Abstract
Objective
Nonalcoholic hepatic steatosis, also known as fatty liver, is a uniform response of the liver to hyperlipidic-hypercaloric diet intake. However, the post-ingestive signals and mechanistic processes driving hepatic steatosis are not well understood. Emerging data demonstrate that protein kinase C beta (PKCβ), a lipid-sensitive kinase, plays a critical role in energy metabolism and adaptation to environmental and nutritional stimuli. Despite its powerful effect on glucose and lipid metabolism, knowledge of the physiological roles of hepatic PKCβ in energy homeostasis is limited.
Methods
The floxed-PKCβ and hepatocyte-specific PKCβ-deficient mouse models were generated to study the in vivo role of hepatocyte PKCβ on diet-induced hepatic steatosis, lipid metabolism, and mitochondrial function.
Results
We report that hepatocyte-specific PKCβ deficiency protects mice from development of hepatic steatosis induced by high-fat diet, without affecting body weight gain. This protection is associated with attenuation of SREBP-1c transactivation and improved hepatic mitochondrial respiratory chain. Lipidomic analysis identified significant increases in the critical mitochondrial inner membrane lipid, cardiolipin, in PKCβ-deficient livers compared to control. Moreover, hepatocyte PKCβ deficiency had no significant effect on either hepatic or whole-body insulin sensitivity supporting dissociation between hepatic steatosis and insulin resistance.
Conclusions
The above data indicate that hepatocyte PKCβ is a key focus of dietary lipid perception and is essential for efficient storage of dietary lipids in liver largely through coordinating energy utilization and lipogenesis during post-prandial period. These results highlight the importance of hepatic PKCβ as a drug target for obesity-associated nonalcoholic hepatic steatosis.
Keywords: Dietary fats, Protein kinase cβ, Hepatic steatosis, Mitochondria respiratory chain
Abbreviations: AKT, protein kinase B; FCCP, trifluoromethoxy carbonyl cyanide phenylhydrazine; HFD, high-fat diet; IRS, insulin receptor substrate; MEK, mitogen-extracellular kinase; mTOR, mechanistic target of rapamycin; OCR, oxygen consumption rate; PKCβ, protein kinase Cβ; PKCβfl/fl, floxed PKCβ mice; PKCβHep−/−, hepatocyte-specific PKCβ-deficient mice; SREBP-1c, sterol response element-binding protein-1c; TG, triglycerides; VLDL, very low-density lipoproteins; WAT, adipose tissue
Highlights
-
•
Hepatocyte-specific PKCβ deficiency protects from diet-induced hepatic steatosis but not either obesity or insulin resistance.
-
•
The underlying mechanism involves modulation of mitochondrial function and lipogenesis.
-
•
Hepatocyte PKCβ does not regulate hepatic insulin signaling.
-
•
Our study provides a link between dietary lipid intake, hepatic protein kinase Cβ, modulation of hepatic lipid metabolism, and development of hepatic steatosis.
1. Introduction
Nonalcoholic fatty liver disease is the most common form of chronic liver disease worldwide [1]. It encompasses a wide spectrum of diseases associated with the over-accumulation of lipids in the liver, ranging from nonalcoholic hepatic steatosis (or nonalcoholic fatty liver) to steatohepatitis, advanced fibrosis, and cirrhosis [2,3]. Despite the high prevalence of hepatic steatosis, the molecular mechanisms of disease progression are not yet completely understood, which limits the development of efficient therapies to counteract this disease and the spectrum of progressive liver disorders. There is increasing interest in developing therapies for liver disorders by targeting factors that sustain hepatic steatosis [4].
It is widely recognized that hyperlipidemic-hypercaloric diets are now frequently consumed in modern societies. Several studies have supported that dietary lipids play important roles in the development of fatty liver disease in both animals and humans [5,6]. It has been hypothesized that the development of nonalcoholic fatty liver disease requires “two hits.” The first hit is the development of hepatic steatosis and the second hit includes cellular stresses such as oxidative stress and elevated levels of inflammatory cytokines in steatotic livers [7]. Hepatic lipid accumulation is proposed to be the root cause of the initiation and progression of nonalcoholic fatty liver disease [5]. Free fatty acids can either enter the mitochondria to undergo beta-oxidation or esterification into triglycerides (TG). These TG can then lead to the formation of lipid droplets in the liver or be secreted as very low-density lipoproteins. The rate at which fat accumulates in liver is determined by several factors, such as the rate of lipid uptake from the circulation and the utilization of lipids within the liver. However, the signaling links between dietary intake of lipids and downstream molecular mechanisms are not fully understood. In particular, diet-induced changes in hepatic signaling pathways by which dietary lipids promote the development and progression of fatty liver remain somewhat obscure. Defining the downstream signaling pathways underlying the control of hepatic and systemic metabolism by dietary lipids is necessary to understand both normal physiology and the pathogenesis of this disease.
The protein kinase C beta (PKCβ), a member of the serine/threonine protein kinase family, is activated by dietary metabolites diacylglycerol (an intermediate in TG biosynthesis and breakdown), cholesterol, phospholipids, hyperglycemia, and oxidative stress. PKCβ is implicated in the pathogenesis of obesity and the development of related metabolic disorders [8,9]. Whole-body PKCβ-deficient mice are protected against diet-induced obesity, hepatic steatosis, and insulin resistance by altering TG and cholesterol homeostasis [[10], [11], [12], [13]]. Moreover, PKCβ polymorphism is linked to insulin resistance in humans [14]. In addition, many in vitro studies have identified a number of substrates for PKCβ, indicating the potential for PKCβ to regulate a wide variety of proteins in various biological pathways relevant to the pathophysiology of nonalcoholic fatty liver disease. For example, PKCβ is reported to regulate phosphorylation and mitochondrial translocation of p66shc to negatively regulate autophagy [15,16]. PKCβ is also reported to phosphorylate retinoblastoma protein, IκB kinase, and nuclear receptors, including vitamin D receptor, farnesoid X receptor, and peroxisome-proliferator activated receptor γ [[17], [18], [19], [20], [21]]. In addition, PKCβ is reported to regulate macrophage differentiation and activation as well as insulin-induced hepatic sterol response element-binding protein-1c (SREBP-1c) expression [[22], [23], [24]]. Moreover, multiple cellular models suggest that PKCβ activation can either interfere with or is required for insulin signaling pathway [[25], [26], [27]]. PKCβ is shown to promote tumor necrosis factor α-induced cell death, which can account for the release of mitochondrial DNA in steatohepatitis [28,29]. Accumulating and emerging evidence thus strongly suggests that PKCβ plays an important role in regulating lipid and glucose homeostasis in response to dietary lipids, although the underlying molecular mechanisms remain unclear. It is possible that the effects of PKCβ deficiency on hepatic steatosis and insulin resistance in vivo are a consequence of the failure of high-fat diet (HFD)-fed PKCβ−/− mice to develop obesity. Moreover, it has also been reported that hepatic function, including steatosis and insulin resistance, may be regulated by PKCβ in muscle and adipose tissue [30,31]. How PKCβ may function in different tissues remains to be investigated.
The liver plays a central role in nutritional metabolism and is one of the first tissues to respond and adapt to nutritional changes. The imbalance between lipid influx and lipid outflux can cause changes in hepatic and serum lipid levels. Considering that dietary fats induce liver and adipose PKCβ expression [11,32,33] and the major role of the liver in regulating metabolism, the goal of this study was to determine the role of hepatic PKCβ in regulating lipid and glucose homeostasis. We found that hepatocyte PKCβ deficiency prevents the development of hepatic steatosis without affecting body weight gain and insulin resistance. PKCβ-deficient mice show a decrease in SREBP-1c transactivation potential, thereby reduced de novo hepatic lipogenesis, and an increase in mitochondrial function. Collectively, our results uncover a key role of hepatocyte PKCβ in the onset of diet-induced hepatic steatosis and support a model in which hepatocyte PKCβ coordinates energy utilization and lipogenesis during the post-prandial period. More importantly, our work identified PKCβ as a novel drug target for the treatment of fatty liver disease.
2. Materials and methods
2.1. Generation of floxed PKCβ (PKCβfl/fl) and hepatocyte-specific PKCβ-deficient (PKCβHep−/−) mouse models
PKCβfl/fl mice were generated through homologous recombination. Exon4 of the PKCβ gene was flanked by two loxP sites. PKCβ conditional knockout mice with C57BL/6 backgrounds were generated at Ohio State University's Comprehensive Cancer Center's Genetically Engineered Mouse Modeling Core Facility using standard embryonic stem (ES) cell technology [34]. The ES JM8.N4 clone EPD0744_4_5H11 was acquired from the International Mouse Phenotyping Consortium. These cells carry the knockout first tm1a Prkcbtm1a(EUCOMM)Wtsi allele (IMPC Project #28059; www.mousephenotype.org). Chimeric males were bred with C57BL/6 albino females and germline transmission was verified by PCR to detect mutants together with wild-type alleles in F1 heterozygous mice. Prior to utilization of the strain for experiments, the mice were crossed with a FLPe ubiquitous strain (ACTB:FLPe B6J, Jackson Laboratory strain #005703) to eliminate lacZ/neo cassettes and obtain clean tm1c alleles according to the breeding schemes recommended by the IMPC.
To inactivate PKCβ in hepatocytes, we crossed PKCβfl/fl mice with albumin-Cre transgenic mice with a C57BL6J genetic background (000664; Jackson Laboratory, Bar Harbor, ME, USA). The littermates were screened by genotyping, and mice with two copies of loxP sites and Cre recombinase were characterized as PKCβHep−/−. These mice were backcrossed 8 generations to C57BL/6J, and their genetic background was verified using SNP genome scanning (143 SNP Panel; Jackson Laboratory). Male mice were used in all of the experiments. PKCβfl/fl and PKCβHep−/− mice were bred and maintained on a 12-h light and 12-h dark cycle with lights on from 7:00 am to 7:00 pm. All of the mice were given standard food pellets (normal chow diet) and water ad libitum. Cohorts of age-matched male mice were used for these studies. Their body weights and food intake were measured weekly. For high-fat diet (HFD)-feeding experiments, the mice were fed diet containing 15% fat supplemented with 1% cholesterol (#D04102102, Research Diets, New Brunswick, NJ, USA) beginning at the age of 6–8 weeks. The Institutional Animal Care and Use Committee at Ohio State University's Care Facility approved all of the studies using animal protocols.
2.2. Mitochondrial isolation and respiration function analysis
Mitochondria were isolated as previously described [35]. Four micrograms of isolated mitochondria from the liver, gastrocnemius/plantaris muscle, BAT, and iWAT were resuspended in respiratory assay buffer composed of 70 mM of sucrose, 220 mM of mannitol, 10 mM of K2HPO4, 5 mM of MgCl2, 2 mM of HEPES, and 1 mM of EGTA, with a pH of 7.4. Electron coupling and electron flow assays were performed using Seahorse Bioanalyzer. Briefly, mitochondria were incubated with the indicated substrates and oxygen consumption rates were determined. Mitochondrial basal respiration in electron coupling assays was determined in a coupling state with 10 mM of succinate initial substrate with 2 μM of rotenone. State 3 respiration was initiated by injecting ADP, state 4 respiration was initiated by injecting oligomycin, and maximal uncoupler-stimulated respiration was initiated by injecting FCCP (trifluoromethoxy carbonyl cyanide phenylhydrazine). Mitochondrial basal respiration in electron flow assays was determined in an uncoupled state with initial substrates of 10 mM of pyruvate and 2 mM of malate in the presence of FCCP. Sequential electron flow throughout the electron transport chain was determined by first injecting rotenone, followed by succinate, antimycin A, and ascorbate and TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine).
2.3. Measurement of SREBP-1c transactivation
(Gal4)5-luciferase reporter plasmid (0.6 μg) was co-transfected with a plasmid encoding either the Gal4-DNA-binding region (Gal4-DBD) or SREBP-1c activation domain linked to Gal4-DNA-binding domain (Gal4-DBD-SREBP-1AD) (0.3 μg) [36] and pCMV-β-galactosidase (0.1 μg) along with constitutively active PKCβ cDNA (0.1 μg) in human hepatoma HepG2 cells in the absence or presence of LY333,531 (5 μM), PD98059 (20 μM), or GSK690693 (1 μM). Fold induction represented luciferase activity on PKCβ transfection relative to basal expression levels in the absence of PKCβ expression vector (taken as 1). Luciferase activity was normalized to β-galactosidase activity.
2.4. Shotgun lipidomic analysis
Cell pellets were homogenized in 0.5 mL of 10 × diluted PBS in 2.0 mL cryogenic vials (Corning Life Sciences) using a digital sonifier (Branson 450). For the shotgun lipidomic analysis, lipid extracts were diluted to a final concentration of ∼500 fmol/μL, and the mass spectrometric analysis was performed on a QqQ mass spectrometer (Thermo Fisher TSQ Quantiva) equipped with an automated nanospray device (TriVersa NanoMate; Advion Bioscience Ltd.) as previously described [37]. Identification and quantification of all of the reported lipid molecular species were performed using an in-house automated software program following the principles for quantification by MS as previously described in detail [38]. Fatty acyl chains of lipids were identified and quantified by neutral loss scans or precursor ion scans of corresponding acyl chains and calculated using the same in-house software program. Data were normalized per milligram of protein. Lipid internal standards were 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (di14:1 PC). All of the lipid internal standards are purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA).
2.5. Histology
Liver, WAT, and BAT from ad libitum-fed mice were isolated and fixed in 4% paraformaldehyde and processed for H&E staining. For Oil Red O staining, liver tissues were fixed in 4% paraformaldehyde overnight and incubated in 12% sucrose for 12 h and then in 18% sucrose overnight before being cryoembedded and sectioned by HistoWiz.
2.6. Plasma and tissue chemistry
Blood was collected using a 1 mL syringe coated in 0.5 M K2EDTA, and serum was collected by centrifugation at 1000g for 20 min. Insulin levels were measured by ELISA. Serum and liver TG, cholesterol, and lipoprotein distribution were measured at the Mouse Metabolic Phenotyping Core Facility at University of Cincinnati's College of Medicine.
2.7. Immunoblotting analysis
Proteins were extracted from the liver tissue of the mice [12,13]. The livers were homogenized in RIPA buffer, 10 mM of NaF, 1 mM of Na3VO4, 1 mM of PMSF, and protease inhibitor tablets (Roche Diagnostics). The protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific), and lysates were analyzed using SDS-polyacrylamide gel electrophoresis and Western blotting analysis on PVDF membranes. Antibody for PKCβ (F-7) was purchased from Santa Cruz Biotechnology, and antibodies for AKT (#4685), P-AKTThr308 (#13038), P-AKTSer473 (#4060), insulin receptor beta (#3025), P-insulin receptor/IGF1R beta (#3021), P-IRS-1Ser307 (#2381), P-IRS-1Ser612 (#3203), P-IRS1Ser318 (#5610), IRS-2 (#4502), P-mTORSer2448 (#5536), P-mTORSer2481 (#2974), mTOR (#2983), rictor (#2114), and GβL (#3274) were purchased from Cell Signaling Technology (Danvers, MA, USA). Phospho-SGK1Ser422 (#55281) and SGK1 (#43606) were purchased from Abcam (Cambridge, MA, USA). Goat anti-mouse and goat anti-rabbit HRP-conjugated secondary antibodies (Bio-Rad) were used.
2.8. In vivo insulin signaling
Following an overnight fast, the mice were anesthetized with 2,2,2-tribromoethanol in PBS and injected with 5 U of regular human insulin (Novolin, Novo Nordisk) via the inferior vena cava [33]. Five min after the insulin bolus, tissues were removed and frozen in liquid nitrogen. Immunoblotting analysis of insulin signaling molecules was performed using liver homogenates prepared in a tissue homogenization buffer that contained 25 mM of Tris-HCl (pH 7.4), 10 mM of Na2VO4, 100 mM of NaF, 50 mM of Na3P2O7, 10 mM of EGTA, 10 mM of EDTA, 2 mM of phenylmethylsulfonyl fluoride, and 1% Nonidet-P40 supplemented with protease inhibitor cocktail (Sigma-Aldrich). All of the protein expression data were quantified by densitometery using NIH Image.
2.9. Insulin tolerance tests
Insulin tolerance tests were performed as previously described [11].
2.10. Determination of membrane DAG content
The livers were homogenized in fractionation buffer (20 mM of HEPES-NaOH, pH 7.4, 250 mM of sucrose, 25 mM of sodium fluoride, 1 mM of sodium pyrophosphate, 0.1 mM of sodium orthovanadate, 2 μM of microcystin LT, and 1 mM of benzamidine). The cell lysates were centrifuged at 800×g for 5 min at 4 °C. Supernatants were further centrifuged at 100,000×g for 20 min at 4 °C. The membrane pellets were solubilized in buffer containing Triton X-100 by bath sonication and centrifuged at 12,000×g for 10 min at 4 °C, and the supernatant was used as the membrane fraction. Purity of the subcellular fractions was assessed by immunoblotting with antibody against the β subunit of insulin-like growth factor-1 receptor (to control the membrane fraction) and against β-actin (as control of cytosolic fraction). Lipids were extracted from the membrane fraction as previously described [39]. DAG content was quantified radioenzymatically by incubating aliquots of the lipid extract with DAG kinase and [32P]ATP. The manufacturer's instructions for a commercially available DAG kit were followed (Abcam ab242293). The 32P-labeled phosphatidic acid was purified using chloroform/methanol/acetic acid (65:15:5, v/v) as a solvent system and quantified.
2.11. Statistical analysis
All of the values are given as mean + SEM. Differences between two groups were assessed using unpaired Student's two-tailed t tests. P < 0.05 was regarded as significant. Statistical analysis was performed using Excel (Microsoft).
3. Results
3.1. Generating floxed PKCβ (PKCβfl/fl) and hepatocyte-specific PKCβ-deficient (PKCβHep−/−) mouse models
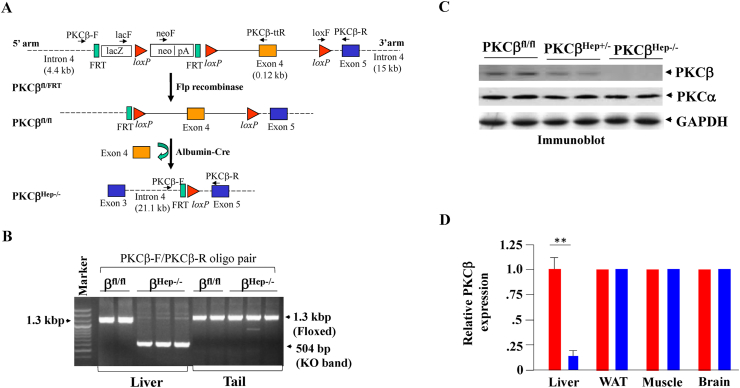
As a vital metabolic organ in the human body, the liver plays a central role in maintaining energy balance. Dietary fat intake also induces hepatic PKC expression in mice (32). To test the role of PKCβ in the liver, PKCβfl/fl mice were created in which exon 4 was flanked with the loxP Cre excision sequence (Figure 1A). Conditional floxed PKCβ alleles were deleted in these mice by breeding them with a transgenic albumin-Cre mouse line that expressed Cre recombinase specifically in hepatocytes. In the resulting PKCβHep−/− mice that expressed Cre recombinase, exon 4 of the PKCβ was excised in hepatocytes (Figure 1B), thereby ablating functional PKCβ expression, as this truncation prevented the translation of the entire kinase [11]. Liver extracts derived from KO mice displayed an 80–90% reduction in PKCβ protein (Figure 1C), consistent with complete deletion of PKCβ in hepatocytes. In view of a recent report that albumin-Cre produced recombination in hepatocytes and hepatic stellate cells [40], we also compared the expression of PKCβ in these cells. Hepatic stellate cells from PKCβfl/fl and PKCβHep−/− mice exhibited comparable PKCβ protein levels (although substantial variability was observed), suggesting either no or partial recombination (results not shown). Additional Western blotting revealed there was no significant difference in PKCβ expression in white adipose tissue, muscle, and brain of the PKCβHep−/− mice compared to livers from the PKCβfl/fl mice, confirming that PKCβ deletion was specific in the liver (Figure 1D).
Figure 1.
Generation and phenotyping of mice with hepatocyte-specific PKCβ deficiency. (A) Targeting strategy used to generate PKCβfl/fl and PKCβHep−/− mice. Maps of the PKCβ genomic locus showing the conditional allele (upper panel) and knockout allele (lower panel). The red arrowheads indicate the loxP sites and the black vertical bars represent the respective exons. (B) Validation of effective DNA recombination by PCR analysis of genomic DNA. (C) PKCβ protein expression as assessed by western blotting in hepatocytes of PKCβfl/fl, PKCβHep+/−, and PKCβHep−/− mice. The same samples were also probed for PKCβ. GAPDH was used as a loading control. (D) Comparison of PKCβ protein level in the liver, white adipose tissue, muscle, and brain of the PKCβfl/fl and PKCβHep−/− mice. Relative expression shows normalized band intensity of PKCβ in the PKCβHep−/− mice compared to controls (taken as 1). Data represent the mean ± SEM. ∗∗p < 0.01.
3.2. Hepatocyte-specific PKCβ deficiency protects against diet-induced hepatic steatosis
When maintained on normal chow ad libitum, the PKCβHep−/− mice exhibited similar body weight compared to control PKCβfl/fl mice (Supplementary Figure 1A). Gross metabolic comparisons between these mice revealed no significant differences in blood phospholipid, triglycerides, and cholesterol (Supplementary Figure 1B). There was a slight but insignificant decrease in hepatic TG content, with no difference in hepatic cholesterol content of the PKCβHep−/− mice compared to the control mice (Supplementary Figures 1C,D). Comparison of blood glucose revealed no differences between genotypes (Supplementary Figure 1E). Thus, loss of the PKCβ in hepatocytes appeared to exert no significant metabolic effect on the mice maintained on normal chow.
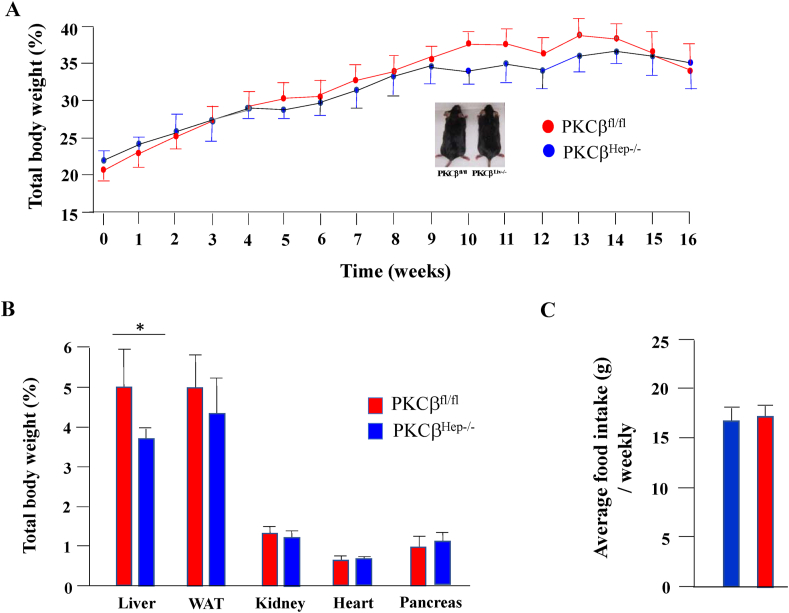
To understand how PKCβ deficiency might influence nutrient handling in mice upon chronic lipid overflow, we fed a HFD to the PKCβfl/fl and PKCβHep−/− mice. After 12–16 weeks on the HFD, the PKCβHep−/− and control mice showed similar weight gains (Figure 2A). Interestingly, the livers were significantly smaller in these mice (Figure 2B), with an insignificant decrease in epididymal white adipose tissue (eWAT) despite similar food intake (Figure 2C). No differences were observed in the kidney, heart, and pancreas weights (Figure 2C).
Figure 2.
Hepatocyte-specific PKC deficiency did not affect diet-induced weight gain. (A) Body weights of the PKCβfl/fl and PKCβHep−/− mice fed an HFD starting from 8 weeks of age (n = 12). (B) Comparison of weights of different tissues from the HFD-fed PKCβfl/fl and PKCβHep−/− mice at 12 weeks (n = 8). (C) Comparison of food intake (g/mouse/weekly) of the PKCβfl/fl and PKCβHep−/− mice fed an HFD. Data represent the mean ± SEM. ∗p < 0.05.
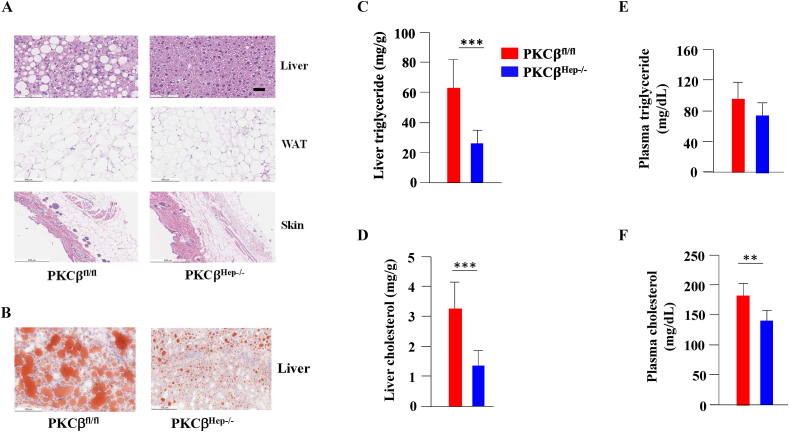
HFD-induced obesity normally leads to lipids accumulation in the liver. Histological examination of the livers revealed reduced numbers and sizes of intracellular vacuoles, an indication of reduced fats, in the PKCβHep−/− mice compared to the control PKCβfl/fl mice (Figure 3A). Oil Red O staining of liver sections verified the deposition of increasing quantities of lipids in the control livers (Figure 3B). Histological analysis of WAT revealed that adipocytes from WAT were slightly smaller than those from the control mice (Figure 3A). There was no difference in the thickness of adipose tissue beneath the dermis between genotypes (Figure 3A). As expected, based on the histological analysis, biochemical measurements confirmed a pronounced decrease in both TG and cholesterol levels in the livers from the HFD-fed PKCβHep−/− mice compared to the control mice (Figures 3C,D). Decreased liver lipid content was accompanied by a significant reduction in plasma cholesterol, with slightly lower (but not significant) plasma triglycerides (Figures 3E,F). Together, these findings demonstrated that hepatocyte PKCβ deficiency protected the mice from developing hepatic steatosis in response to caloric excess.
Figure 3.
Hepatocyte-specific PKCβ deficiency protected the mice from HFD-induced hepatic steatosis. (A and B) Representative sections of the liver, eWAT, and skin from the PKCβfl/fl and PKCβHep−/− mice were stained with either H&E or Oil Red O. Scale bar, 50 μm. (C and D) Comparison of liver triglyceride and cholesterol contents in the PKCβfl/fl and PKCβHep−/− mice fed an HFD for 12 weeks. (E and F) Comparison of the plasma TG and cholesterol levels in the mice. Data represents the mean ± SEM. ∗p < 0.05; ∗∗p < 0.05; ∗∗∗p < 0.01.
3.3. Hepatocyte-specific PKCβ deficiency does not protect against diet-induced insulin resistance
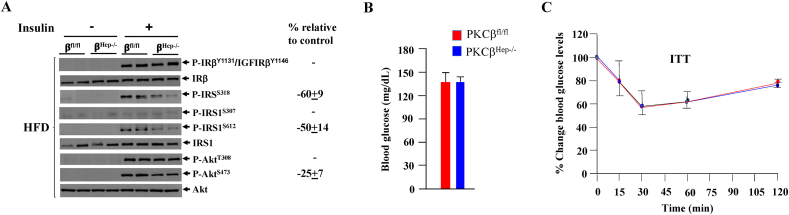
There have been reports that PKCβ can phosphorylate insulin receptor, insulin receptor substrate-1 (IRS1), and protein kinase B (AKT) in various cell culture models [[25], [26], [27]]. These in vitro experimental results have been conflicting, suggesting both negative and positive regulatory roles. To investigate the role of PKCβ in insulin signaling in the liver, fasted mice were injected with insulin or saline and analyzed for changes in phosphorylation of insulin-signaling components. Insulin-induced tyrosyl phosphorylation of the insulin receptor was comparable in livers of the control and PKCβ mice (Figure 4A). No apparent effect of hepatocyte PKCβ deficiency on insulin-induced low-level phosphorylation of IRS1-Ser307 was observed, whereas insulin-stimulated IRS1-Ser318 and -Ser612 phosphorylation normalized to the expression of IRS1 was lower in the PKCβHep−/− livers compared to the controls. Insulin-stimulated phosphorylation of AKT-Thr308 was similar, whereas mildly reduced phosphorylation of AKT-Ser473 was observed in the PKCβHep−/− livers.
Figure 4.
Hepatocyte PKCβ deficiency did not affect glucose homeostasis. (A) Western blotting of pooled liver lysates following IP insulin injection and probing for the indicated proteins. Western blots are representative of three separate experiments. Percentage change shows the normalized band intensity of the PKCβHep−/− mice compared to controls. (B) Comparison of fasted blood glucose levels in the HFD-fed PKCβfl/fl and PKCβHep−/− mice. (C) Insulin tolerance test (ITT) on the mice. Data represents the mean ± SEM. n = 4, ∗p < 0.05, and ∗∗∗p < 0.001.
There are several mechanisms possible for PKCβ to regulate AKT-Ser473 phosphorylation. One possibility is that PKCβ acts as an AKT kinase or activates the mechanistic target of rapamycin (mTORC) 2 to phosphorylate AKT on serine 473. mTORC1 and mTORC2 share mTOR protein that can be phosphorylated at several residues, including Thr2446, Ser2448, and Ser2481. Phosphorylation of mTOR at Ser2481 distinguishes activated mTORC2 from activated mTORC1 [41]. To evaluate mTORC2 activity, we investigated the phosphorylation status of mTORC2 and its substrate SGK1 in the livers of the mice treated with insulin. Unlike AKT-Ser473 phosphorylation, no differences were observed in the phosphorylation levels of mTOR-Ser2446 and -Ser2481 (Supplementary Figure 2) and phospho-SGK1-Ser422 between genotypes (results not shown). We also did not observe any significant difference in expression of mTORC2 components rictor and GβL. Our results support that hepatic PKCβ is not essential for AKT-Ser473 phosphorylation but may be required for its maximal activation in the liver in response to insulin.
We next investigated whether hepatic PKCβ deficiency has any effect on glucose homeostasis in vivo. Blood glucose levels were similar between genotypes (Figure 4B), suggesting no major effect of hepatocyte PKCβ deficiency on glucose homeostasis. Consistent with these results, no differences were observed in insulin-tolerance tests (ITTs) between the control and PKCβHep−/− mice (Figure 4C).
Diacylglycerol (DAG), an activator of PKCs, has been proposed to mediate lipid-induced hepatic insulin resistance [42]. However, the importance of DAG in lipid-induced hepatic insulin resistance remains controversial. A recent report connected membrane diacylglycerol levels through PKC to insulin resistance in NAFLD [43] by comparing membrane DAG levels in the livers of these mice. No significant changes in membrane DAG levels (87 ± 24 vs 82 ± 19 pmol/mg protein, n = 4, p > 0.05) were observed between genotypes. In short, these findings indicated that disruption of hepatocyte PKCβ has no major effect on insulin signaling and glucose homeostasis.
3.4. Hepatocyte-specific PKCβ deficiency attenuates SREBP-1c transactivation and improves mitochondrial function
As a central regulator of lipid homeostasis, the liver is responsible for orchestrating the synthesis of new fatty acids, their export and subsequent redistribution to other tissues, and their utilization as energy substrates. Altered lipid homeostasis in the liver is the pathophysiological hallmark of hepatic steatosis. The disruption of one or more of these pathways may precipitate the retention of fat within the liver and the subsequent development of hepatic steatosis.
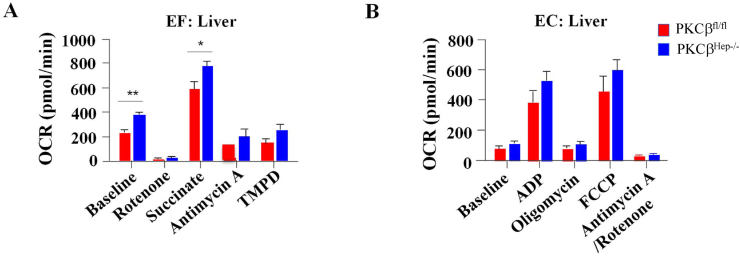
Healthy mitochondria are crucial for adequately controlling lipid metabolism in the liver. To gain insight into the molecular impact of hepatocyte-specific PKCβ deficiency on mitochondrial metabolism, energetics in mitochondria isolated from livers of the control and PKCβHep−/− mice fed an HFD were compared using a Seahorse XF analyzer. Under uncoupling conditions, baseline oxygen consumption rates (OCRs) were significantly increased in the liver mitochondria from the PKCβHep−/− mice and in the presence of succinate (Figure 5A). Under coupling conditions, there was an increase in liver mitochondria OCR with both adenosine diphosphate (ADP) and trifluoromethoxy carbonyl cyanide phenylhydrazine (FCCP) compared to the controls, although this was not significant (Figure 5B).
Figure 5.
Hepatocyte-specific PKCβ deficiency improved mitochondrial function in the liver. OCRs were measured in triplicate in isolated mitochondria from the liver of the PKCβfl/fl and PKCβHep−/− mice on an HFD in the absence or presence of the indicated substrate, inhibitor, or modulator of the electron transport chain. EF, electron flow assay; EC, electron coupling assay. Data are presented as mean ± SEM, n = 6. ∗p < 0.05, and ∗∗p < 0.01.
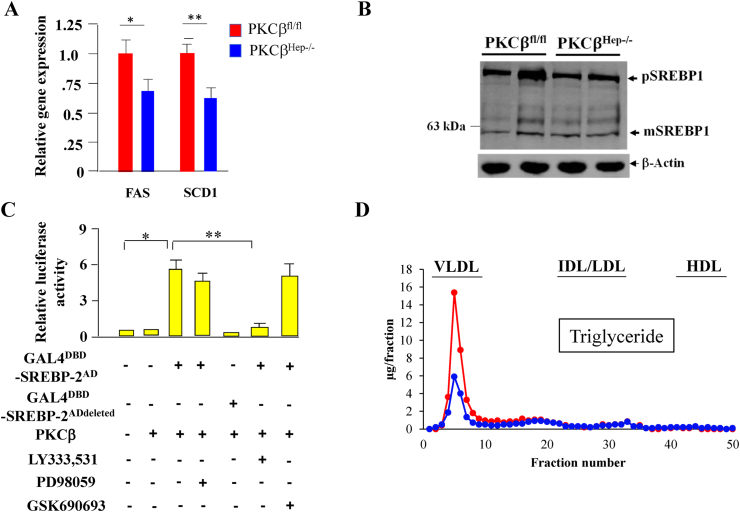
We observed that improvement in mitochondrial function was also accompanied by reduced levels of fatty acid synthase and stearoyl CoA desaturase transcripts in the liver of the PKCβHep−/− mice compared to the controls (Figure 6A). The transcription factor sterol regulatory element-binding protein-1c (SREBP-1c) plays a central role in de novo fatty acid synthesis gene expression. To first assess whether PKCβ deficiency affected SREBP-1c processing, we compared precursor and nuclear forms of endogenous SREBP-1 in the liver of the control and PKCβHep−/− mice. We observed a slight reduction in the expression of precursor SREBP-1, but nuclear levels of SREBP-1 were similar in the PKCβHep−/− livers compared to the control livers (Figure 6B). To determine whether PKCβ deficiency affected the activation of hepatic SREBP-1c, we used plasmids in which the activation domain of SREBP-1c was fused to Gal4-DBD and evaluated the activation of a Gal4-responsive reporter plasmid by overexpressed PKCβ in the absence or presence of the indicated inhibitor. Interestingly, PKCβ increased the activation of SREBP-1c plasmid, and this activation was blocked by a specific inhibitor of PKCβ LY333,531, but not by MEK inhibitor PD98059 or AKT inhibitor GSK690,693. These results support that PKCβ activates SREBP-1c through its amino terminal (Figure 6C).
Figure 6.
Effect of hepatocyte-specific PKCβ deficiency on hepatic fatty acid synthase expression, SREBP-1c processing, SREBP-1c transactivation potential, and plasma VLDL levels. (A) Comparison of hepatic fatty acid synthase and stearoyl coenzyme desaturase 1 expression between the PKCβfl/fl and PKCβHep−/− mice on an HFD. (B) Immunoblotting of pooled total cell extracts from livers of the mice using antibody to SREBP-1 and β-actin. Western blots are representative of two separate experiments. Percentage change shows normalized band intensities of both bands of the PKCβHep−/− liver compared to controls. (C) PKCβ activated the transcriptional activation potential of SREBP-1c independent of ERK-1/2 and AKT. (D) Triglyceride-lipoprotein VLDL distribution in pooled plasma from the control and PKCβHep−/− mice fed an HFD for 12 weeks. Analysis of lipids in lipoprotein fractions was performed after separating pooled plasma samples by fast-performance liquid chromatography with a Superose 6 10/300 GL high-performance column (GE Healthcare Life Sciences). Fractions were assayed for total triglycerides. n = 6–8, ∗p < 0.05, and ∗∗p < 0.01.
To determine the potential effect of hepatocyte PKCβ deficiency on very low-density lipoprotein (VLDL) levels, we compared plasma levels between genotypes. There was a significant reduction in plasma VLDL levels in PKCβHep−/− mice compared to the controls, suggesting that an increase in its production and secretion did not contribute to reduced hepatic steatosis in these mice (Figure 6D).
It is safe to conclude that the loss of hepatocyte PKCβ increased mitochondrial respiratory chain and lowered SREBP-1c transactivation, which may have accounted for the reduced liver fat content in the PKCβHep−/− mice compared to the control mice.
3.5. Hepatocyte-specific PKCβ deficiency leads to elevated liver cardiolipin and reduced acylcarnitine levels commonly associated with fatty liver disease
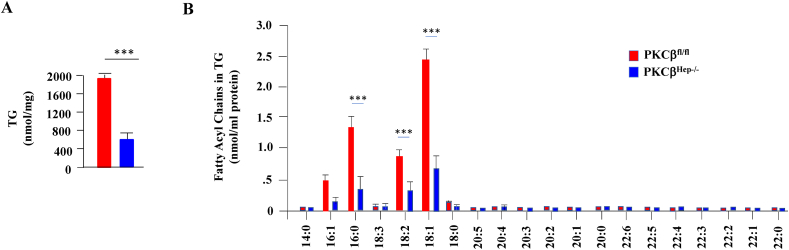
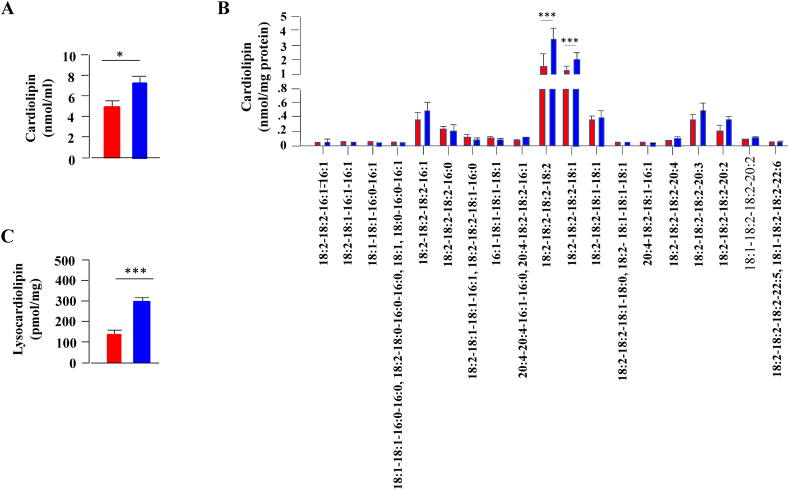
Recent studies underscored the importance of membrane lipids in mitochondrial function and the pathophysiology of hepatic steatosis [44]. To identify lipids discriminating the pathophysiological status of the liver in response to PKCβ deficiency, we performed shotgun lipidomics on the livers from the WT and PKCβHep−/− mice to compare fatty acyls, TG, acylcarnitines, cardiolipin, lysocardiolipin, and various phospholipids (phosphatidic acid, phosphatidylcholine, lysophosphatidylcholine, phosphatidylethanolamine, lysophosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, and phosphatidylserine). Congruent with our biochemical study, shotgun lipidomics identified a significant decrease in hepatic TG content in the PKCβHep−/− livers compared to the control livers (Figure 7A). There were no specific changes in TG molecular species composition of TG between the PKCβHep−/− mice and their WT counterparts. The decrease was significantly observed in TG molecular species (16:1), (16:0), (18:2), and (18:1) in the PKCβHep−/− livers compared to the control livers (Figure 7B). Similarly, livers from mice lacking PKCβ exhibited a reduction in levels of acylcarnitine (4.22 ± 1.37 PKCβfl/fl vs 2.89 ± 0.68 pmol/mg protein PKCβHep−/−, n = 4, p = 0.18), which did not reach statistical significance. There were no other significant changes in liver sphingomyelin, phosphatidylethanolamine, lysophosphatidylethanolamine, phosphatidylcholine, lysophosphatidylcholine, phosphatidylglycerol, and phosphatidylinositol levels in the hepatic fatty acid fractions of the control and PKCβHep−/− mice (results not shown). However, hepatocyte PKCβ deficiency caused marked increases in both cardiolipin and lysocardiolipin (Figure 8A). Importantly, cardiolipin comprises 10–20% of the mass of total mitochondrial phospholipids, and recent studies correlated positively higher cardiolipin levels to improved mitochondrial membrane potential and respiration [45,46]. The biological function of this essential mitochondrial lipid is determined by the composition of its 4 fatty acyl chains as they control mitochondrial architecture and function [46]. We next compared cardiolipin acyl composition in the same liver tissues. Ablation of hepatocyte PKCβ predominantly elevated most abundant cardiolipin molecular species (18:2–18:2–18:2–18:2) and (18:2–18:2–18:2–18:1), whereas lysocardiolipin molecular species (18:2–18:2–18:2–18:1) specifically showed a significant increase (105.27 ± 25.9 PKCβfl/fl vs 247 ± 7.23 pmol/mg protein PKCβHep−/−, n = 4, ∗∗∗p < 0.001) and not lysocardiolipin (41.20 ± 12.69 PKCβfl/fl vs 52.03 ± 1.91 pmol/mg protein PKCβHep−/−, n = 4, p = 0.212) (Figure 8B).
Figure 7.
Lipidomic analysis of livers in the PKCβfl/fl and PKCβHep−/− mice on an HFD. Comparison of liver mass contents of TG (A) and individual molecular species of TG (B) in the liver samples of the HFD-fed PKCβHep−/− mice relative to the PKCβfl/fl mice. Mass content of lipids was determined by multidimensional mass spectrometric array analyses by comparing the peak intensity of each individual ion to that of the selected internal standards after correction for 13C isotopomer distribution differences as described in the Materials and methods section. Data are mean ± SE, n = 4, and ∗∗p < 0.001.
Figure 8.
Comparison of liver mass content of cardiolipin and lysocardiolipin the liver of the PKCβfl/fl and PKCβHep−/− mice on an HFD. Comparison of contents of cardiolpin (A), individual molecular species of cardiolipin (B), and total lysocardiolipin (C) in the liver samples of the PKCβHep−/− mice relative to the PKCβfl/fl mice. Mass content of lipids was determined by multidimensional mass spectrometric array analyses by comparing the peak intensity of each individual ion to that of the selected internal standards after correction for 13C isotopomer distribution differences as described in the Materials and Methods section. Data are mean ± SE, n = 4, and ∗∗p < 0.001.
4. Discussion
The liver plays a central role in lipid metabolism, and the imbalance between lipid influx and lipid outflux can cause changes in hepatic and serum lipid levels. In this study, we demonstrate that in vivo deficiency of hepatocyte PKCβ prevents the development of nonalcoholic hepatic steatosis and may be sufficient to obtain therapeutic effects in the context of diet-induced adiposity. This protection in PKCβHep−/− mice might be attributed to the combination of at least two mechanisms: reduced de novo lipogenesis and enhanced mitochondrial function. Our results demonstrated that hepatocyte PKCβ downregulates the transactivation potential of transcription factor SREBP-1c, resulting in reduced de novo lipogenesis and triglyceride synthesis in PKCβHep−/− mice. It is currently well accepted that SREBP-1c induced by insulin mediates the transcriptional effect on enzymes involved in fatty acid synthesis and triglyceride synthesis, including fatty acid synthase and stearoyl-coenzyme A desaturase 1. Excessive accumulation of hepatic TG can occur as a result of increased hepatic PKCβ expression, leading to elevated fat synthesis. Previous studies in humans and rodents also demonstrated that liver TG accumulation is mainly linked to enhanced de novo lipid synthesis via the lipogenic pathway in the liver [47]. Therefore, the beneficial effects of PKCβ deficiency are at least partially due to the suppression of hepatic de SREBP-1 function. In the nucleus, transcriptional activities of nuclear forms of SREBPs are regulated by recruiting transcriptional cofactors, such as CBP/p300 and the mediator complexes [48,49]. PKCβ may influence transactivation of SREBP-1c possibly through direct phosphorylation of SREBP-1c or phosphorylation of cofactors involved in SREBP-1c transactivation. Of note, phosphorylation of p300 at serine 89 by PKC is shown to repress its function as a transcriptional coactivator [50].
We also found that hepatocyte PKCβ is a negative regulator of the mitochondrial respiratory chain because hepatocyte-specific PKCβ deficiency promoted oxidative phosphorylation by improving the efficiency of mitochondrial respiratory chain. Our study suggests that an important factor in hepatocyte-specific PKCβ deficiency increases the activity of the oxidative phosphorylation chain and its uncoupling to dissipate excess incoming metabolic energy and reduce lipid accumulation. As a result, diet-induced hepatic PKCβ activation can severely impair mitochondrial oxidative phosphorylation, leading to steatosis. This is defined as the OxPhos funnel effect in which efficient transport chains and ATP synthase complexes consume increased amounts of NADH and FADH2, which drive the oxidation of fatty acids and shift the balance away from lipogenesis and release of lipids into the circulation from the liver. Consequently, increased mitochondrial function rates can result in a reduction of serum and hepatic TG and cholesterol levels. PKCβ may influence oxidative phosphorylation complex activity, stability, or assembly possibly through direct phosphorylation of one or more subunits of these complexes or through downstream kinases or phosphatases. The effect may also be indirect through other proteins associated with oxidative phosphorylation. Direct phosphorylation of complex I by PKCβ has not been shown. However, increases in serine and tyrosine phosphorylation of the 18 kDa subunit of complex I and increased threonine phosphorylation of cytochrome oxidase following activation of another PKC isoform, PKCε, have been reported in hippocampal synaptosomes [51]. Alternatively, two lipids that have been closely linked with mitochondrial function are altered and may promote mitochondrial respiratory chains in PKCβHep−/− livers. Our lipidomic study showed that the loss of PKCβ specifically in hepatocytes resulted in an increase in cardiolipin and was accompanied by a reduction in acylcarnitine levels. Interestingly, cardiolipin is a complex mitochondrial-specific phospholipid that regulates numerous enzyme activities, especially those related to oxidative phosphorylation and coupled reactions [44]. The significance of cardiolipin in the organization of components of the electron transport chain into higher order assemblies, called respiratory super-complexes, is well established [44]. Depletion of cardiolipin results in severe mitochondrial dysfunction and is implicated in mitochondrial dysfunction in fatty liver disease [52]. Cardiolipin is also shown to restrict pumped protons within its head group domain, providing a structural basis for mitochondrial membrane potential and supplying protons to the ATP synthase. Notably, we previously reported that inhibition of PKCβ increases mitochondrial membrane potential in cell culture models [16]. Furthermore, the total acylcarnitine content was significantly reduced in PKCβHep−/− livers, reflecting an imbalance between CPTI-mediated acylcarnitine production and their disposal via β oxidation. Aberrant acylcarnitine levels have been linked with obesity and steatohepatitis; accumulation of acylcarnitine is a sign of mitochondrial stress, mitochondrial dysfunction, and impaired fatty acid oxidation [51]. We propose that increased levels of cardiolipin in PKCβHep−/− livers may help preserve efficient mitochondrial function despite HFD intake, which may cause simultaneous reductions in levels of acylcarnitines. Consistent with this mechanism, mitochondrial oxidative phosphorylation activities decrease in patients with fatty liver disease, and livers from these patients exhibit reduced cardiolipin and elevated hepatic acylcarnitine levels [[53], [54], [55], [56]]. There are two possible mechanisms for PKCβ to regulate cellular cardiolipin levels. One possibility is that PKCβ promotes cardiolipin catabolism by activating phospholipase D activity because the PKCβ isoform is reported to activate this enzyme [[57], [58], [59]]. Alternatively, PKCβ negatively regulates cardiolipin synthase as phosphatidylglycerol required for cardiolipin synthesis is a potent and selective activator of PKCβ [60,61], thereby controlling the feed-forward loop. Induction of autophagy by PKCβ deficiency may serve as a homeostatic mechanism to protect the liver from diet-induced mitochondrial damages [16]. Disruption of macroautophagy and mitophagy is thought to contribute to hepatic steatosis by increasing the accumulation of dysfunctional organelles [62]. In conclusion, our study provides putative links between hepatic PKCβ, the mitochondrial respiratory chain, and the progression of hepatic steatosis. Measurement of hepatocellular lipid metabolism (i.e., rates of synthesis and oxidation of endogenous/exogenous lipids) would provide better insight into the relative contribution of each mechanism to differences shown in Figure 3 between the control and PKCβHep−/− mice.
Of note, the PKCβHep−/− mice developed insulin resistance but were protected against diet-induced hepatic steatosis, supporting the notion that hepatic steatosis can be disconnected from insulin resistance. This was also observed in many other mouse models [63]. PKCβ has been reported to phosphorylate several components of the insulin-signaling cascade with divergent effects. In contrast to a signal promoting AKT-Ser473 phosphorylation observed in a cell type- and stimulus-specific manner in a prior study [64], the insulin receptor itself as well as IRS-1 may be a site of negative regulation by PKCβ [[25], [26], [27]]. IRS1 phosphorylation at serine sites 636 and 639 has been shown to confer inhibition of insulin signaling [65,66]. PKCβ can thus promote and at the same time inhibit the conductance of metabolic insulin signaling. These studies were based on in vitro phosphorylation or overexpression of PKCβ and potential substrates in different cultured cells and therefore may require careful in vivo investigation. We therefore compared hepatic insulin signaling and whole-body insulin sensitivity using newly generated mouse models. Our results, in combination with previous in vitro studies [25,26], provide the first in vivo evidence that both IRS-1 and AKT are indeed physiological substrates of PKCβ in the liver, and PKCβ is not an absolute requirement for AKT-Ser473 phosphorylation, but is required for its maximal activation by insulin in the liver. Our results also suggested that PKCβ exerts effect on AKT-Ser473 phosphorylation independent of mTORC2, in agreement with a previous study showing that inhibiting PKCβ had no effect on mTORC2 activity [67]. We were unable to detect any significant changes in insulin-stimulated glucose disposal between the control and PKCβHep−/− mice, suggesting that the loss of PKCβ phosphorylation of IRS1-Ser318 or -Ser612, coupled with a subtle reduction in AKT-Ser473 phosphorylation, may not have significant functional consequences on insulin-induced glucose homeostasis. Lack of any role of AKT-Ser473 phosphorylation on its activity was also reported by others [68]. Thus, our results do not support a critical role of hepatic PKCβ in hepatic insulin signaling and glucose homeostasis.
In summary, we generated a liver-specific knockout PKCβ gene mouse model that will be valuable for further understanding diet-induced energy homeostasis. Of note, although albumin promoter driving Cre mice is generally used for hepatocyte-specific deletion, hepatic stem cells have recently been reported to possess some Cre-recombinase activity [40], so this cell type must be considered in the phenotype observed. In view of the relatively low PKCβ expression in this cell type and lack of any significant recombination observed in our study, it is likely that hepatocytes are the main cell type contributing to the PKCβHep−/− phenotype. Another consideration is that albumin is expressed quite early in liver development and thus mice would grow with silenced liver PKCβ, which may have some carry over effects into adulthood. Nonetheless, this newly generated PKCβ mouse model should be useful for understanding in vivo roles of this gene in the pathophysiology of liver diseases. Nonalcoholic fatty liver disease represents a major challenge, given its high levels of incidence worldwide [1]. Despite its increasing prevalence and burden to healthcare systems, there are currently no FDA-approved NAFLD therapeutics. Our results suggest that PKCβ plays a key role in increasing susceptibility to developing hepatic steatosis. Alterations in hepatocyte PKCβ can contribute to the pathophysiology of hepatic steatosis, and targeting its inactivation might be a promising therapeutic strategy for treating fatty liver disease and its related metabolic disorders.
Grant support
This research was supported by grants from the NIH (5R01 HL138198) and OSU Center for Clinical and Translational Sciences.
Author contributions
YS and FH conducted the diet, immunoblotting, and animal experiments. KDM and MO generated floxed and hepatocyte-specific PKCβ-deficient mouse models. XH performed lipidomics. NK analyzed the data. KKB conducted mitochondrial function studies. KDM supervised the work, wrote the manuscript, and interpreted the results.
Acknowledgments
We thank Dr. Arthur Burghes for helpful advice during generation of the hepatocyte-specific PKCβ-deficient mouse model. We are grateful to Dr. Tim Osborne for providing plasmids encoding Gal4-SREBP-1c. We also acknowledge that the University of Cincinnati's Mouse Metabolic Phenotyping Center measured plasma lipids, lipoprotein profiles, glucose and insulin, and liver lipid contents. This research was supported by grants from the NIH (5R01 HL138198) and the OSU Center for Clinical and Translational Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101133.
Conflict of interest
The authors disclose the following: Neil K. Mehta is a co-founder of Instacare Therapeutics. The remaining authors disclose no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplementary Figure 1. Phenotyping of mice with hepatocyte-specific deficiency of PKCβ fed a chow diet. Comparison of body weights at different ages (A), plasma phospholipid, cholesterol, and triglyceride levels (B), hepatic triglyceride levels (C), hepatic cholesterol levels (D), and blood glucose levels (E) of the PKCβfl/fl and PKCβHep−/- mice fed a chow diet. Supplementary Figure 2. Western blotting showing loss of PKCβ expression had no significant effects on phospho-mTOR levels or the expression of mTOR, GβL, and raptor proteins in the livers of the HFD-fed mice injected with insulin.
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Arab J.P., Arrese M., Trauner M. Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annual Review Pathology. 2018;13:321–350. doi: 10.1146/annurev-pathol-020117-043617. [DOI] [PubMed] [Google Scholar]
- 3.Samuel V.T., Shulman G.I. Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metabalism. 2018;27:22–41. doi: 10.1016/j.cmet.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younossi Z.M., Loomba R., Rinella M.E., Bugianesi E., Marchesini G., Neuschwander-Tetri B.A. Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2018;68:361–371. doi: 10.1002/hep.29724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ioannou G.N. The role of cholesterol in the pathogenesis of NASH. Trends in Endocrinology and Metabolism. 2016;27:84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Wree A., Broderick L., Canbay A., Hoffman H.M., Feldstein A.E. From NAFLD to NASH to cirrhosis—new insights into disease mechanisms. Nature Review Gastroenterology & Hepatology. 2013;10:627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 7.Day C.P., James O.F. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 8.Mehta D., Mehta K.D. PKCβ: expanding role in hepatic adaptation of cholesterol homeostasis to dietary fat/cholesterol. American Journal of Physiology & Liver Physiology. 2017;312:G266–G273. doi: 10.1152/ajpgi.00373.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta N.K., Mehta K.D. Protein kinase C-beta: an emerging connection between nutrient excess and obesity. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;312:1. doi: 10.1016/j.bbalip.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Bansode R.R., Huang W., Roy S.K., Mehta M., Mehta K.D. Protein kinase Cβ deficiency increases fatty acid oxidation and reduces fat storage. Journal of Biological Chemistry. 2008;283:231–236. doi: 10.1074/jbc.M707268200. [DOI] [PubMed] [Google Scholar]
- 11.Huang W., Bansode R., Mehta M., Mehta K.D. Loss of protein kinase Cβ function protects mice again diet-induced obesity and development of hepatic steatosis and insulin resistance. Hepatology. 2009;49:1525–1536. doi: 10.1002/hep.22815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W., Bansode R.R., Xie Y., Rowland L., Mehta M., Davidson N.O. Disruption of the murine protein kinase Cβ gene promotes gallstone formation and alters biliary lipid and hepatic cholesterol metabolism. Journal of Biological Chemistry. 2011;286:22795–22805. doi: 10.1074/jbc.M111.250282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W., Bansode R.R., Bal N.C., Mehta M., Mehta K.D. Protein kinase Cβ deficiency attenuates obesity syndrome of ob/ob mice by promoting white adipose tissue remodeling. Journal of Lipid Research. 2012;53:368–378. doi: 10.1194/jlr.M019687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterhoff M.A., Heuer S., Pfeiffer M., Tasic J., Kaiser S., Isken F. Identification of a functional protein kinase Cβ promoter polymorphism in humans related to insulin resistance. Molecular Genetic Metabolism. 2008;893:210–215. doi: 10.1016/j.ymgme.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Pinton P., Rimessi A., Marchi S., Orsini F., Migliaccio E., Giorgio M. Protein kinase C β and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 16.Patergnani S., Marchi S., Rimessi A., Bonora M., Giorgi C., Mehta K.D. PRKCB/protein kinase C, beta and the mitochondrial axis as key regulators of autophagy. Autophagy. 2013;9:1–19. doi: 10.4161/auto.25239. [DOI] [PubMed] [Google Scholar]
- 17.Huang W., Mehta D., Sif S., Kent L.N., Jacob S.T., Ghoshal K. Dietary fat/cholesterol-sensitive PKCβ-RB signaling: potential role in NASH/HCC axis. Oncotarget. 2017;8:73757–73765. doi: 10.18632/oncotarget.17890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su T.T., Guo B., Kawakami Y., Sommer K., Chae K., Humphries L.A. PKC-β controls IκB kinase lipid raft recruitment and activation in response to BCR signaling. Nature Immunology. 2002;3:780–786. doi: 10.1038/ni823. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh J.C., Jurutka P.W., Galligan M.A., Terpening C.M., Haussler C.A., Samuels D.S. Human vitamin D receptor is selectively phosphorylated by protein kinase C on serine 51, a residue crucial to its trans-activation function. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:9315–9319. doi: 10.1073/pnas.88.20.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gineste R., Sirvent A., Paumelle R., Helleboid S., Aquilina A., Darteil R. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Molecular Endocrinology. 2008;22:2433–2447. doi: 10.1210/me.2008-0092. [DOI] [PubMed] [Google Scholar]
- 21.Sotiropoulos K.B., Clermont A., Yasuda Y., Rask-Madsen C., Mastumoto M., Takahashi J. Adipose-specific effect of rosiglitazone on vascular permeability and protein kinase C activation: novel mechanism for PPARγ agonist's effects on edema and weight gain. FASEB Journal. 2006;20:1203–1205. doi: 10.1096/fj.05-4617fje. [DOI] [PubMed] [Google Scholar]
- 22.Tonetti D.A., Henning-Chubb C., Yamanishi D.T., Huberman E. Protein kinase C-β is required for macrophage differentiation of human HL-60 leukemia cells. Journal of Biological Chemistry. 1994;269:23230–23235. [PubMed] [Google Scholar]
- 23.Asehnoune K., Strassheim D., Mitra S., Yeol Kim J., Abraham E. Involvement of PKCα/β in TLR4 and TLR2 dependent activation of NF-κB. Cellular Signalling. 2005;17:385–394. doi: 10.1016/j.cellsig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto T., Watanabe K., Inoue N., Nakagawa Y., Ishigaki N., Matsuzaka T. Protein kinase Cbeta mediates hepatic induction of sterol-regulatory element binding protein-1c by insulin. Journal of Lipid Research. 2010;51:1859–1870. doi: 10.1194/jlr.M004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizuka T., Kajita K., Natsume Y., Kawai Y., Kanoh Y., Miura A. Protein kinase C (PKC) β modulates serine phosphorylation of insulin receptor substrate-1 (IRS-1) - effect of overexpression of PKCβ on insulin signal transduction. Endocrinology Research. 2004;30:287–299. doi: 10.1081/erc-120039580. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami Y., Nishimoto H., Kitaura J., Maeda-Yamamoto M., Kato R.M., Littman D.R. Protein kinase C βII regulates Akt phosphorylation on Ser-473 in a cell type- and stimulus-specific fashion. Journal of Biological Chemistry. 2004;279:47720–47725. doi: 10.1074/jbc.M408797200. [DOI] [PubMed] [Google Scholar]
- 27.Bossenmaire B., Mosthaf L., Mischak H., Ullrich A., Haring H.U. Protein kinase C isoforms beta 1 and beta 2 inhibit the tyrosine kinase activity of the insulin receptor. Diabetologia. 1997;40:863–866. doi: 10.1007/s001250050761. [DOI] [PubMed] [Google Scholar]
- 28.Wang F., Liu H., Irwin M.G., Xia Z., Huang Z., Ouyang J. Role of protein kinase C β2 activation in TNF-α-induced human vascular endothelial cell apoptosis. Canadian Journal of Physiology and Pharmacology. 2009;87:221–229. doi: 10.1139/y09-004. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Martinez I., Santoro N., Chen Y., Hoque R., Ouyang X., Caprio S. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. Journal of Clinical Investigation. 2016;126:859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hennige A.M., Heni M., Machann J., Staiger H., Sartorius T., Hoene M. Enforced expression of protein kinase C in skeletal muscle causes physical inactivity, fatty liver and insulin resistance in the brain. Journal of Cellular and Molecular Medicine. 2010;14:903–913. doi: 10.1111/j.1582-4934.2008.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinton P., Pavan C., Zavan B. PKC-β activation and pharmacologically induced weight gain during antipsychotic treatment. Pharmacogenomics. 2011;12:453–455. doi: 10.2217/pgs.11.25. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Vienberg S.G., Bezy O., O'Neill B.T., Kahn C.R. Role of PKCδ in insulin sensitivity and skeletal muscle metabolism. Diabetes. 2015;64:4023–4032. doi: 10.2337/db14-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang W., Mehta K.D. Modulation of hepatic protein kinase Cβ expression in metabolic adaptation to a lithogenic diet. Cellular & Molecular Gastroenterology & Hepatology. 2015;1:395–405. doi: 10.1016/j.jcmgh.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caserta E., Egriboz O., Wang H., Martin C., Koivisto C., Pecot T. Noncatalytic PTEN missense mutation predisposes to organ-selective cancer development invivo. Genes & Development. 2015;29:1707–1720. doi: 10.1101/gad.262568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskin K.K., Grueter C.E., Kusminski C.M., Holland W.L., Bookout A.L., Satapati S. MED13-dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. EMBO Molecular Medicine. 2014;6:1610–1621. doi: 10.15252/emmm.201404218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth J.I., Datta S., Athanikar J.N., Freedman L.P., Osborne T.F. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Molecular and Cellular Biology. 2004;24:8288–8300. doi: 10.1128/MCB.24.18.8288-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X., Yang J., Cheng H., Ye H., Gross R.W. Towards fingerprinting cellular lipidomics directly from biological samples by two-dimensional electronspray ionization mass spectrometry. Analytical Biochemistry. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Han X., Yang J., Cheng H., Yang K., Abendschein D.R., Gross R.W. Shotgun lipidomics identifies cardiolipin depletion in diabetic myocardium linking altered substrate utilization with mitochondrial dysfunction. Biochemistry. 2005;44:16684–16694. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- 39.Miele C., Paturzo F., Teperino R., Sakane F., Fiory F., Oriente F. Glucose regulates diacylglycerol intracellular levels and protein kinase C activity by modulating diaylglycerol kinase subcellular localization. Journal of Biological Chemistry. 2007;282:31835–31843. doi: 10.1074/jbc.M702481200. [DOI] [PubMed] [Google Scholar]
- 40.Newberry E.P., Xie Y., Lodeiro C., Solis R., Moritz W., Kennedy S. Hepatocyte and stellate cell depletion of liver fatty acid binding protein reveals distinct roles in fibrogenic injury. FASEB Journal. 2019;33:4610–4625. doi: 10.1096/fj.201801976R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copp J., Manning G., Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Research. 2009;69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erion D.M., Shulman G.I. Diacylglycerol-mediated insulin resistance. Nature Medicine. 2010;16:400–402. doi: 10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyu K., Zhang Y., Zhang D., Kahn M., ter Horst K.W., Rodrigues M.R.S. A membrane-bound diacylglycerol species induces PKCε-mediated hepatic insulin resistance. Cell Metabolism. 2020;32:1–11. doi: 10.1016/j.cmet.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vrecken P., Valianpour F., Nijjtmans I.G., Grivell L.A., Plecko B., Wanders R.J. Defective remodeling of cardiolipin and phosphatidylgycerol in Barth syndrome. Biochemical and Biophysical Research Communications. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y., Anjaneyulu M., Donelian A., Yu W., Greemberg M.L., Ren M. Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:11235–11240. doi: 10.1073/pnas.1900890116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oemer G., Koch J., Wohlfarter Y., Alam M.T., Lackner K., Sailer S. Phospholipid acyl chain diversity controls the tissue-specific assembly of mitochondrial cardiolipins. Cell Reports. 2020;30:4281–4291. doi: 10.1016/j.celrep.2020.02.115. [DOI] [PubMed] [Google Scholar]
- 47.Postic C., Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. Journal of Clinical Investigation. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enmeier C.C., Taatjes D.J. Activator-mediator binding regulates mediator-cofactor interactions. Proceedings of the National Academy of Sciences of the United States of America. 2010;25:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliner J.D., Andresen J.M., Hansen S.K., Zhou S., Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes & Development. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 50.Yuan W., Gambee J.E. Phosphorylation of p300 at serine 89 by protein kinase C. Journal of Biological Chemistry. 2000;275:40946–40951. doi: 10.1074/jbc.M007832200. [DOI] [PubMed] [Google Scholar]
- 51.Dave K.R., DeFazio A., Raval A.P., Torraco A., Saul I., Barrientos A. Ischemic preconditioning targets the respiration of synaptic mitochondria via protein kinase Cε. Journal of Neuroscience. 2008;28:4172–4182. doi: 10.1523/JNEUROSCI.5471-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chicco A.J., Sparagna G.C. Role of cardiolipin alterations in mitochondrial dysfunction and disease. American Journal of Physiology - Cell Physiology. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 53.Enooku K., Nakagawa H., Fujiwara N., Kondo M., Minami T., Hoshida Y. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Science Report. 2019;9:1–663. doi: 10.1038/s41598-019-47216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng K.Y., Watt M.J., Rensen S., Greve J.W., Huynh K., Jayawardana K.S. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. Journal Lipid Research. 2018;59:1977–1986. doi: 10.1194/jlr.M085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begriche K., Massart J., Robin M.A., Bonnet F., Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology. 2013;58:1497–1507. doi: 10.1002/hep.26226. [DOI] [PubMed] [Google Scholar]
- 56.Petrosillo G., Portincasa P., Grattagliano I., Casanova G., Matera M., Ruggiero F.M. Mitochondrial dysfunction in rat with nonalcoholic fatty liver involvement of complex I, reactive oxygen species and cardiolipin. Biochemistry Biophus Acta. 2007;1767:1260–1267. doi: 10.1016/j.bbabio.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 57.Pai J.K., Pachter J.A., Weinstein B., Bishop W.R. Overexpression of protein kinase C β1 enhances phospholipase D activity and diacylglycerol formation in phorbol ester-stimulated rat fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:598–602. doi: 10.1073/pnas.88.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han J.S., Shin I. Ceramide does not inhibit protein kinase C β-dependent phospholipase D activity stimulated by anti-Fas monoclonal antibody in A20 cells. Cellular Signalling. 2000;12:731–736. doi: 10.1016/s0898-6568(00)00125-x. [DOI] [PubMed] [Google Scholar]
- 59.Dieter P., Fitzke E. Differential regulation of phospholipase D and phospholipase C by protein kinase C-β and -δ in liver macrophages. Cellular Signalling. 1995;7:687–694. doi: 10.1016/0898-6568(95)00038-q. [DOI] [PubMed] [Google Scholar]
- 60.Murray N.R., Fields A.P. Phosphotidylglycerol is a physiologic activator of nuclear protein kinase C. Journal of Biological Chemistry. 1998;273:11514–11520. doi: 10.1074/jbc.273.19.11514. [DOI] [PubMed] [Google Scholar]
- 61.Bailey L.J., Choudhary V., Bollag W.B. Possible role of phosphatidylglycerol-activated protein kinase C-βII in keratinocyte differentiation. Open Dermatology. 2017;11:59–71. doi: 10.2174/1874372201711010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allaire M., Rautou P.E., Codogno P., Lotersztajn S. Autophagy in liver disease: time for translation? Journal of Hepatology. 2019;70:985–998. doi: 10.1016/j.jhep.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 63.Sun Z., Lazar M.A. Dissociating fatty liver and diabetes. Trends in Endocrinology and Metabolism. 2013;24:4–12. doi: 10.1016/j.tem.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawakami Y., Nishimoto H., Kitaura J., Maeda-Yamamoto M., Kao R.M., Littman D.R. Protein kinase C βII regulates Akt phosphorylation on Ser-473 in a cell type- and stimulus-specific fashion. Journal of Biological Chemistry. 2004;279:47720–47725. doi: 10.1074/jbc.M408797200. [DOI] [PubMed] [Google Scholar]
- 65.Mothe I., Obberghen E.V. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731, modulates insulin action. Journal of Biological Chemistry. 1996;271:11222–11227. doi: 10.1074/jbc.271.19.11222. [DOI] [PubMed] [Google Scholar]
- 66.Luo M., Langlais P., Yi Z., Lefort N., De Fillippis E.A.D., Hwang H. Phosphorylation of human insulin receptor substrate-1 at serine 629 plays a positive role in insulin signaling. Endocrinology. 2007;148:4895–4905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleiman E., Carter G., Ghansah T., Patel N.A., Cooper D.R. Developmentally spliced PKCβII provides a possible link between mTORC2 and Akt kinase to regulate 3T3-L1 adipocyte insulin-stimulated glucose transport. Biochemical and Biophysical Research Communications. 2009;388:554–559. doi: 10.1016/j.bbrc.2009.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vincent E.E., Elder D.J.E., Thomas E.C., Philips L., Morgan C., Pawade J. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. British Journal of Cancer. 2011;104:1755–1761. doi: 10.1038/bjc.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Phenotyping of mice with hepatocyte-specific deficiency of PKCβ fed a chow diet. Comparison of body weights at different ages (A), plasma phospholipid, cholesterol, and triglyceride levels (B), hepatic triglyceride levels (C), hepatic cholesterol levels (D), and blood glucose levels (E) of the PKCβfl/fl and PKCβHep−/- mice fed a chow diet. Supplementary Figure 2. Western blotting showing loss of PKCβ expression had no significant effects on phospho-mTOR levels or the expression of mTOR, GβL, and raptor proteins in the livers of the HFD-fed mice injected with insulin.