Abstract
Background:
Tissue differentiation-inducing non-protein coding RNA (TINCR) has been shown to play a crucial role in pathogenesis of various types of human cancer including breast cancer (BC). The purpose of this study was to determine the potential prognostic value of serum lncRNA TINCR in BC.
Methods:
Quantitative reverse transcription PCR (qRT-PCR) was performed to detect serum lncRNA TINCR levels in 72 triple-negative BC (TNBC) patients, 105 non-TNBC patients, 60 benign breast disease patients and 86 healthy subjects.
Results:
The results showed that serum lncRNA TINCR level was significantly increased in BC, especially in TNBC. High circulating lncRNA TINCR was significantly correlated with worse clinicopathological features and clinical outcome of TNBC. Multivariate analysis revealed that serum lncRNA TINCR was an independent prognostic factor for overall survival of TNBC. However, little association was found between serum lncRNA TINCR and the prognosis of non-TNBC.
Conclusions:
Taken together, our findings demonstrate that serum lncRNA TINCR might be a useful novel and non-invasive biomarker for the prognosis prediction of TNBC.
Keywords: serum lncRNA TINCR, triple-negative breast cancer, biomarker, prognosis prediction, survival
Introduction
Breast cancer (BC) is among the most frequent cancer in women worldwide and the leading cause of cancer-related death among women. The number of newly diagnosed BC cases in 2018 reached 2.1 million, accounting for 24.2% of all new female cancer cases.1 The initiation and development of BC is a result of the interplay between genetic, epigenetic and lifestyle factors.2,3 The improvement of the early BC diagnosis and the significant advances in the multidisciplinary treatment methods have greatly improved the clinical outcome of BC. However, the prognosis of triple-negative breast cancer (TNBC) remains unsatisfactory. TNBC, accounting for about 10-15% of all breast cancer cases, refers to any breast cancer that does not express the estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor receptor 2 (HER2). Considering the distinctive tumor biology of TNBC, it is critical to develop novel biomarkers for effectively predicting the prognosis and the risk of recurrence for better management of TNBC.
Long non-coding RNAs (lncRNAs) are broadly defined as non-coding RNA molecules longer than 200 nucleotides.4 Despite not being translated into proteins, accumulating evidence has demonstrated that lncRNAs are key modulators of many important biological processes such as proliferation, differentiation, development, survival and apoptosis.5 A normal expression of lncRNAs is closely associated with the initiation and development of many types of human cancer including BC.6,7 For instance, LINC00673 was overexpressed in BC tissues, and upregulation of LINC00673 was correlated with unfavorable clinicopathological parameters. In addition, knockdown of LINC00673 suppressed cell proliferation and metastasis of BC cells, and vice versa, indicating LINC00673 might play a tumor suppressive role in BC.8 Recently, circulating lncRNAs has become attractive biomarkers for the diagnosis and prognosis of BC. The levels of plasma H19 was significantly increased in BC patients, and it showed great promise for the early detection and prognosis prediction of BC.9
LncRNAs have also been shown to be closely involved in TNBC tumorigenesis and exhibits great potential for predicting the prognosis of TNBC. For instance, Mitobe et al reported that knock-down of lncRNA TMPO-AS1 inhibited the proliferation and migration of TNBC cells in vitro as well as suppressed tumor growth and metastasis in vivo, indicating TMPO-AS1 might be a potential therapeutic target of TNBC.10 Similarly, the expression level of LINC00993 was significantly decreased in TNBC, and reduced LINC00993 was associated with better survival of TNBC patients, suggesting that LINC00993 acts as a tumor suppressor in TNBC.11
Tissue differentiation-inducing non-protein coding RNA (TINCR) is a 3.7-kb lncRNA that located at human chromosome 19. It plays a crucial role in the induction of key differentiation genes.12 Recently, aberrant expression of lncRNA TINCR has been demonstrated to be associated with progression of various types of cancer such as BC, hepatocellular carcinoma and cervical squamous cell carcinoma.13-15 However, the clinical significance of serum lncRNA TINCR in BC remains poorly known. Our preliminary findings showed that serum lncRNA TINCR was especially higher in TNBC than non-TNBC. Therefore, we aimed to determine the prognostic value of serum lncRNA TINCR in TNBC and non-TNBC separately.
Materials and Methods
Study Population
This study included 177 patients with histological diagnoses of BC, 60 patients with benign breast disease (BBD) and 86 healthy individuals. A total of 105 BC cases were in the non-TNBC group, while the remaining 72 BC patients were in the TNBC group. We classified the BC into 4 subtypes according to ER, PR, and HER2/neu status: 1) TNBC, 2) luminal A, 3) luminal B, 4) HER2-positive. For non-TNBC, there were 61 cases in the luminal A subgroup, 23 cases in the luminal B subgroup and 21 cases in the HER2-positive group. All the TNBC and non-TNBC patients received surgical resection, and modified radical mastectomy was the most common type of surgery. The radiotherapy, chemotherapy or targeted therapy were given based on the patients’ clinical conditions. The basic characteristics of BC cases including age, menopause, smoking, histological subtype, tumor size, lymph node metastasis, TNM stage and grade were retrieved from the patients’ medical records. The detailed information of the participants of this study cohort was summarized in Table 1. Overall survival (OS) was defined as the time from treatment initiation to death from any cause. Recurrence-free survival (RFS) was defined as the time from treatment initiation to first recurrence (loco-regional or distant metastasis) or death due to any cause. The study was approved by the Ethics Committee of the First Affiliated Hospital of Dalian Medical University, and written Informed consent was obtained from all participants.
Table 1.
The Basic Characteristics of the Study Cohort.
| Parameters | TNBC, n = 72 | Non-TNBC, n = 105 | BBD, n = 60 | HCs, n = 86 |
|---|---|---|---|---|
| Age | ||||
| <50 | 46 | 62 | 38 | 53 |
| ≥50 | 26 | 43 | 22 | 33 |
| Menopause | ||||
| No | 44 | 59 | 36 | 48 |
| Yes | 28 | 46 | 24 | 38 |
| Smoking | ||||
| No | 43 | 72 | 38 | 49 |
| Yes | 29 | 33 | 22 | 27 |
| Histological subtype | / | / | ||
| Ductal | 61 | 85 | ||
| Lobular | 11 | 20 | ||
| Tumor size (cm) | / | / | ||
| ≤2 | 29 | 36 | ||
| >2 and ≤5 | 29 | 59 | ||
| >5 | 14 | 10 | ||
| LNM | / | / | ||
| Negative | 37 | 56 | ||
| Positive | 35 | 49 | ||
| TNM | / | / | ||
| I-II | 42 | 59 | ||
| III-IV | 30 | 46 | ||
| Grade | / | / | ||
| G1 | 27 | 30 | ||
| G2 | 31 | 37 | ||
| G3 | 14 | 38 |
Note: TNBC, triple-negative breast cancer; TNM, tumor, node, metastasis; LNM, lymph node metastasis; BBD, benign breast disease; HCs, healthy controls.
Serum Sample Collection
The serum samples were collected before receiving any treatment including surgery. A venous blood sample (at least 6 mL) was collected into a serum tube and centrifuged at 3,000 rpm for 5 min at 4°C to separate the serum from cellular components. Serum was then immediately frozen and stored at −80°C until further use.
RNA Extraction and Quantitative Reverse Transcription PCR (RT-qPCR)
Total RNA was extracted from the serum samples using TRIzol reagents (Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer’s instructions. The RNA concentrations were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was obtained by the process of reverse transcription using a cDNA Reverse Transcription Kit (Promega, Madison, WI, USA). The cDNA samples were amplified on an ABI 7500 system (Applied Biosystems, Foster City, CA, USA) using SYBR Green under the following conditions: 2 min at 95°C, followed by 40 cycles of 95°C for 10 s, 60°C for 35 s, and 72°C for 30 s. The relative expression levels of serum lncRNA TINCR were calculated using the comparative cycle threshold (2− ΔΔCT) method. GAPDH was chosen as the endogenous control for data normalization in our study.
Statistical Analysis
The statistical analyses were performed with the GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). The Mann-Whitney U test and Kruskal-Wallis test were used to compare differences of serum lncRNA TINCR in different groups. The discriminative power of serum lncRNA TINCR between TNBC and non-TNBC was assessed using the receiver operating characteristic (ROC) curve. The Kaplan-Meier method and log-rank test were used to evaluate the correlation between serum lncRNA TINCR level and survival of TNBC or non-TNBC. Multivariate analysis was performed to identify the independent prognostic factors. P < 0.05 was considered statistically significant.
Results
Serum lncRNA TINCR Was Significantly Increased in BC, Especially in TNBC
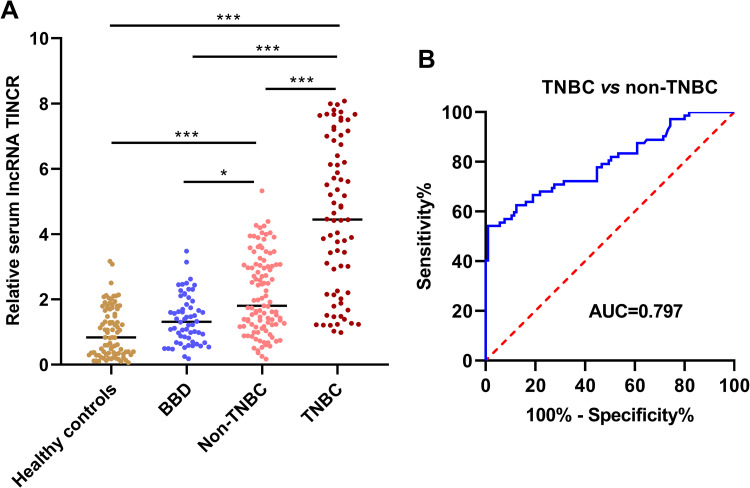
The expression levels of serum lncRNA TINCR were measured in the clinical samples of 105 patients with non-TNBC, 72 patients with TNBC, 60 patients with BBD and 86 healthy controls. The results showed that serum lncRNA TINCR was markedly higher in non-TNBC patients and TNBC patients than in patients with BBD and healthy controls. In addition, significant higher level of serum lncRNA TINCR was observed in TNBC compared to non-TNBC. No dramatic significance was found for serum lncRNA TINCR between patients with BBD and healthy controls (*P < 0.05, ***P < 0.001) (Figure 1). As revealed in Figure 1B, the area under the curve (AUC) value of serum TINCR for identifying TNBC cases from non-TNBC cases was 0.797.
Figure 1.
Serum lncRNA TINCR was increased in BC. (A) The expression level of serum lncRNA TINCR was significantly in BC, especially in TNBC (Kruskal-Wallis test, *P < 0.05, ***P < 0.0001). (B) Serum lncRNA TINCR discriminated TNBC from non-TNBC with good performance (ROC curve analysis, AUC = 0.797).
Association Between Serum TINCR Expression and Clinicopathological Features of Non-TNBC and TNBC
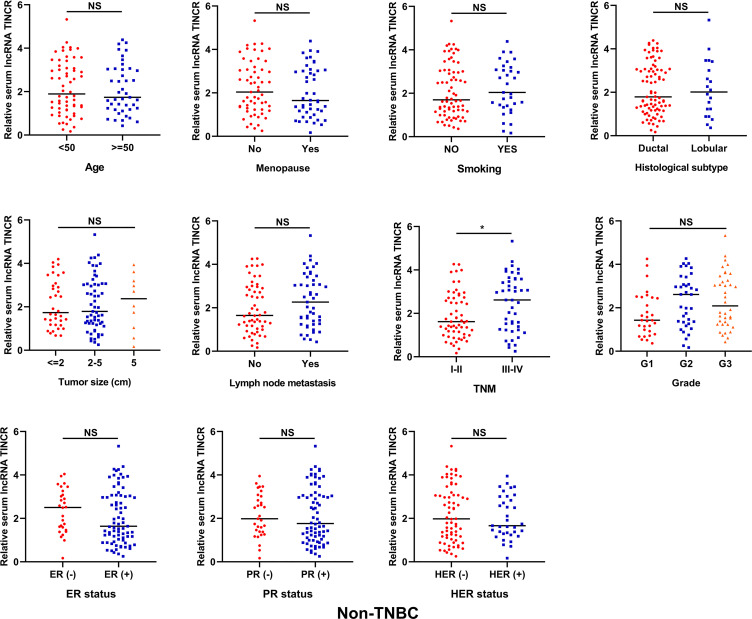
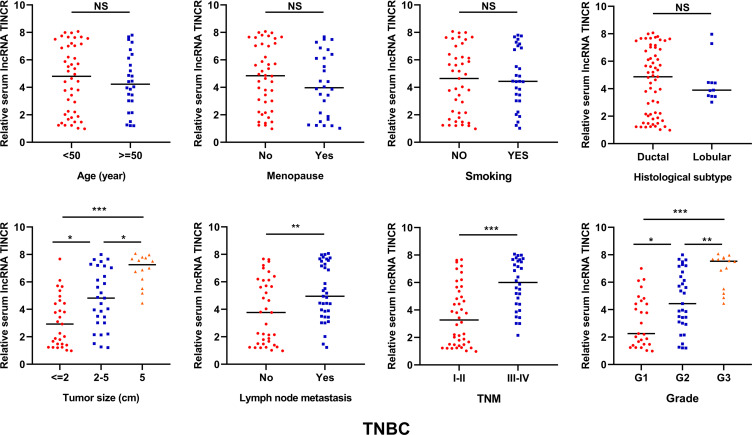
As shown in Figure 2, no significant difference was detected for serum lncRNA TINCR in non-TNBC patients stratified by different clinicopathological parameters including age, menopause, smoking, histological subtype, tumor size, lymph node metastasis, grade and ER/PR/HER2 status. The non-TNBC patients at the advanced clinical stages had higher serum lncRNA TINCR level than those at the early clinical stages (*P < 0.05). For the TNBC patients, the cases with larger tumor size, or with lymph node metastasis, or at the advanced clinical stage or with higher tumor grade had higher serum lncRNA TINCR levels than their respective controls (*P < 0.05, **P < 0.01, ***P < 0.001). However, no obvious correlations were observed between serum lncRNA TINCR expression levels and age, menopause, smoking, and histological subtype (Figure 3).
Figure 2.
The correlation between serum lncRNA TINCR and clinicopathological parameters in non-TNBC (Mann-Whitney U test, *P < 0.05).
Figure 3.
The correlation between serum lncRNA TINCR and clinicopathological parameters in TNBC (Mann-Whitney U test, *P < 0.05, **P < 0.01, ***P < 0.0001).
The Pre-Treated Serum lncRNA TINCR Level Was Strongly Associated With Recurrence in TNBC
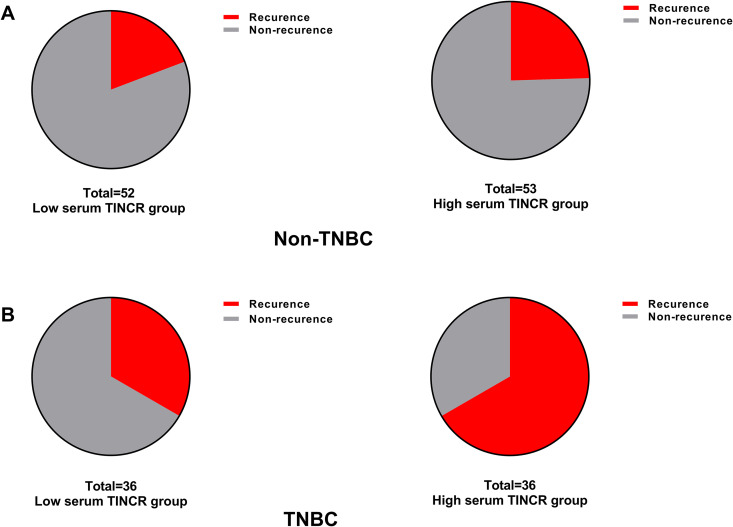
The median expression of serum lncRNA TINCR in all the TNBC cases was used to divided the TNBC patients into high (n = 36) and low (n = 36) serum lncRNA TINCR group. Similarly, the median expression of serum lncRNA TINCR in all the non-TNBC cases was used to divided the non-TNBC patients into high (n = 53) and low (n = 52) serum lncRNA TINCR group. For non-TNBC, 12 cases (23.08%) and 13 cases (24.53%) relapsed in the low and high serum lncRNA TINCR group, respectively (Figure 4A). For TNBC, 12 cases (33.33%) and 24 cases (66.67%) relapsed in the low and high serum lncRNA TINCR group, respectively (Figure 4B).
Figure 4.
The level of serum lncRNA TINCR in the pre-treated samples was strongly associated with recurrence in TNBC. (A) No significant difference for the percentage of recurrent cases was found between high and low serum lncRNA TINCR group in non-TNBC. (B) A higher percentage of recurrent cases was found in the high serum lncRNA TINCR group compared to the low serum lncRNA TINCR group.
The Prognostic Significance of Serum lncRNA TINCR in Non-TNBC and TNBC
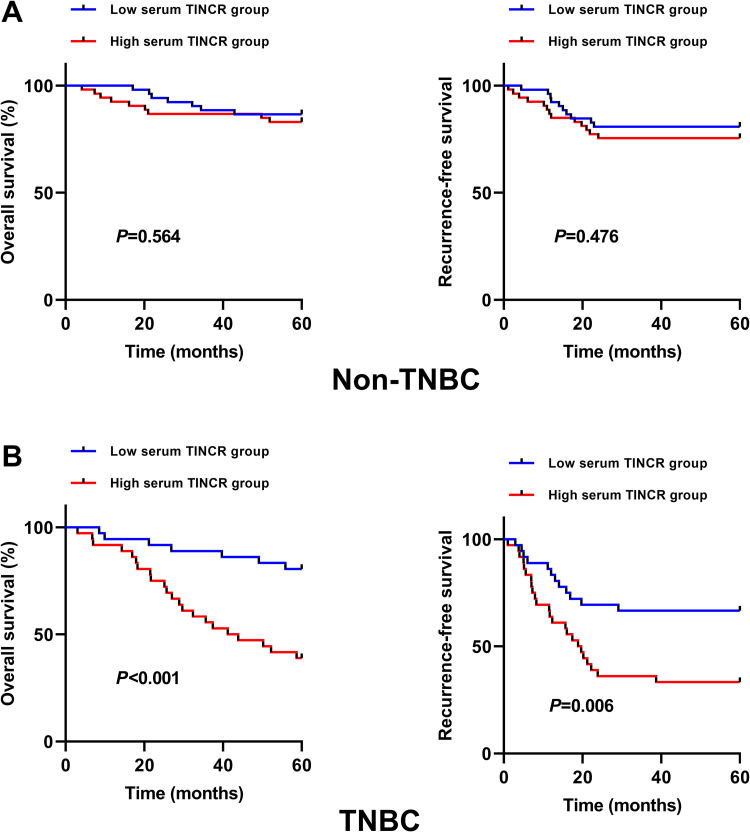
As displayed in Figure 5A, for non-TNBC, no significant difference was observed for OS and RFS between high and low serum lncRNA TINCR group. We have also analyzed the prognostic impact of serum lncRNA TINCR in different subtypes of non-TNBC. The results showed that no strong association was found between serum lncRNA TINCR and OS/RFS in different subtypes of non-TNBC (Supplementary Figure 1). For TNBC, the patients in the high serum lncRNA TINCR group had poorer OS (P < 0.001) and RFS (P = 0.006) than those in the low serum lncRNA TINCR group (Figure 5B). For non-TNBC, the multivariate analysis showed that TNM stage and grade were independent prognostic factors (Table 2). For TNBC, lymph node metastasis, TNM stage and serum lncRNA TINCR (HR = 2.540, 95%CI = 1.348-5.527, P = 0.003) were independent prognostic factors (Table 3).
Figure 5.
The correlation between serum lncRNA TINCR and OS/RFS in non-TNBC and TNBC. (A) No significant difference for OS (P = 0.564) and RFS (P = 0.476) was found between high and low serum lncRNA TINCR group in non-TNBC (Kaplan-Meier method + log-rank test). (B) The TNBC patients in the high serum lncRNA TINCR group had worse OS (P < 0.001) and RFS (P = 0.006) than those in the low serum lncRNA TINCR group (Kaplan-Meier method + log-rank test).
Table 2.
Multivariate Analysis Results of the Prognostic Factors in Non-TNBC.
| Parameters | HR | 95% CI | P |
|---|---|---|---|
| Age | 1.172 | 0.758-2.142 | 0.578 |
| Menopause | 0.894 | 0.632-1.471 | 0.669 |
| Smoking | 1.423 | 0.926-2.709 | 0.501 |
| Histological subtype | 0.995 | 0.712-2.250 | 0.743 |
| Tumor size | 1.496 | 0.931-2.840 | 0.324 |
| Lymph node metastasis | 1.732 | 0.982-3.725 | 0.102 |
| TNM stage | 2.608 | 1.327-6.529 | 0.008 |
| Grade | 2.228 | 1.163-4.876 | 0.021 |
| Serum TINCR | 1.669 | 0.965-3.558 | 0.157 |
Note: HR, hazard ration.
Table 3.
Multivariate Analysis Results of the Prognostic Factors in TNBC.
| Parameters | HR | 95% CI | P |
|---|---|---|---|
| Age | 0.931 | 0.825-2.041 | 0.771 |
| Menopause | 1.001 | 0.843-2.355 | 0.341 |
| Smoking | 1.438 | 0.923.3.210 | 0.796 |
| Histological subtype | 1.180 | 0.868-2.519 | 0.373 |
| Tumor size | 1.628 | 0.964-3.841 | 0.251 |
| Lymph node metastasis | 1.916 | 1.054-4.236 | 0.042 |
| TNM stage | 3.249 | 1.621-7.532 | <0.001 |
| Grade | 1.842 | 0.992-3.968 | 0.075 |
| Serum TINCR | 2.540 | 1.348-5.527 | 0.003 |
Note: HR, hazard ration.
Discussion
The prognosis of TNBC remains unfavorable, thus it is very important to identify novel and reliable biomarkers for predicting the clinical outcome of TNBC. In this study, we first time demonstrated that serum lncRNA TINCR was obviously upregulated in BC, especially in TNBC. In addition, serum lncRNA TINCR differentiated TNBC from non-TNBC with relatively good performance. These findings suggest that serum lncRNA TINCR might be a promising biomarker for predicting the prognosis of TNBC. Then we found that serum lncRNA TINCR was strongly associated with unfavorable clinicopathological variables and survival of TNBC. Moreover, serum lncRNA TINCR served as an independent prognostic factor of TNBC. However, little correlation was found between serum lncRNA TINCR and the prognosis of non-TNBC. Taken together, serum lncRNA TINCR might be a robust prognostic biomarker for TNBC.
Consistent with our findings, lncRNA TINCR has been shown to be a key player in the tumorigenesis of BC. For instance, Xu et al identified a cluster of overall survival-related lncRNAs in BC tissues, and bioinformatic analysis of these oncogenic lncRNAs revealed that many importance cancer related pathways were significantly affected. Downregulation of lncRNA TINCR, DSCAM-AS1 or HOTAIR suppressed the oncogenic activities of BC cells.16 A lncRNA-based prognostic model which included HOTAIR, SNHG16, HCP5, and TINCR demonstrated good performance for risk assessment of BC.17 Liu et al reported that lncRNA TINCR was overexpressed in both BC tissues and cell lines. In addition, high level of tissue lncRNA TINCR was significantly correlated with poor prognosis of BC.13 LncRNA TINCR overexpression promoted trastuzumab resistance by directly targeting HER-2 and inducing epithelial-mesenchymal transition process via targeting Snail-1. High expression of lncRNA TINCR was associated with shorter survival time in BC receiving trastuzumab therapy.18
Similarly, lncRNA TINCR also plays a tumor promoting role in other types of cancer. For example, Xu et al showed that the expression of lncRNA TINCR was significantly upregulated in gastric cancer (GC) tissues and cell lines, and might serve as a prognostic marker in GC.19 LncRNA TINCR was overexpressed in esophageal squamous cell carcinoma (ESCC) tissues. Ectopic expression of lncRNA TINCR enhanced ESCC cell proliferation, migration and invasion.20 On the contrary, it has also been reported that lncRNA TINCR might function as tumor suppressor in cancer development. LncRNA TINCR was decreased in prostate cancer tissues and cell lines. In addition, reduced lncRNA TINCR expression was closely correlated with unfavorable clinical outcome of prostate cancer.21 Zhang et al showed that lncRNA TINCR was reduced in colorectal cancer (CRC) tissues and cell lines compared with their counterparts. Loss of lncRNA TINCR promoted CRC proliferation and metastasis.22
Intriguingly, the alteration of lncRNA TINCR expression in lung cancer remains controversial. Liu et al showed that the expression level of lncRNA TINCR was significantly decreased in lung cancer tissues and cell lines.23 However, Zhu and He reported that lncRNA TINCR was overexpressed in non-small cell lung cancer tissues and cell lines.24 Such differences may be due to the heterogeneity of lung cancer, which may lead to the upregulation or downregulation of lncRNA TINCR in different subtypes or stages of the same cancer. Further studies are warranted to elucidating the contradictory findings.
A limitation of our study was the relatively small sample size. Further studies in a larger cohort are urgently needed to validate the clinical significance of serum lncRNA TINCR in TNBC. In addition, the underlying molecular mechanisms for the tumor promoting role of lncRNA TINCR in TNBC warrants deeper investigation. Admittedly, although serum lncRNA TINCR shows great promise for the predicting the prognosis of TNBC, a single biomarker is not sufficient for accurately monitoring the progression of TNBC. Therefore, combining serum lncRNA TINCR, other known biomarkers and clinicopathological information might help robustly predicting the prognosis of TNBC and improve its clinical management
In summary, our study has shown that that serum lncRNA TINCR expression was significantly higher in BC patients, especially in TNBC. In addition, high serum lncRNA TINCR is strongly associated with unfavorable clinicopathological features and survival of TNBC. Moreover, serum lncRNA TINCR is an independent prognostic factor in TNBC. Taken together, serum lncRNA TINCR might be a promising and robust biomarker for predicting the prognosis of TNBC.
Supplemental Material
Supplemental Material, Supplementary_Figure_1 for Serum lncRNA TINCR Serve as a Novel Biomarker for Predicting the Prognosis in Triple-Negative Breast Cancer by Xiaojie Wang, Shuang Li, Huiyu Xiao and Xiaoqin Deng in Technology in Cancer Research & Treatment
Footnotes
Authors’ Note: The study was approved by the Ethics Committee of the First Affiliated Hospital of Dalian Medical University, Dalian (committee approval number: DLMU2017011010).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Xiaoqin Deng  https://orcid.org/0000-0003-1877-2255
https://orcid.org/0000-0003-1877-2255
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102(10):680–691. [DOI] [PubMed] [Google Scholar]
- 3. Golubnitschaja O, Debald M, Yeghiazaryan K, et al. Breast cancer epidemic in the early twenty-first century: evaluation of risk factors, cumulative questionnaires and recommendations for preventive measures. Tumour Biol. 2016;37:12941–12957. [DOI] [PubMed] [Google Scholar]
- 4. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin L, Chang HY. Uncovering the role of genomic “dark matter” in human disease. J Clin Invest. 2012;122(5):1589–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng C, Zhang B, Zhang Y, et al. A long non-coding RNA OLBC15 promotes triple-negative breast cancer progression via enhancing ZNF326 degradation. J Clin Lab Anal. 2020;34(8):e23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xia E, Shen Y, Bhandari A, et al. Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am J Cancer Res. 2018;8(7):1273–1287. [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang K, Luo Z, Zhang Y, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17(2):187–194. [DOI] [PubMed] [Google Scholar]
- 10. Mitobe Y, Ikeda K, Sato W, et al. Proliferation-associated long noncoding RNA, TMPO-AS1, is a potential therapeutic target for triple-negative breast cancer. Cancer Sci. 2020;111(7):2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo S, Jian L, Tao K, Chen C, Yu H, Liu S. Novel breast-specific long non-coding RNA LINC00993 acts as a tumor suppressor in triple-negative breast cancer. Front Oncol. 2019;9:1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10(10):1597–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Du Y, Hu X, Zhao L, Xia W. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer. 2018;18(1):367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2017;37(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hou A, Zhang Y, Zheng Y, Fan Y, Liu H, Zhou X. LncRNA terminal differentiation-induced ncRNA (TINCR) sponges miR-302 to upregulate cyclin D1 in cervical squamous cell carcinoma (CSCC). Hum Cell. 2019;32(4):515–521. [DOI] [PubMed] [Google Scholar]
- 16. Xu S, Kong D, Chen Q, Ping Y, Pang D. Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer. 2017;16(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Gao C, Liu C, et al. Four lncRNAs associated with breast cancer prognosis identified by coexpression network analysis. J Cell Physiol. 2019;234(8):14019–14030. [DOI] [PubMed] [Google Scholar]
- 18. Dong H, Hu J, Zou K, et al. Activation of LncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer. 2019;18(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648–5661. [DOI] [PubMed] [Google Scholar]
- 20. Xu Y, Qiu M, Chen Y, et al. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29(8):950–958. [DOI] [PubMed] [Google Scholar]
- 21. Dong L, Ding H, Li Y, Xue D, Liu Y. LncRNA TINCR is associated with clinical progression and serves as tumor suppressive role in prostate cancer. Cancer Manag Res. 2018;10:2799–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang ZY, Lu YX, Zhang ZY, et al. Loss of TINCR expression promotes proliferation, metastasis through activating EpCAM cleavage in colorectal cancer. Oncotarget. 2016;7(16):22639–22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu X, Ma J, Xu F, Li L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed Pharmacother. 2018;99:9–17. [DOI] [PubMed] [Google Scholar]
- 24. Zhu ZJ, He JK. TINCR facilitates non-small cell lung cancer progression through BRAF-activated MAPK pathway. Biochem Biophys Res Commun. 2018;497(4):971–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplementary_Figure_1 for Serum lncRNA TINCR Serve as a Novel Biomarker for Predicting the Prognosis in Triple-Negative Breast Cancer by Xiaojie Wang, Shuang Li, Huiyu Xiao and Xiaoqin Deng in Technology in Cancer Research & Treatment