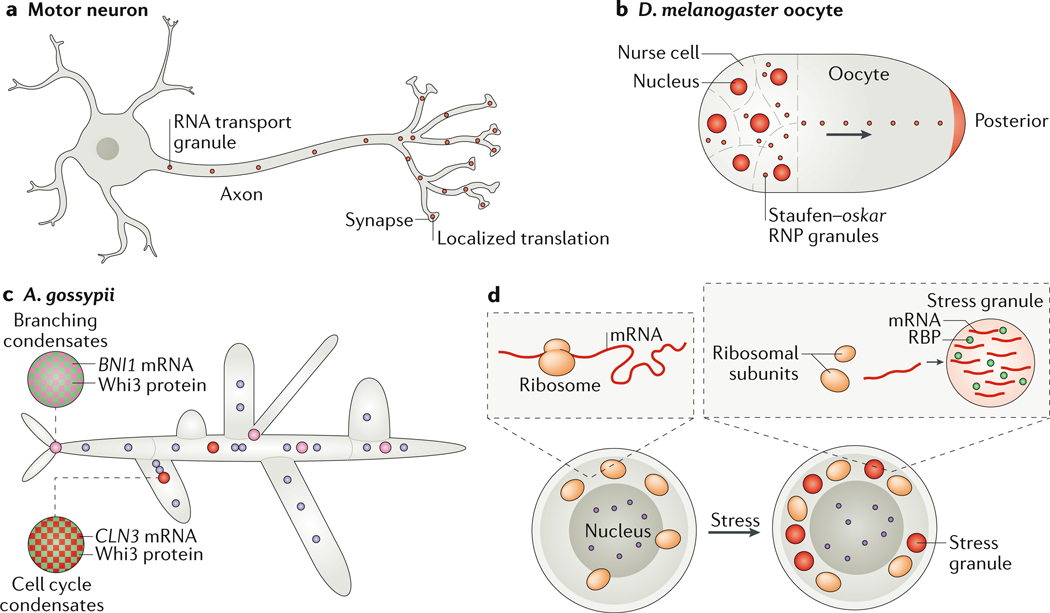
Figure 5: Examples of cytoplasmic RNA–protein condensates.
Compartmentalization of cytoplasmic RNA is prevalent in large (A and B) or multinucleate cells (C and D). (A) In motor neurons, RNA transport granules move mRNAs on microtubules (not shown) towards synapses, to allow their localized translation away from the cell body. (B) Oocytes compartmentalize and store maternal RNA. For example, in Drosophila melanogaster oocytes, the oskar mRNA is transported along microtubules to the posterior pole of the embryo by the RNA-binding protein (RBP) Staufen. (C) Ashbya gossypii is a multinucleate filamentous fungus that utilizes condensates to compartmentalize its cytoplasm. The RBP Whi3 is used to define sites of branching, by binding to the SPA2 and BNI1 mRNAs, and to control nuclear cell cycle asynchrony by binding G1 cyclin RNA (CLN3). CLN3 and BNI1–SPA2 condensates are immiscible; condensate immiscibility is controlled by RNA sequence and structure. Note that the nuclei are not drawn to scale — condensates are roughly 1/10–1/5 the size of nuclei. (D)Cells in conditions of acute stress undergo translation shutdown and release mRNAs from the translation machinery. The mRNAs form stress granules with RBPs. The purple circle is the nucleus.
In neurons, active transport of RNA–protein condensates in the axon111–113 is crucial for neuronal function, and multiple neuron-specific RBPs are localized to condensates that deliver RNA to sites of local translation to enable rapid cellular responses (Figure 5A). Mutations in proteins that are enriched in RNA transport condensates can lead to neurodegenerative diseases such as ALS114. Notably, RNA condensate transport is evolutionary ancient and used in filamentous fungi to transport specific mRNAs to the apical-tip cell using the same machinery as in neurons. For example, mRNP condensates in corn smut (Ustilago maydis) require the RBP Rrm4 and microtubules for establishment of polarity115–119. Thus, active transport of condensates has essential and conserved roles in compartmentalizing the cell cytoplasm.
A special case of compartmentalization occurs in syncytial cells, which comprise several nuclei sharing the same cytoplasm. These cells can form as a result of cell fusion, such as in muscle cells or syncytial trophoblasts, or through endocytosis without cytokinesis. In syncytial cells, each transcriptionally active nucleus sets a sphere of influence within the cytoplasm, and the different nuclei communicate to perform normal cellular functions. Condensates pattern the cytoplasm of syncytial cells, for example in muscle cells (formed by cell fusion45, 120) and in A. gossypii, which is a multinucleate filamentous fungus (formed by asynchronous endomitosis) (Figure 5C). Many other polyploid and syncytial cells exist121, which may utilize condensates to compartmentalize their contents109.
Supporting catalytic processes
Almost all eukaryotic cell types concomitantly form multiple RNA–protein condensates, such as P bodies, Cajal bodies, PML bodies [G], paraspeckles, speckles, nucleoli, nuclear speckles, and transcription-related condensates122, 123. Many of these condensates form in interphase nuclei and dissolve during mitosis. Why form multiple condensates in the same nucleoplasm or cytoplasm? One reason is that different RNAs perform different functions in the cell and, in the absence of regulatory proteins, RNA has the tendency to aggregate in cellular salt concentrations in a cell free system26. Thus, one function of these condensates may be to sequester RNAs of different types and prevent irreversible aggregation. Indeed, recent work on the conserved stress granule protein G3BP1 provides evidence that it may have a role in limiting entanglement of long RNAs124. It is likely that another function of RNA condensates is to increase the rate of biochemical reactions involving RNA or the condensates RNAs help seed, by locally increasing the concentration of reaction components. This has been most easily ascribed to condensates that perform a specific catalytic functions on target RNAs such as transcription125, splicing in the spliceosome (not to be confused with nuclear speckles, which function to store and modify splicing factors)126, ribosome assembly in the nucleolus127, mRNA polyadenylation128, and mRNA decay in P bodies129. Condensates can also concentrate essential non-condensate components 130, 131 and regulate and localize translation132. Emergent RNA-encoded biophysical properties are essential for sorting RNAs to distinct compartments in some cases5. Further work is needed to understand how condensate identity and sorting is achieved and maintained in the same cell compartment, given the liquid-like nature of many condensates.
Storage and inheritance of specific molecules
In germ cells, condensates can store RNA and proteins temporally or propagate condensate formation in daughter cells through multiple rounds of division. Germline granules, which have been reviewed extensively elsewhere, are an excellent example of storage of RNAs for propagation through cell divisions133, 134. Long-term storage is useful in cell types that do not frequently divide, as frequently dividing cells may rapidly dilute condensate contents. The influence of cellular lifespan on condensate lifespan is of particular interest given the wide variety of cellular lifespans that exist in different cell types in the same organism, from neurons that persist for decades to neutrophils, which have a half-life of only 6–8 hours. In long-lived cells, condensates were proposed to act as a ‘wastebasket’ for storage of problematic proteins and RNAs135.
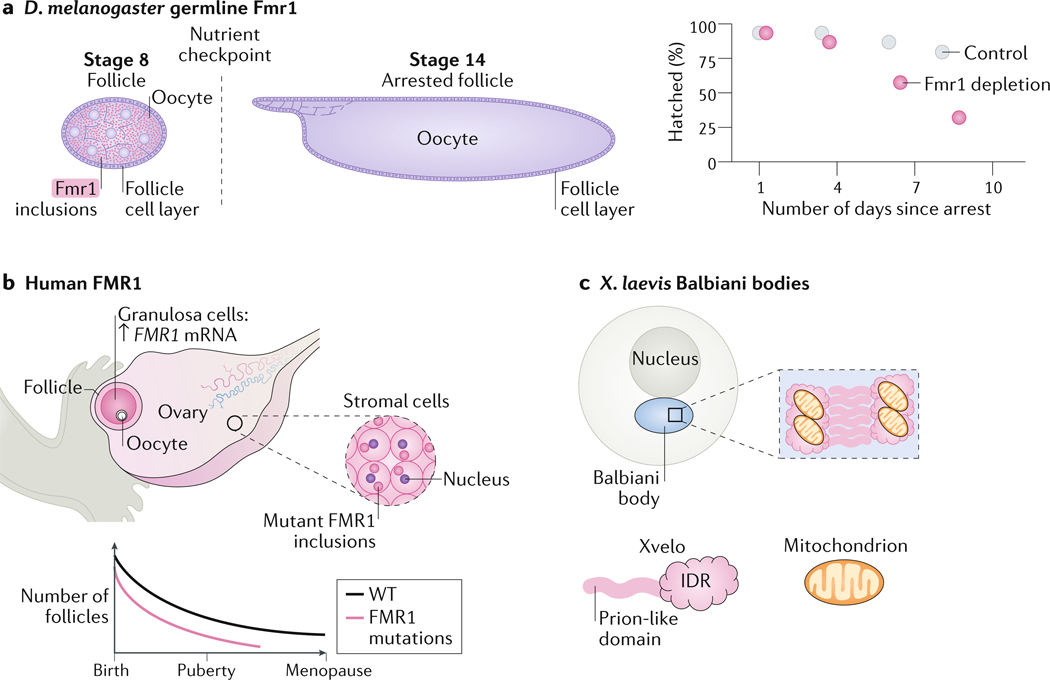
The role of condensates in long-term storage within a single cell has been most commonly observed in oocytes. In mammals, oogenesis is completed during development and the oocytes must be stored until puberty and fertilization, which can take decades. Maternal RNA must be preserved during this time and until the activation of zygotic gene expression following fertilization. One way to preserve the maternal RNA is through storage in condensates. The RBP FMR1 was proposed to perform this function in different organisms. In D. melanogaster oocytes, FMR1 may form condensates consisting of long maternal RNA, and loss of FMR1 reduces the functionality lifetime of oocytes136 (Figure 6A). Similarly, in humans, FMR1 mutations, which cause fragile X syndrome are associated the formation of FMR1 inclusions and with infertility in women137, 138, suggesting that RNA storage in oocytes is a conserved regulatory mechanism (Figure 6B). Finally, in X. laevis, the IDR- and prion-domain-containing protein Xvelo (also known as velo1) is crucial for the formation of the Balbiani body (a condensate containing proteins, RNA, mitochondria and other organelles) in the oocyte through assembly of an amyloid–like network with RNA and mitochondria139 (Figure 6C). What governs the dissolution of long-term storage condensates is still poorly understood, but some examples include the disruption of interactions between condensate components140, modification of the scaffolding RBP141–143, and engagement of a chaperone144.
Figure 6: Oocytes may use condensates for long-term RNA storage.
(A) Loss of fragile X mental retardation protein 1 (Fmr1) in Drosophila melanogaster oocytes accelerates the reduction in hatch rate in arrested oocytes over the fly life time. Fmr1 is expressed and forms condensates in the cytoplasm of stage 8 oocytes. Starvation of female flies leads to arrest of oocyte development at stage 14. Fmr1 depletion leads to a reduction in hatch rate over time.
(B) In humans, mutations in FMR1 that cause fragile X syndrome are associated with female infertility. The FMR1 protein forms inclusions in ovarian stromal cells. The number of follicles in women with fragile X syndrome decreases overtime more than in healthy women, leading to premature ovarian failure.
(C) In Xenopus laevis oocytes, the protein Xvelo, which contains an intrinsically disordered region (IDR) and a prion-like domain is required for the formation of Balbiani bodies through assembly of an amyloid–like network with RNA and mitochondria.
Buffering noise and responding to stress
Condensate formation can help cells to rapidly respond to environmental cues and thus promote survival. The buffering capacity of condensates can serve two purposes: suppressing naturally occurring transcriptional and translational heterogeneity between cells, and responding to acute stress. Single cell RNA-seq and proteomics analyses have revealed that no two cells are exactly identical and that the transcriptional profile of the cell often has little to do with its protein expression profile145. How do cells retain their identity in conditions of transcriptional and translational noise? One mechanism may be through the formation of RNA–protein storage condensates146. If cellular condensates obey physical principals of LLPS, in the case of a simple two-component system, then above a critical concentration for demixing, the soluble concentration will remain constant while the volume fraction of the condensed phase continues to grow7. However it is important to note, that in cells, such a simple prediction is unlikely to be correct because of the compositional complexity of condensates147. In this way, condensate formation could ensure maintaining a constant concentration of soluble molecules despite fluctuations in their number. Alternatively, excess of cellular components could be stored in condensates, thereby allowing responding to cues more rapidly than can be achieved by changes in transcription and/or translation. Indeed, properties of a long-lived condensate can be altered rapidly following the recognition of a specific stimulus; for example, nucleic acid cues facilitate LLPS of the innate immunity factor cGAS and thus promote its activity148. Finally, condensates can be formed to deal with acute stress. For example, acute cellular stresses such as heat shock, trigger translation shutdown, release of mRNAs from the translation machinery and the formation of mRNA-containing stress granules, which sequester mRNA and proteins until the stress is resolved149, 150 (Figure 5D). P bodies are another type of RNA–protein condensate that forms in response to acute cellular stresses129. Condensates can also form from RBPs that are removed from RNA during stress, as in the case of polyadenylate-binding protein in budding yeast151.
Conclusion and future directions
The formation of membraneless, RNA-dependent condensates is a tightly regulated process that depends on RNA sequence (identity, length, modification), on various sequence encoded properties (RNA structure, RNA–RNA interactions, RNA–protein interactions) and on RNA and protein expression levels. Collectively, these RNA encoded features confer specific condensate biophysical properties, which are essential for condensate functions in homeostasis. The consequence of altering condensate biophysical properties can be loss of cell function and disease. Following a decade of research focus on the protein components of condensates, many exciting, unexplored avenues exist for future study and methodological advances. Although RNA is challenging to study, we hope to have inspired the reader to engage with the expansive potential of RNA in condensate biology.
Acknowledgments:
We thank Dr. Christiane Iserman for critical review of this manuscript. This work has been funded by: R01-GM081506 (NIH-NIGMS) (A.S.G.), HHMI Faculty Scholars program (A.S.G.), T32 CA 9156–43 (C.R.), F32 F32GM136164 (NIH-NIGMS) (C.R.) and the L’Oreal Women in Science Fellowship (C.R). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Glossary:
- P granules
presumptive germ cells granules; a non-membrane bound.
- Stress granules
a membraneless condensate consisting of RNA and protein that appears in cells in conditions of stress.
- G-quadruplex
a nucleic acid four-stranded structure formed by guanine-rich sequences.
- Paraspeckles
a membraneless condensate consisting of RNA and protein in interphase nuclei; scaffolded by the long non-coding RNA NEAT1.
- Microspeckles
a membraneless condensate consisting of RNA and protein in interphase nuclei; scaffolded by the long non-coding RNA NEAT1 splicing isoform 1–1.
- Speckles
A membraneless condensate in the nucleus that stores and modifies splicing factors.
- Cajal bodies
also known as coiled bodies. Membraneless nuclear bodies containing RNA and protein that are sites of post-transcriptional modification of small nuclear and nucleolar RNAs.
- PML bodies
membraneless bodies consisting of RNA and protein and scaffolded by the promyelocytic leukemia (PML) protein. May have roles in apoptosis, cell division and response to viral infection.
- Phase diagrams
a chart used to show the state of a mixture at equilibrium, depending on particular conditions; for biomolecular condensates, this state is often a function of concentration of mixture components such as protein and RNA.
- Condensed phase
Refers to solid or liquid condensates, which form by electrostatic interactions and de-mixing from solution.
- Dilute phase
The molecules remaining in solution, unincorporated into either solid or liquid condensates.
- Re-entrant phase behaviour
This behavior is seen when sufficiently high concentrations of a component, such as RNA, prevents condensation, in many cases due to charge repulsion effects.
- Intrinsically disordered regions
(IDRs) Sequences of RNA or protein that lack defined three-dimensional order or structure.
Footnotes
Conflict of Interest:
The authors declare no competing financial interests
Peer review information
Nature Reviews Molecular Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Pak CW et al. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin EW et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmon TS, Holehouse AS, Rosen MK & Pappu RV Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langdon EM et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ries RJ et al. m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbaum-Garfinkle S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 112, 7189–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H. et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell 60, 220–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maharana S. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milin AN & Deniz AA Reentrant Phase Transitions and Non-Equilibrium Dynamics in Membraneless Organelles. Biochemistry 57, 2470–2477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putnam A, Cassani M, Smith J. & Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat Struct Mol Biol 26, 220–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CS et al. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J. et al. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentze MW, Castello A, Schwarzl T. & Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19, 327–341 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Franzmann T. & Alberti S. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J Biol Chem (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitrea DM et al. Methods for Physical Characterization of Phase-Separated Bodies and Membrane-less Organelles. J Mol Biol 430, 4773–4805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa S, Naganuma T, Shioi G. & Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193, 31–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa S. et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141, 4618–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Standaert L. et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 20, 1844–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A, Behm-Ansmant I, Schweizer D. & Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 27, 3970–81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DD & Gurdon JB Absence of Ribosomal Rna Synthesis in the Anucleolate Mutant of Xenopus Laevis. Proc Natl Acad Sci U S A 51, 139–46 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin S. et al. Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. J Neurochem 125, 175–84 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Tucker KE et al. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 154, 293–307 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeynaems S. et al. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci U S A 116, 7889–7898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunde BM, Moore C. & Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8, 479–90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Treeck B. et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A 115, 2734–2739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schisa JA, Pitt JN & Priess JR Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128, 1287–98 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Trcek T. et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 6, 7962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler JR, Matheny T, Jain S, Abrisch R. & Parker R. Distinct stages in stress granule assembly and disassembly. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brangwynne CP, Mitchison TJ & Hyman AA Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 108, 4334–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler JR, Jain S, Khong A. & Parker R. Isolation of yeast and mammalian stress granule cores. Methods 126, 12–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubstenberger A. et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68, 144–157 e5 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Khong A. et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820 e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han TW et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–79 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Gellert M, Lipsett MN & Davies DR Helix formation by guanylic acid. Proc Natl Acad Sci U S A 48, 2013–8 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bang I. Untersuchungen über die guanylsäre. Biochemische Zeitschrift 26, 293–311 (1910). [Google Scholar]
- 37.Nilsen TW & Graveley BR Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–63 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Hsu PJ, Chen YS & Yang YG Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 28, 616–624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clemson CM et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33, 717–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R, Harvey AR, Hodgetts SI & Fox AH Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 23, 872–881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mito M, Kawaguchi T, Hirose T. & Nakagawa S. Simultaneous multicolor detection of RNA and proteins using super-resolution microscopy. Methods 98, 158–165 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Jain A. & Vale RD RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee YB et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5, 1178–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estan MC et al. Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy. Nat Commun 10, 797 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith JA, et al. FXR1 splicing is important for muscle development and biomolecular condensates in muscle cells. J. Cell Biol. 219, e201911129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roundtree IA, Evans ME, Pan T. & He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert WV, Bell TA & Schaening C. Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominissini D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–6 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Meyer KD & Jaffrey SR The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15, 313–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaccara S, Ries RJ & Jaffrey SR Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20, 608–624 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Zhuang YFX m6A-binding YTHDF proteins promote stress granule formation. Nature Chemical Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salter JD, Bennett RP & Smith HC The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem Sci 41, 578–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamura Y. et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet 73, 693–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rice GI et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44, 1243–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bansal H. et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 28, 1171–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonkhout N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gokhale NS & Horner SM RNA modifications go viral. PLoS Pathog 13, e1006188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solomon O. et al. RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat Commun 8, 1440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butcher SE & Pyle AM The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res 44, 1302–11 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Ganser LR, Kelly ML, Herschlag D. & Al-Hashimi HM The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Biol 20, 474–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siegfried NA, Busan S, Rice GM, Nelson JA & Weeks KM RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11, 959–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smola MJ & Weeks KM In-cell RNA structure probing with SHAPE-MaP. Nat Protoc 13, 1181–1195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Z, Gong J. & Zhang QC PARIS: Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution. Methods Mol Biol 1649, 59–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding Y, Chan CY & Lawrence CE Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res 32, W135–41 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding Y, Chan CY & Lawrence CE RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11, 1157–66 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorenz R. et al. ViennaRNA Package 2.0. Algorithms Mol Biol 6, 26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gruber AR, Lorenz R, Bernhart SH, Neubock R. & Hofacker IL The Vienna RNA websuite. Nucleic Acids Res 36, W70–4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alberti S. et al. A User’s Guide for Phase Separation Assays with Purified Proteins. J Mol Biol 430, 4806–4820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volkov V. Quantitative description of ion transport via plasma membrane of yeast and small cells. Front Plant Sci 6, 425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharyya D, Mirihana Arachchilage G. & Basu S. Metal Cations in G-Quadruplex Folding and Stability. Front Chem 4, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y. et al. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res 47, 11746–11754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strulson CA, Boyer JA, Whitman EE & Bevilacqua PC Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA 20, 331–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamagami R, Bingaman JL, Frankel EA & Bevilacqua PC Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nat Commun 9, 2149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denesyuk NA & Thirumalai D. Crowding promotes the switch from hairpin to pseudoknot conformation in human telomerase RNA. J Am Chem Soc 133, 11858–61 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Dupuis NF, Holmstrom ED & Nesbitt DJ Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc Natl Acad Sci U S A 111, 8464–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kilburn D, Roh JH, Guo L, Briber RM & Woodson SA Molecular crowding stabilizes folded RNA structure by the excluded volume effect. J Am Chem Soc 132, 8690–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakano S, Karimata HT, Kitagawa Y. & Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc 131, 16881–8 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Lee HT, Kilburn D, Behrouzi R, Briber RM & Woodson SA Molecular crowding overcomes the destabilizing effects of mutations in a bacterial ribozyme. Nucleic Acids Res 43, 1170–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernhardt HS & Tate WP Primordial soup or vinaigrette: did the RNA world evolve at acidic pH? Biol Direct 7, 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mariani A, Bonfio C, Johnson CM & Sutherland JD pH-Driven RNA Strand Separation under Prebiotically Plausible Conditions. Biochemistry 57, 6382–6386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smola MJ, Rice GM, Busan S, Siegfried NA & Weeks KM Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10, 1643–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Munder MC et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee B. et al. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23, 169–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rouskin S, Zubradt M, Washietl S, Kellis M. & Weissman JS Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun L. et al. RNA structure maps across mammalian cellular compartments. Nat Struct Mol Biol 26, 322–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cordin O, Banroques J, Tanner NK & Linder P. The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Rajkowitsch L. et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4, 118–30 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Hondele M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim Y. & Myong S. RNA Remodeling Activity of DEAD Box Proteins Tuned by Protein Concentration, RNA Length, and ATP. Mol Cell 63, 865–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buratti E. & Baralle FE Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 24, 10505–14 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Treeck B. & Parker R. Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trcek T, et al. Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters. Molecular Cell 10.1016/j.molcel.2020.05.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma E, Sterne-Weiler T, O’Hanlon D. & Blencowe BJ Global Mapping of Human RNA-RNA Interactions. Mol Cell 62, 618–26 (2016). [DOI] [PubMed] [Google Scholar]
- 95.Engreitz JM et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mustoe AM, Lama NN, Irving PS, Olson SW & Weeks KM RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc Natl Acad Sci U S A 116, 24574–24582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rehmsmeier M, Steffen P, Hochsmann M. & Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–17 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berry J, Weber SC, Vaidya N, Haataja M. & Brangwynne CP RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 112, E5237–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shelkovnikova TA, Robinson HK, Southcombe JA, Ninkina N. & Buchman VL Multistep process of FUS aggregation in the cell cytoplasm involves RNA-dependent and RNA-independent mechanisms. Hum Mol Genet 23, 5211–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murakami T. et al. ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88, 678–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin Y, Protter DS, Rosen MK & Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molliex A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patel A. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–77 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Kiledjian M. & Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J 11, 2655–64 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamazaki T. et al. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol Cell 70, 1038–1053 e7 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Lee C, Occhipinti P. & Gladfelter AS PolyQ-dependent RNA-protein assemblies control symmetry breaking. J Cell Biol 208, 533–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee C. et al. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell 25, 572–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heraud-Farlow JE & Kiebler MA The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci 37, 470–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Langdon EM & Gladfelter AS A New Lens for RNA Localization: Liquid-Liquid Phase Separation. Annu Rev Microbiol 72, 255–271 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Ferrandon D, Elphick L, Nusslein-Volhard C. & St Johnston D. Staufen protein associates with the 3’UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–32 (1994). [DOI] [PubMed] [Google Scholar]
- 111.Liao YC et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 179, 147–164 e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kanai Y, Dohmae N. & Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–25 (2004). [DOI] [PubMed] [Google Scholar]
- 113.Kiebler MA & Bassell GJ Neuronal RNA granules: movers and makers. Neuron 51, 685–90 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Conlon EG & Manley JL RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev 31, 1509–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Becht P, Konig J. & Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci 119, 4964–73 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Zarnack K. & Feldbrugge M. mRNA trafficking in fungi. Mol Genet Genomics 278, 347–59 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Konig J. et al. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J 28, 1855–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baumann S, Pohlmann T, Jungbluth M, Brachmann A. & Feldbrugge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci 125, 2740–52 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Baumann S, Konig J, Koepke J. & Feldbrugge M. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15, 94–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vogler TO et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Orr-Weaver TL When bigger is better: the role of polyploidy in organogenesis. Trends Genet 31, 307–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jordina Guillé n-Boixet AK, Holehouse Alex S., Pappu Rohit V., & Simon Alberti TMF RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 18, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu H. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Galganski L, Urbanek MO. & Krzyzosiak WJ Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45, 10350–10368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Falahati H, Pelham-Webb B, Blythe S. & Wieschaus E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol 26, 277–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fang X. et al. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo Y, Na Z. & Slavoff SA P-Bodies: Composition, Properties, and Functions. Biochemistry 57, 2424–2431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nott TJ et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saha S. et al. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell 166, 1572–1584 e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tsang B. et al. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci U S A (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang JT & Seydoux G. P granules. Curr Biol 24, R637–R638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seydoux G. & Braun RE Pathway to totipotency: lessons from germ cells. Cell 127, 891–904 (2006). [DOI] [PubMed] [Google Scholar]
- 135.Congdon EE & Duff KE Is tau aggregation toxic or protective? J Alzheimers Dis 14, 453–7 (2008). [DOI] [PubMed] [Google Scholar]
- 136.Greenblatt EJ & Spradling AC Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 361, 709–712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marozzi A. et al. Association between idiopathic premature ovarian failure and fragile X premutation. Hum Reprod 15, 197–202 (2000). [DOI] [PubMed] [Google Scholar]
- 138.Sullivan AK et al. Association of FMR1 repeat size with ovarian dysfunction. Hum Reprod 20, 402–12 (2005). [DOI] [PubMed] [Google Scholar]
- 139.Boke E. et al. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 166, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mugler CF et al. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rai AK, Chen JX, Selbach M. & Pelkmans L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 (2018). [DOI] [PubMed] [Google Scholar]
- 142.Wang JT et al. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wippich F. et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013). [DOI] [PubMed] [Google Scholar]
- 144.Walters RW, Muhlrad D, Garcia J. & Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21, 1660–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Y, Beyer A. & Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 165, 535–50 (2016). [DOI] [PubMed] [Google Scholar]
- 146.Klosin A. et al. Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468 (2020). [DOI] [PubMed] [Google Scholar]
- 147.Riback JA et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Du M. & Chen ZJ DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Protter DSW & Parker R. Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Decker CJ & Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4, a012286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Riback JA et al. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 168, 1028–1040 e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]