Abstract
Chest CT is used in the COVID-19 diagnosis process as a significant complement to the reverse transcription polymerase chain reaction (RT–PCR) technique. However, it has several drawbacks, including long disinfection and ventilation times, excessive radiation effects, and high costs. While X-ray radiography is more useful for detecting COVID-19, it is insensitive to the early stages of the disease. We have developed inference engines that will turn X-ray machines into powerful diagnostic tools by using deep learning technology to detect COVID-19. We named these engines COV19-CNNet and COV19-ResNet. The former is based on convolutional neural network architecture; the latter is on residual neural network (ResNet) architecture. This research is a retrospective study. The database consists of 210 COVID-19, 350 viral pneumonia, and 350 normal (healthy) chest X-ray (CXR) images that were created using two different data sources. This study was focused on the problem of multi-class classification (COVID-19, viral pneumonia, and normal), which is a rather difficult task for the diagnosis of COVID-19. The classification accuracy levels for COV19-ResNet and COV19-CNNet were 97.61% and 94.28%, respectively. The inference engines were developed from scratch using new and special deep neural networks without pre-trained models, unlike other studies in the field. These powerful diagnostic engines allow for the early detection of COVID-19 as well as distinguish it from viral pneumonia with similar radiological appearances. Thus, they can help in fast recovery at the early stages, prevent the COVID-19 outbreak from spreading, and contribute to reducing pressure on health-care systems worldwide.
Keywords: Novel coronavirus, SARS-CoV-2, Pneumonia, CXR radiographs, Residual network, Convolutional neural network
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), officially known as COVID-19, is a severe acute respiratory syndrome disease [1, 2]. It belongs to the same family as the Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV. The occurrence of this disease was first reported in Wuhan, China, in December 2019. It quickly spread to other regions in China and then to other countries.
The estimates on the prevalence of COVID-19 have been continuously updated since the initial outbreak. As of August 24 2020, the number of confirmed cases was 23,279,683 globally, and the number of confirmed deaths was 805,902. At the time of writing this paper, the five countries with the highest number of cases are, in order, the USA (5,612,163), Brazil (3,582,362), India (3,106,348), the Russian Federation (956,749), and South Africa (607,045) [3].
The RT-PCR test is not reliable enough for the early diagnosis and treatment of this disease [4, 5]. Additionally, a RT-PCR test takes approximately 6 h to complete. Considering this, the CXR film is relevant for COVID-19 diagnosis and identification. If the RT-PCR or CXR result appears normal but the clinical suspicion is still high, then a chest CT scan must be conducted.
CT helps detect possible complications that are not seen in CXR [6]. Radiologists play a vital role in this process by accurately reporting their radiological findings from both the CXR and CT scans. These reports provide important information on the absence or presence of a coronavirus infection. Findings that cause serious suspicion relevant to “COVID-19 are ground glass patterned areas, which, even in the initial stages, affect both lungs, in particular the lower lobes, and especially the posterior segments, with a fundamentally peripheral and subpleural distribution” [7].
COVID-19 and other pneumonia types have similar imaging features, which makes it difficult to differentiate them. Although CT is an effective imaging method for the early detection and diagnosis of COVID-19, its implementation poses several difficulties and disadvantages. If the process is not conducted in a safe manner by considering the rate and form of the spread of COVID-19, the risk of transmitting the disease to health workers and other patients increases. For this reason, the American College of Radiology (ACR) has released recommendations to be followed when performing imaging for suspected COVID-19 infection. The ACR statement states the following: “CT should be used sparingly and reserved for hospitalized, symptomatic patients with specific clinical indications for CT. Appropriate infection control procedures should be followed before scanning subsequent patients. Facilities may consider deploying portable radiography units in ambulatory care facilities for use when chest X-rays are considered medically necessary. The surfaces of these machines can be easily cleaned, avoiding the need to bring patients into radiography rooms.” [8].
Additionally, the average effective dose of radiation is 0.1 mSv for CXR, but this quantity is 70 times more for a chest CT. Such a high level of radiation has a significant negative effect on DNA, causing mutations that may lead to cancer [9].
On the other hand, the cost of chest CT scans is higher than that of CXRs. Considering the budgets that countries have allocated for fighting COVID-19, the imaging cost for CT scans for every suspected patient results in notable expenses.
Artificial intelligence (AI) technologies produce successful results, especially in the differentiation and diagnosis of diseases with similar symptoms and uncertainty. Deep learning (DL) involves neural network models learning to make classifications based on images or different types of data sources. DL models have promising potential for prognostic and diagnostic accuracy due to their competency in comparison to human experts. Deep neural networks use labels to discover hidden patterns in inputted complex data [10]. This technology can be used to develop real-time systems that detect different types of diseases [11, 12]. Using a pre-trained model for a new problem is called transfer learning. There are many pre-trained models in DL that are used to solve various problems by making simple modifications. Xception, VGG16, VGG19, ResNet, ResNetV2, InceptionV3, InceptionResNetV2, MobileNet, MobileNetV2, DenseNet, and NASNet are well-known ImageNet pre-trained models. There are a limited number of DL studies on the detection of COVID-19 from CXR images, and models pre-trained using natural images have been used in these studies [6, 13–18].
Ayrton [15] used ResNet50-based deep transfer learning technique on a small dataset of 339 images (39 COVID-19 positive and 300 normal) and reached a validation accuracy of 96.2%. Further, Wang and Wong [18] proposed a DL model named COVID-Net for the detection of COVID-19. This model has 92.4% accuracy in classifying normal, pneumonia, and COVID-19 classes. Hemdan et al. [14] used 50 CXR images (25 COVID-19 and 25 normal) to build the COVIDX-Net framework, which includes seven different pre-trained models: VGG19, DenseNet121, InceptionV3, ResNetV2, Inception-ResNetV2, Xception, and MobileNetV2. The VGG19 and DenseNet models were each found to have an accuracy of 90%, while the Inceptionv3 model has the worst classification performance at an accuracy of 50%.
Asif et al. [19] proposed a CNN model based on Inceptionv3 with transfer learning for the detection of COVID-19. The researchers used a dataset that consists of 864 COVID-19, 1345 viral pneumonia, and 1341 normal chest X-ray images. Before using this dataset, they increased the number of images by 10 times for each class through data augmentation methods. They obtained a training accuracy of 97% and a validation accuracy of 93% after the completion of 4000 epochs in the proposed model.
ImageNet pre-trained models show insufficient performance for some tasks. Natural images and medical images differ significantly, and transfer learning models are mainly trained using datasets that contain natural images. Therefore, these models do not perform well when medical images are used as the dataset. Hence, transfer learning cannot be considered more efficient than training a network from scratch, as the networks learn different high-level features [20]. Besides, using a pre-trained model in DL applications is quite simple; it merely requires writing a few lines of code. In contrast, creating a model from scratch requires expert knowledge and experience in the DL field.
Unlike the abovementioned researchers, we developed two deep neural networks from scratch: COV19-CNNet based on CNN and COV19-ResNet based on ResNet. The number of required hidden layers is a significant aspect to be considered for obtaining good results. It changes based on the complexity of the dataset and the problem. Therefore, the number of hidden layers must be well-defined in order to successfully train a model. We attempted to find an optimal number of hidden layers to solve the COVID-19 detection problem. We used the Python programming language to create our deep neural networks and the computation power of a Google Colaboratory Linux server with an Ubuntu 16.04 operating system a Tesla K80 GPU graphics card to train them. The networks were trained with a dataset that contained only CXR images (210 COVID-19, 350 viral pneumonia, and 350 normal). The majority of previously conducted studies given in the literature have made use of limited datasets containing less than 100 COVID-19 images. Due to this, we decided to employ a larger dataset to develop deep neural networks which demonstrate reliable performance. Our inference engine, COV19-ResNet, reached an accuracy of 100%, a precision rate of 100%, a recall rate of 100%, a specification of 100%, and an F1 score of 100% for the detection of COVID-19. However, the accuracy values obtained for normal and viral pneumonia classes were 95.71% and 97.14%, respectively.
Methods
Patient and Dataset
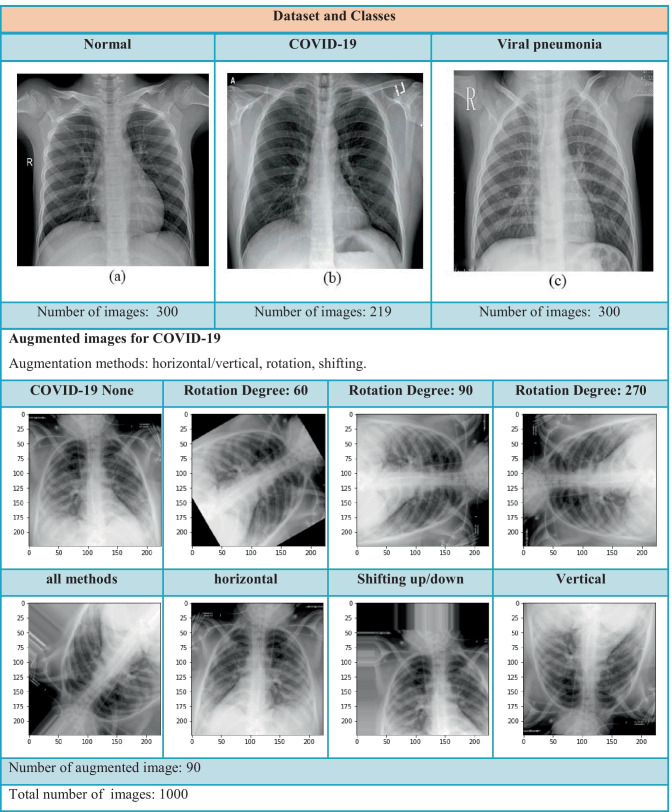
The present study was a retrospective one. The database used consists of 210 COVID-19, 350 viral pneumonia, and 350 normal CXR images that were created using two different data sources. Figure 1 presents the sample CXR images for each class. The first data source was created by a team of researchers from Qatar University (Doha, Qatar) and Dhaka University (Dhaka, Bangladesh) along with collaborators from Pakistan and Malaysia. The dataset consists of 219 COVID-19 positive, 1341 normal, and 1345 viral pneumonia CXR images, and it is constantly updated with new CXR images [21].
Fig. 1.
Dataset and classes
The second data source consists of 5863 CXR images (pneumonia and normal) selected from retrospective cohorts of patients from the Guangzhou Women and Children’s Medical Center [22].
Our dataset initially consisted of 910 X-ray images (PNG format), and we produced 90 additional COVID-19 samples using data augmentation methods such as horizontal/vertical, rotation, and shifting. The well-known 60/20/20 rule, which is used as a common strategy to split data in machine learning, was enforced in our study; 60% of the dataset was assigned for training, 20% for validation, and the final 20% for testing.
COV19-CNNet
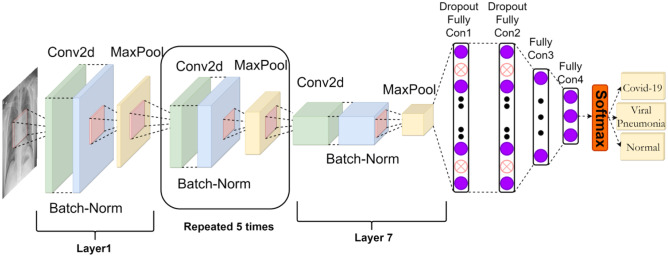
A CNN is a special type of neural network that has shown excellent performance in computer vision competitions. This network gains the ability to recognize different objects after training on a big image dataset. This architecture works in a similar manner to human vision—both use correlations between different images. Some diseases that are difficult to detect by experts can be detected in seconds by the systems created with this algorithm. However, considerable skill and experience in DL is required to select suitable hyperparameters, such as the learning rate, kernel sizes of convolutional filters, and number of layers, when creating a CNN from scratch for a new task. In this study, we developed a CNN-based interference engine called COV19-CNNet, which was built from scratch to detect COVID-19, using our expertise in and experiences with DL. It consists of seven convolutional layers for feature extraction and four dense layers for classification (Fig. 2).
Fig. 2.
COV19-CNNet architecture
The initial convolutional layers of the network tends to respond to basic features such as oriented lines and textures, whereas intermediate layers of the network respond to more complex features. By the time information reaches the higher layers of the network, the feature maps of these layers are able to extract specific findings.
Having multi-convolutional layers in the network makes it possible to extract useful information from images. The large number of parameters required is one of the biggest problems in training a deep convolutional network. This problem was solved for this model by using a pooling operation. Outputs of the convolutional layers were summarized using the 2D max-pooling operation. Batch normalization was used to rescale outputs of some particle layers.
A flatten layer was used in the transition from the convolution layers to the four dense layers. The dense layers which are also known as fully connected layers help learn non-linear combinations of the high-level features outputted by the convolutional layers. The last dense layer has three neurons to categorize images as COVID-19, viral pneumonia, and normal. It uses Softmax as an activation function.
We used specific strategies in the training phase to prevent overfitting: In each step of the training process, random unit sets in the dense layers were selected and ignored using the dropout technique. This is an efficient strategy that makes it possible to train various networks using different data units. Thus, if any part of the network becomes too sensitive to noise in the data, the other parts will compensate for it. The early stopping method was also used to prevent overfitting. Through this, the training of the model ceases when performance improvement is stopped on the validation dataset.
COV19-ResNet
Deep neural networks are useful for dealing with complex tasks. However, adding too many layers generally causes vanishing gradient problems, and the weights of the early layers cannot be updated by backpropagation. This can be solved by adding an identity connection. The structure of ResNet allows the gradients to flow backward directly through an identity connection from the later layers to the initial filters. ResNet is a continuation of deep networks. It improves CNN architecture with residual learning and offers an effective methodology for the training of deep networks.
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
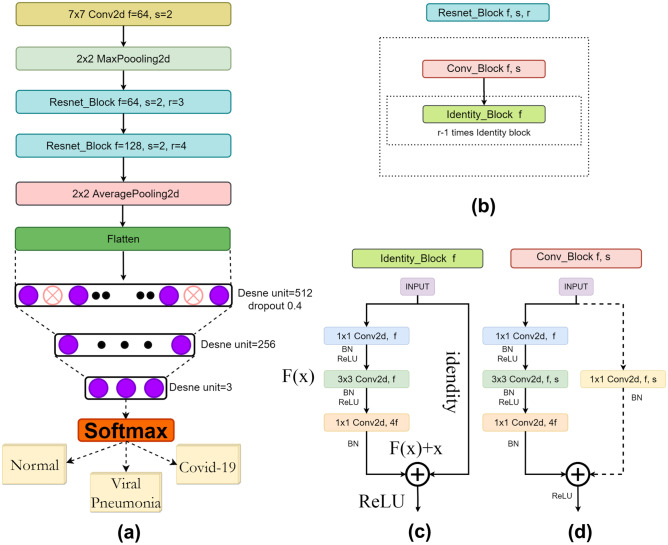
The given mathematical formula depicts the main idea behind the ResNet structure (see Fig. 3c). Function F(x) transforms the input data and the function “Identity,” which allows a direct connection from the previous layer to the current layer. X is the input, W is the weight, and B is the bias vector.
Fig. 3.
a COV19-ResNet architecture, b Resnet_Block, c Identity_block, and d Conv_Block
In this research, we developed a deep neural network from scratch entitled COV19-ResNet based on ResNet architecture (Fig. 3a). It takes an image (224 × 224 × 1) as the input. The network initially performs 7 × 7 convolution and 2 × 2 max-pooling functions. After these operations, two “resnet_block” types (Fig. 3b) are used: the first “resnet_block” contains one “conv_block” and three “identity_block,” while the second one contains one “conv_block” and three “identity_block.” An “identity_block” has three layers and one identity connection (shown as a solid arrow, shown in Fig. 3c). These consist of 1 × 1, 3 × 3, and 1 × 1 convolutions. A “conv_block” (Fig. 3d) has three convolution layers stacked one over the other and one downsampling layer.
These three convolution layers are of 1 × 1, 3 × 3, and 1 × 1 kernel sizes. The stride parameter of the 3 × 3 convolution layer is set as 2. This layer is responsible for reductions in the input size. The downsampling layer consists of 1 × 1 convolution and stride 2 to prevent mismatching in size. Finally, the average pooling layer, a flatten layer, and three fully connected layers are used respectively. The detailed structure of the COV19-ResNet is presented in Fig. 3a.
Results
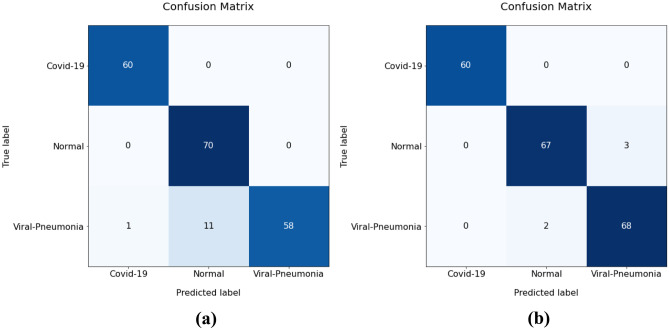
Our dataset consisted of 1000 CXR images. We organized the dataset into three folders (60% training, 20% test, 20% validation) containing subfolders for each image category (300 COVID-19, 350 normal, and 350 viral pneumonia images). The confusion matrices of COV19-CNNet and COV19-ResNet were calculated to investigate their performances. As shown in Fig. 4a, the COVID-19 and normal classes achieved perfect classification with the use of COV19-CNNet, whereas the viral pneumonia class had 12 misclassified samples within the 70 test samples. On the other hand, as seen in Fig. 4b, COVID-19 achieved the perfect classification with the use of COV19-ResNet; the normal class had three misclassified samples and the viral pneumonia class had two misclassified samples within the 70 test samples. According to these confusion matrixes, the accuracy performances for COV19-ResNet and COV19-CNNet are 97.61% and 94.28%, respectively.
Fig. 4.
Confusion matrix of a COV19-CNNet and b COV19-ResNet
The most common measures of performance in diagnosis are accuracy, precision, recall, specificity, and F1 score [23]. Table 1 shows a comparison of our two inference engines in terms of these performance measures. The formulas of the performance measures are as follows:
Table 1.
The performance measures of COV19-CNNet and COV19-ResNet on test data
| Class | Accuracy | Precision | Recall/Sensitivity | Specificity | F1-score | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M1 | M2 | M1 | M2 | M1 | M2 | M1 | M2 | |
| COVID-19 | 100.00 | 100.00 | 98.36 | 100.00 | 100.00 | 100.00 | 99.28 | 100.00 | 99.17 | 100.00 |
| Normal | 100.00 | 95.71 | 86.42 | 97.10 | 100.00 | 95.71 | 91.53 | 98.46 | 92.72 | 96.40 |
| Viral Pneumonia | 82.86 | 97.14 | 100.00 | 95.80 | 83.00 | 97.14 | 100.00 | 97.69 | 90.71 | 96.47 |
| Average | 94.28 | 97.61 | 94.93 | 97.63 | 94.33 | 97.61 | 96.94 | 98.72 | 94.20 | 97.62 |
| M1: COV19-CNNet; M2: COV19-ResNet | ||||||||||
| 6 |
| 7 |
| 8 |
| 9 |
| 10 |
| 11 |
TP, TN, FN, and FP represent the number of true positives, true negatives, false negatives, and false positives, respectively.
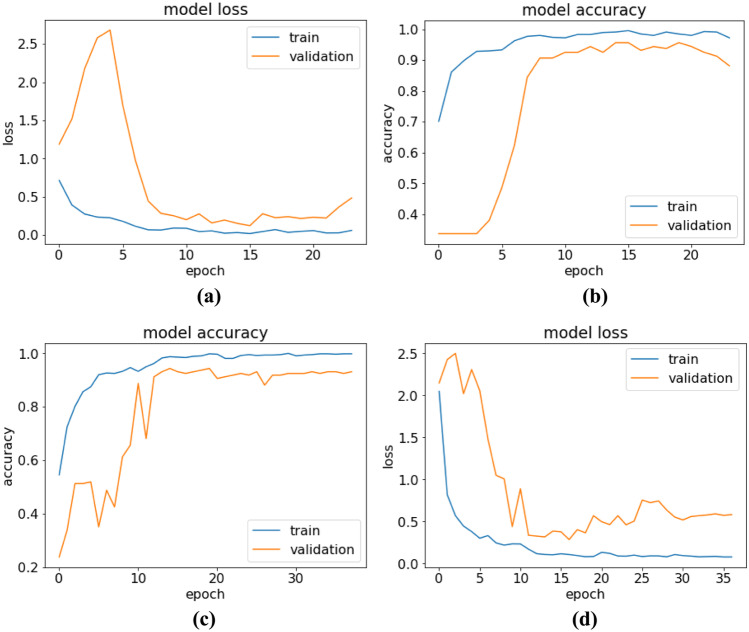
The training-validation accuracy and loss values of COV19-CNNet and COV19-ResNet in each epoch are shown in Fig. 5. In each model, the early stopping technique with different patience values and validation loss are monitored to prevent overfitting. During training, the best weights are stored. When the training phase is stopped, the best weights are loaded to the model.
Fig. 5.
Training-testing curves of accuracy and loss for a–b COV19-CNNet and c–d COV19-ResNet
Discussion
DL is an AI technology that has been applied successfully in the medical field due to its high capability of feature extraction from images [5, 24]. Some of these applications differentiate the types of viral pneumonia or detect the presence of pneumonia from CXR images [16, 25]. They are also used in the detection of breast cancer [26], brain tumor segmentation [27], brain abnormalities [28], and Alzheimer’s disease as well as in the diagnosis and classification of skin lesions [29, 30].
It is necessary to detect COVID-19 in suspected patients as early as possible and a proper treatment process initiated. CXR and CT scans are commonly used for detecting COVID-19 [6, 13, 31, 32]. However, according to the American College of Radiology (ACR), CT scans should not be used to screen or as the first-line imaging to diagnose COVID-19, and CT has several drawbacks, which were mentioned in the introduction section. Therefore, X-ray imaging is the preferred method for diagnosis, and it plays a vital role in clinical care and epidemiological researches [33]. X-ray imaging is a more accessible, faster, cheaper, easier to disinfect, more portable, and less harmful method than CT, but the sensitivity of CXR is lower than that of CT [33]. In the early stages of COVID-19, medical images usually show negative radiological signs. However, it is seen that DL makes a huge contribution to COVID-19 detection by removing this disadvantage when the studies in Table 2 are examined carefully. The accuracy performance in the binary classification task for normal and COVID-19 samples was quite satisfactory. However, pneumonia caused by COVID-19 is actually a subset of pneumonia; the two have similar features and are difficult to differentiate from each other. Thus, the success of the classifier in the multi-class classification problem is low as compared with the binary class.
Table 2.
DL applications on detecting COVID-19 from chest X-ray images
| Literature | Data set without augmentation | DL tool | Accuracy | Specificity | Sensitivity | F1 score |
|---|---|---|---|---|---|---|
| Loey et al. [16] |
3 Classes N/C/P 69/79/79 |
ALexnet | 85.19 | - | 85.19 | 85.19 |
| Googlenet | 81.48 | - | 81.48 | 81.46 | ||
| Resnet18 | 81.48 | - | 81.48 | 84.66 | ||
| Apostolopoulos and Mpesiana [13] |
3 classes Dataset_1 N/C/BP 504/224/700 |
VGG19 | 93.48 | 98.75 | 92.85 | |
| MobileNet (v2) | 92.85 | 97.09 | 99.10 | |||
| Inception | 92.85 | 99.70* | 12.94* | |||
| Xception | 92.85 | 99.99* | 0.08* | |||
| Inception ResNet v2 | 92.85 | 99.83* | 0.01* | |||
|
3 classes Dataset_2 N/C/BP+VP 504/224/714 |
MobileNet (v2) | 94.72 | 96.46 | 98.66 | - | |
| Ucar and Korkmaz [6] |
3 classes N/C/P 1583/76/4290 |
SqueezeNet with raw dataset | 76.37 | 79.93 | - | 98.25 |
|
3 classes N/C/P 1536/1536/1536 |
SqueezeNet with augmented dataset | 98.26 | 99.13 | - | 98.25 | |
| Ozturk et al. [17] |
3 Classes N/C/P 500/127/500 |
Darknet | 87.02 | 92.18 | 85.35 | 87 |
| Proposed study |
3 Classes N/C/VP 350/210/350 |
COV19-CNNet | 94.28 | 96.94 | 94.33 | 94.20 |
| COV19-ResNet | 97.61 | 98.72 | 97.61 | 97.62 | ||
|
N: Normal (healthy) C: COVID-19 NC: Non-COVID-19 |
P: Pneumonia VP: Viral pneumonia BP: Bacterial pneumonia |
|||||
Apostolopoulos and Mpesiana [13] used two different datasets. They used MobileNet (v2) and reached an accuracy rate of 97.40% for the binary class and 92.85% for multi-class. However, they encountered meaningless sensitivity and specificity values for Inception, Xception, and Inception ResNet v2 models because of imbalances in the data. These values are denoted using asterisks (*) in Table 2. In this study, we used only 90 augmented samples for overcoming the problem of imbalanced data. Thus, our inference engines have accuracy, sensitivity, specificity, and F1 score values of over 90%.
Loey et al. [16] used a limited number of X-ray images in their study. Their dataset consisted of four categories and a total of 307 images (79 COVID-19, 69 normal, and 79 pneumonia). They were used in the deep transfer learning CNN models such as Alexnet, ResNet18, and GoogLeNet. In addition to this, they utilized a Generative Adversarial Network (GAN) to overcome the overfitting problem. They increased the number of dataset images to 30 times than that of the original dataset. The number of images in the dataset reached 8100 images after using the GAN network. They tested these pre-trained models for a different number of categories. They used 10% from the original dataset in the testing phase and 20% of the total generated images in the validation phase. The validation and testing accuracies of all pre-trained models for binary classification were nearly 100%. ResNet18 was the best pre-trained model for three-class and four-class classification. In the three-class classification, its validation and test accuracy were 99.6% and 81.5%, respectively. In the four-class classification, its validation and test accuracy were 99.6% and 66.7%, respectively. However, significant differences between the test and validation accuracies were observed; the validation accuracies were found to be higher than the test accuracies. Thus, it should be kept in mind that the data used in this study is highly augmented and the validation samples were composed of the same. In our study, we augmented COVID-19 images to be only 0.5 times larger than the raw images to balance the three categories. However, we used only raw images during the testing phase. We found the validation (94%) and test accuracy (94%) values of the COV19-CNNet model to be close to each other. Likewise, the COV19-ResNet model was also found to have close values of validation (97%) and test accuracy (98%).
Ozturk et al. [17] used the Darknet classifier, which forms a real-time object detection system named YOLO (you only look once) [34]. Their proposed model was evaluated using the five-fold cross-validation procedure for both the binary and multi-classification problem. The Dark-CovidNet model achieved accuracy levels of 98.08% and 87.02% for binary and multi-classification, respectively.
Ucar and Korkmaz [6] used the pre-trained SqueezeNet model for the diagnosis of COVID-19. They used the dataset (1583 normal, 4290 pneumonia, and 76 COVID-19 CXR images) in two different ways: raw and augmented. The offline augmentation model used by them enhanced the COVID-19 class approximately 20 times. Thus, they reached 1536 images for the COVID-19 class and tried to balance the dataset by fixing this number of samples for all classes. They used 80% of the dataset for training, 10% for validation, and 10% for testing. The classification accuracy levels obtained by them for the raw dataset and augmented data were 76.37% and 98.26%, respectively. Working with highly augmented data increased the accuracy value, but this value decreased in the actual dataset. 76 images were used in their study, whereas 210 COVID-19 CXR images were used in ours.
The SqueezeNet model [6] has an accuracy of 100%, a precision rate of 99.35%, a recall rate of 100%, a specification of 96.67%, and an F1 score of 99.67% for COVID-19, while the values of all these metrics are 100% in our COV19-ResNet engine. In addition, the accuracy values of the SqueezeNet model for normal and viral pneumonia are 98.04% and 96.73%, respectively. The COV19-ResNet accuracy values for normal and viral pneumonia are 95.71% and 97.14%, respectively. The COV19-CNNet accuracy levels for COVID-19, normal, and viral pneumonia are 100%, 100%, and 82.86%, respectively. In light of these findings, it is evident that our COV19-ResNet model performs better than the SqueezeNet model in classifying COVID-19 and viral pneumonia. Additionally, the best classifier for the normal class among the three models is our COV19-CNNet engine, which has an accuracy rate of 100%.
ImageNet pre-trained models are not sufficient for classifying medical images as they are trained using natural images. Medical and natural images are different in many aspects. Therefore, we developed inference engines with new and special deep neural networks that were trained from scratch. We focused on the problem of multi-class classification in this study, a rather difficult task for the diagnosis of COVID-19. Table 2 presents the performances of the pre-trained models [6, 13, 16, 17] and the diagnostic inference engines we developed for this difficult task. Different combinations were obtained from different datasets, and the data was augmented using special functions. We chose the samples that were close together in the classes to avoid hypersensitivity against a certain class. To this end, we increased the augmentation methods as well as the number of COVID-19 images (90 samples). In fact, it is more appropriate to use a large dataset for deep neural network training from scratch in the medical field. However, it is difficult to create large datasets, especially with respect to emerging diseases such as COVID-19. This makes training deep neural networks from scratch relatively more difficult than transfer learning. It should also be noted that, besides the abovementioned ones, there are several other studies on detecting COVID-19 using CXRs; however, they are not peer-reviewed articles. As a result, these studies have not been included in Table 2.
Conclusion
X-ray images can be used to develop an effective diagnostic tool with DL technologies for detecting COVID-19 and viral pneumonia. COV19-CNNet and COV19-ResNet were developed with special architectures from scratch without using a pre-trained DL model. These inference engines were trained using CXR images and created with an optimum number of hidden layers and parameters. Both engines can accurately detect COVID-19 cases, but COV19-ResNet has a higher accuracy rate.
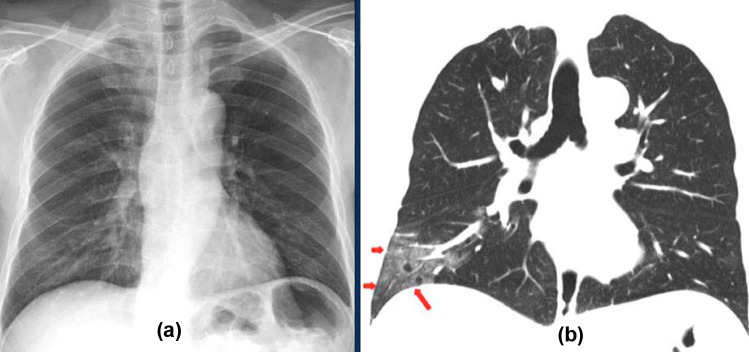
The disadvantage of the CXR imaging modality is that it shows negative radiological signs in the early stages of COVID-19. However, the inference engines we developed with DL technology can detect COVID-19 patients at an early stage. For instance, the CT and CXR images shown in Fig. 6 belong to a patient in the early stage of COVID-19 [35]. The ground-glass opacities in the right lower lobe of the CT image (red arrows) (Fig. 6b) are not visible in the CXR image (Fig. 6a). Nevertheless, using the CXR image, our diagnostic inference engines diagnosed this patient as having COVID-19 with 100% accuracy at an early stage. Early diagnosis makes a significant contribution to the treatment and quarantine process, ensuring that the primary treatment of chronic patients at high risk can be initiated. Thus, intensive care units and other health resources can be used in a more efficient manner.
Fig. 6.
Comparison of chest radiological images: a X-ray and b CT thorax images
Several studies have shown us that DL has the potential to be applied in the diagnosis and treatment of diseases. Considering this, DL models trained with large medical datasets need to be developed in the future. These models can then be used to solve various problems by being integrated successfully into many devices, including medical imaging tools.
This study, alongside other studies in this area, indicates that AI-aided research can make sizeable contributions to fight the coronavirus outbreak. All data about COVID-19 should be shared without any restrictions around the world so that this information can be actively used in similar studies. Additionally, the developed AI-aided systems need to be tested in health institutions. However, it should not be overlooked that doctors show resistance regarding the use of AI-aided systems in the diagnosis and treatment process. It is necessary to emphasize that these systems are powerful tools that can support doctors in the diagnosis and treatment of diseases. We developed and tested two diagnostic inference engines based on CNN and the architecture of ResNet in this study. We plan to develop a radiological system equipped with comprehensive features and use these intelligent inference engines as a part of it.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
There is no source of funding for this article.
Contributor Information
Ayturk Keles, Email: ayturkk@hotmail.com.
Mustafa Berk Keles, Email: musberkk@gmail.com.
Ali Keles, Email: alakmus@hotmail.com.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;6736(20):1–10. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO coronavirus disease (COVID-19) dashboard. 2020. https://covid19.who.int. Accessed 24 Aug 2020.
- 4.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung NC, Mayo JR, et al. A guideline for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society. Radiology. 2017;284(1):228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 5.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020;295(3):715–772. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ucar, F, Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images Med Hypotheses 2020 10.1016/j.mehy.2020.109761 [DOI] [PMC free article] [PubMed]
- 7.Imaging the coronavirus disease COVID-19. 2020. https://healthcare-in-europe.com/en/news/imaging-the-coronavirus-disease-covid-19.html. Accessed 20 Aug 2020.
- 8.ACR releases CT and chest X-ray guidance amid COVID-19 pandemic. 2020. https://www.diagnosticimaging.com/view/acr-releases-ct-and-chest-x-ray-guidance-amid-covid-19-pandemic. Accessed 20 May 2020.
- 9.Mettler FA, Hunda W, Yoshizumi TT, Mahesh M. Effective doses in radiology and diagnostic nuclear medicine: a catalog. Radiology. 2008;248:254–263. doi: 10.1148/radiol.2481071451. [DOI] [PubMed] [Google Scholar]
- 10.Riordon J, Sovilj D, Sanner S, Sinton D, Young EWK. Deep learning with microfluidics for biotechnology. Trends Biotechnol. 2019;37(3):310–324. doi: 10.1016/j.tibtech.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik. 2019;29(2):102–127. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Maier A, Syben C, Lasser T, Riess CA. A gentle introduction to deep learning in medical image processing. Zeitschrift für Medizinische Physik. 2019;29(2):86–101. doi: 10.1016/j.zemedi.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Apostolopoulos ID, Bessiana T. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020 doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemdan EED, Shouman MA, Karar ME. COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. https://arxiv.org/abs/2003.11055 Accessed 26 May 2020.
- 15.Joaquin AS. Using deep learning to detect pneumonia caused by NCOV-19 from X-ray images. 2020 https://towardsdatascience.com/using-deep-learning-to-detect-ncov-19-from-x-ray-images-1a89701d1acd. Accessed 26 May 2020.
- 16.Loey M, Smarandache F, Khalifa MNE. Within the lack of COVID-19 benchmark dataset: a novel GAN with deep transfer learning for corona-virus detection in CXR images. Symmetry. 2020;12(4):651–669. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 17.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. 2020 https://arxiv.org/abs/2003.09871. Accessed 24 May 2020. [DOI] [PMC free article] [PubMed]
- 19.Asif S, Wenhui Y, Jin H, Tao Y, Jinhai S. Classification of COVID-19 from chest X-ray CXR images using deep convolutional neural networks. 2020 https://www.medrxiv.org/content/10.1101/2020.05.01.20088211v2. Accessed 30 May 2020.
- 20.He K, Girshick R, Dollar P. Rethinking ImageNet pre-training, 2019. In IEEE/CVF International Conference on Computer Vision (ICCV). Seoul, South Korea. 2019 10.1109/ICCV.2019.00502.
- 21.Rahman T. COVID-19 radiography database (Winner of the COVID-19 Dataset Award by Kaggle Community) 2020. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 10 March 2020.
- 22.Labeled optical coherence tomography (OCT) and chest X-ray images for classification. 2018. https://data.mendeley.com/datasets/rscbjbr9sj/2. Accessed 15 March 2020.
- 23.Goutte C, Gaussier E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In: Losada DE, Fernández-Luna JM, editors. Advances in information retrieval. ECIR 2005. Lecture Notes in Computer Science (3408). Heidelberg: Springer, Berlin. pp. 345–359.
- 24.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation, In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Lecture Notes in Computer Science, Springer, Cham; 2015;9351; 234-241. 10.1007/978-3-319-24574-4_28
- 25.O Stephen, M Sain, UJ Maduh, DU Jeong, An efficient deep learning approach to pneumonia classification in healthcare. J Healthc Eng. 2019 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed]
- 26.Ragab DA, Sharkas SM, Ren J. Breast cancer detection using deep convolutional neural networks and support vector machines. Peer J. 2019;7:e6201. doi: 10.7717/peerj.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35(5):1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 28.Talo M, Baloglu UB, Yıldırım O, Acharya UR. Application of deep transfer learning for automated brain abnormality classification using MR images. Cogn Syst Res. 2019;1(54):176–88. doi: 10.1016/j.cogsys.2018.12.007. [DOI] [Google Scholar]
- 29.Ayan E, Ünver HM. Data augmentation importance for classification of skin lesions via deep learning. Paper presented at: Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT) 2018;1–4.
- 30.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernheim A, Huang XM, Yang Y, Fayad ZA, Diao NK, Li BXKS, et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3):685–691. doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanne JP. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology. 2020;295(1):16–17. doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas C, Mulholland EK, Carlin JB, Ostensen H, Amin R, Campo M, et al. Standardized interpretation of pediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ. 2005;83(5):353–359. [PMC free article] [PubMed] [Google Scholar]
- 34.Redmon J, Farhadi A. Yolo9000: better, faster, stronger. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, 2017, pp. 6517–6525, 10.1109/CVPR.2017.690.
- 35.Ng MY, Lee EY, Yang J, Yang F, Li X, Wang H, et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol Cardiothorac Imaging. 2020;2:e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]