Abstract
Liver fibrosis is an increasing health problem worldwide, for which no effective antifibrosis drugs are available. Although the involvement of aerobic glycolysis in hepatic stellate cell (HSC) activation has been reported, the role of pyruvate kinase M2 (PKM2) in liver fibrogenesis still remains unknown. We examined PKM2 expression and location in liver tissues and primary hepatic cells. The in vitro and in vivo effects of a PKM2 antagonist (shikonin) and its allosteric agent (TEPP-46) on liver fibrosis were investigated in HSCs and liver fibrosis mouse model. Chromatin immunoprecipitation sequencing and immunoprecipitation were performed to identify the relevant molecular mechanisms. PKM2 expression was significantly up-regulated in both mouse and human fibrotic livers compared with normal livers, and mainly detected in activated, rather than quiescent, HSCs. PKM2 knockdown markedly inhibited the activation and proliferation of HSCs in vitro. Interestingly, the PKM2 dimer, rather than the tetramer, induced HSC activation. PKM2 tetramerization induced by TEPP-46 effectively inhibited HSC activation, reduced aerobic glycolysis, and decreased MYC and CCND1 expression via regulating histone H3K9 acetylation in activated HSCs. TEPP-46 and shikonin dramatically attenuated liver fibrosis in vivo. Our findings demonstrate a nonmetabolic role of PKM2 in liver fibrosis. PKM2 tetramerization or suppression could prevent HSC activation and protects against liver fibrosis.
Liver fibrosis is the pathologic accumulation of extracellular matrix and fibrous scar tissues associated with chronic liver injury. Persistent liver injury and progressive fibrosis lead to cirrhosis, which is an increasing health concern worldwide.1, 2, 3, 4 Moreover, the hepatic stellate cells (HSCs) play a critical role in the pathogenesis of liver fibrosis.5, 6, 7 On chronic liver injury, quiescent HSCs are activated and transdifferentiate from vitamin A–and retinoid lipid droplet–storing cells into proliferating and fibrogenic myofibroblasts expressing α-smooth muscle actin (α-SMA).5,7, 8, 9 In recent years, the functional importance of HSC activation as the main collagen-producing cells in liver fibrosis has been convincingly identified.7,10, 11, 12 But as yet, our understanding on the regulation of HSCs still remains far from clear.
Aerobic glycolysis (alias Warburg effect), in which cells generate ATP through glycolysis, is regarded as a hallmark of malignant tumors,13, 14, 15, 16 including hepatocellular carcinoma.17, 18, 19 However, current evidence suggests that it is also a characteristic metabolic trait of activated HSCs during liver fibrogenesis.20,21 The pyruvate kinase M1/2 (PKM) gene encodes two protein isoforms, PKM1 and PKM2, which are generated by mutually exclusive alternative splicing.22,23 It has been reported that PKM2, but not PKM1, is up-regulated in tumor cells and correlates with aerobic glycolysis.24,25 PKM1 is constitutively expressed as a high-pyruvate kinase (PK) activity form that converts phosphoenolpyruvic acid to pyruvate and drives the tricarboxylic acid cycle, whereas PKM2 may assemble both in a high-PK activity tetrameric form and in a low-PK activity dimeric form.26 Tetrameric PKM2 possesses the PKM1-like kinetic ability to convert phosphoenolpyruvic acid to pyruvate, whereas the PKM2 dimer has protein kinase activity and can translocate to the nucleus,27 where it takes part in histone modification events and regulates downstream genes. Previous evidence showed that in tumor cells the nuclear PKM2 dimer up-regulates proglycolysis enzymes, thus promoting aerobic glycolysis, by activating the expression of specific transcription factors, including MYC and CCND1.28,29
Although an important function of the PKM2 dimer has been reported in tumor cells, it is still unclear whether it affects HSC activation. In the present study, we investigated the molecular function, and the role of dimeric PKM2 in particular, in the activation of HSCs, providing evidence that PKM2 tetramerization (dimer-to-tetramer transformation) protects against HSC activation and liver fibrosis.
Materials and Methods
Chemicals
For in vitro experiments, TEPP-46 (MCE, Monmouth Junction, NJ), DASA-58 (MCE), and shikonin (Selleck Chemicals, Houston, TX) were dissolved in dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO) and further diluted to the required concentration. For in vivo experiments, TEPP-46 and shikonin suspensions were prepared in 0.5% carboxymethyl cellulose sodium normal saline solution.
Human Liver Samples
A total of 21 normal liver, 23 fibrotic liver, and 43 hepatocellular carcinoma formalin-fixed, paraffin-embedded tissue samples were used in this study. Normal liver samples were obtained from individuals who underwent liver resection for benign hepatic lesions. Fibrotic liver and hepatocellular carcinoma tissues were obtained from patients who received liver biopsy or resection. All of the procedures involving human samples were approved by the Integrated Hospital of Traditional Chinese Medicine of Southern Medical University Office for Protection of Human Subjects.
Mouse Liver Fibrosis Model
All animal protocols were approved by the Animal Care and Use Committee of Southern Medical University. The animals were housed under a controlled temperature (20°C ± 2°C) with 12-hour light/12-hour dark cycles and ad libitum access to food and water.
Six-week–old male C57BL/6 mice (Experimental Animal Center of Southern Medical University, Guangzhou, China) received 0.1 mL 40% carbon tetrachloride (CCl4; 2:5 v/v in olive oil) or vehicle (olive oil only) gavage three times a week for 8 weeks. A subset of mice received daily gavage of TEPP-46 (20 mg/kg) or shikonin (10 mg/kg) or vehicle (0.5% carboxymethyl cellulose sodium normal saline solution) during weeks 5 to 8 (N > 5 for all groups). The animals were sacrificed 3 days after the final treatment. The liver and serum were harvested for histologic, biochemical, and molecular analyses.
Cell Cultures
LX-2 cells and the isolated primary mouse cells were grown in high-glucose Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum, 100 U/mL of penicillin, and 100 μg/mL of streptomycin and maintained in a 5% CO2/water-saturated incubator at 37°C.
Primary Mouse Cell Isolation
Primary mouse hepatocytes, HSCs, macrophages, and liver sinusoidal endothelial cells were isolated and cultured as previously reported.30 After the mice were anesthetized with 2% pentobarbital sodium, the abdominal cavity was opened, and the liver was perfused and digested in situ. Next, the liver was harvested, minced, homogenized under sterile conditions, and further digested ex vivo. Then, the digested liver was filtered through a 70-μm steel mesh, and the cells were separated by density gradient centrifugation.
For HSC isolation, the liver was successively perfused in situ with EGTA solution, collagenase D (Roche, Basel, Switzerland) solution, and pronase (Sigma-Aldrich) solution, and was digested ex vivo with a buffer containing collagenase D, pronase, and DNase (Roche). For hepatocyte isolation, the pronase was removed from the in situ and ex vivo digestive solution. For immunocytes, macrophages and liver sinusoidal endothelial cells were separated via magnetic beads (Miltenyi Biotec, Cologne, Germany) after density gradient centrifugation.
RNA Interference and Lentivirus
Specific siRNA against human or mouse PKM2 was used in this study.31 The siRNA was synthesized by Ribobio Co (Guangzhou, China), and was transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA).
The PKM-overexpression lentivirus was constructed by Genechem Co (Shanghai, China). The viruses were used to infect LX-2 cells with 8 μg/mL Polybrene (Sigma-Aldrich) and then were selected using puromycin (Invitrogen) for 7 days. All siRNA or shRNA sequences are shown in Table 1.
Table 1.
Sequence-Based Reagents Used in the Study
| Name | Sequence | Supplier |
|---|---|---|
| PKM2 mouse qPCR forward primer | 5′-AGGCTGCCATCTACCACTTG-3′ | Life Technologies (Grand Island, NY) |
| PKM2 mouse qPCR reverse primer | 5′-CCAGACTTGGTGAGCACGAT-3′ | Life Technologies |
| α-SMA mouse qPCR forward primer | 5′-TCCCTGGAGAAGAGCTACGAACT-3′ | Life Technologies |
| α-SMA mouse qPCR reverse primer | 5′-AAGCGTTCGTTTCCAATGGT-3′ | Life Technologies |
| Col1a1 mouse qPCR forward primer | 5′-CCTGGCAAAGACGGACTCAAC-3′ | Life Technologies |
| Col1a1 mouse qPCR reverse primer | 5′-GCTGAAGTCATAACCGCCACTG-3′ | Life Technologies |
| TIMP1 mouse qPCR forward primer | 5′-CCAGAACCGCAGTGAAGAGT-3′ | Life Technologies |
| TIMP1 mouse qPCR reverse primer | 5′-GTACGCCAGGGAACCAAGAA-3′ | Life Technologies |
| TGF-β1 mouse qPCR forward primer | 5′-ATTCAGCGCTCACTGCTCTT-3′ | Life Technologies |
| TGF-β1 mouse qPCR reverse primer | 5′-ATGTCATGGATGGTGCCCAG-3′ | Life Technologies |
| GAPDH mouse qPCR forward primer | 5′-GGACCTCATGGCCTACATGG-3′ | Life Technologies |
| GAPDH mouse qPCR reverse primer | 5′-TAGGGCCTCTCTTGCTCAGT-3′ | Life Technologies |
| PAI-1 mouse qPCR forward primer | 5′-GACACCCTCAGCATGTTCATC-3′ | Life Technologies |
| PAI-1 mouse qPCR reverse primer | 5′-AGGGTTGCACTAAACATGTCAG-3′ | Life Technologies |
| PDGFRβ mouse qPCR forward primer | 5′-AGCCAGAAGTAGCGAGAAGC-3′ | Life Technologies |
| PDGFRβ mouse qPCR reverse primer | 5′-GGCAGTATTCCGTGATGATG-3′ | Life Technologies |
| PKM2 human qPCR forward primer | 5′-AGCATGATCAAGAAGCCCCG-3′ | Life Technologies |
| PKM2 human qPCR reverse primer | 5′-TCTGTGGGGTCGCTGGTAA-3′ | Life Technologies |
| α-SMA human qPCR forward primer | 5′-TGAGAAGAGTTACGAGTTGCCTG-3′ | Life Technologies |
| α-SMA human qPCR reverse primer | 5′-GTTAGCATAGAGGTCCTTCCTGATG-3′ | Life Technologies |
| Col1a1 human qPCR forward primer | 5′-CAGCCGCTTCACCTACAGC-3′ | Life Technologies |
| Col1a1 human qPCR reverse primer | 5′-TCAATCACTGTCTTGCCCCA-3′ | Life Technologies |
| TIMP1 human qPCR forward primer | 5′-TGTTGTTGCTGTGGCTGATAGC-3′ | Life Technologies |
| TIMP1 human qPCR reverse primer | 5′-TCTGGTGTCCCCACGAACTT-3′ | Life Technologies |
| TGF-β1 human qPCR forward primer | 5′-GGCCAGATCCTGTCCAAGC-3′ | Life Technologies |
| TGF-β1 human qPCR reverse primer | 5′-GTGGGTTTCCACCATTAGCAC-3′ | Life Technologies |
| GAPDH human qPCR forward primer | 5′-TGCACCACCAACTGCTTAGC-3′ | Life Technologies |
| GAPDH human qPCR reverse primer | 5′-GGCATGGACTGTGGTCATGAG-3′ | Life Technologies |
| PAI-1 human qPCR forward primer | 5′-AGTGGACTTTTCAGAGGTGGA-3′ | Life Technologies |
| PAI-1 human qPCR reverse primer | 5′-GCCGTTGAAGTAGAGGGCATT-3′ | Life Technologies |
| PDGFRβ human qPCR forward primer | 5′-GCCCTTATGTCGGAGCTGAAGA-3′ | Life Technologies |
| PDGFRβ human qPCR reverse primer | 5′-GTTGCGGTGCAGGTAGTCCA-3′ | Life Technologies |
| Myc mouse ChIP forward primer | 5′-CTGGGTTGGCAATTCACTCC-3′ | Life Technologies |
| Myc mouse ChIP reverse primer | 5′-GTGGGGAAAATCAAGGCGCT-3′ | Life Technologies |
| Ccnd1 mouse ChIP forward primer | 5′-GATACCCCTCTGGCCCTTTG-3′ | Life Technologies |
| Ccnd1 mouse ChIP reverse primer | 5′-GATGCCAGACGAGCCCTAAG-3′ | Life Technologies |
| Human PKM2 siRNA target sequence | 5′-CCATAATCGTCCTCACCAA-3′ | Ribobio |
| Human PKM1 siRNA target sequence | 5′-GCGTGGAGGCTTCTTATAA-3′ | Ribobio |
| Mouse PKM2 siRNA target sequence 1 | 5′-CCCTGTGCTGTGTAAGGAT-3′ | Ribobio |
| Mouse PKM2 siRNA target sequence 2 | 5′-GATGTCGACCTTCGTGTAA-3′ | Ribobio |
| Mouse PKM2 siRNA target sequence 3 | 5′-TCCTATCATTGCCGTGACT-3′ | Ribobio |
Ccnd1, cyclin D1; ChIP, chromatin immunoprecipitation; Col1a1, collagen type I alpha 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PAI-1 (SERPINE1), plasminogen activator inhibitor type-1; PDGFRβ (PDGFRB), platelet-derived growth factor receptor-β; PKM2, pyruvate kinase M2; qPCR, quantitative PCR; α-SMA, α-smooth muscle actin; TGF-β1 (TGFB1), transforming growth factor-β1; TIMP1, tissue inhibitor of metalloproteinases 1.
Liver Function Test and Hydroxyproline Assay
Serum levels of several biochemical markers were measured to assess liver function and liver injury. Serum alkaline phosphatase, alanine aminotransferase, aspartate aminotransferase, total bilirubin, albumin, and glucose were measured using the Roche Cobas 6000 Analyzer (Roche). Levels of hydroxyproline were detected using a colorimetric hydroxyproline assay kit (Abcam, Cambridge, UK), according to the manufacturer's instructions.
Histology, Immunohistochemistry, and Immunofluorescence
Histology, immunohistochemistry, and immunofluorescence were performed as previously reported.32 Paraffin-embedded liver tissue samples were sectioned at 4 mm thickness. After routine processing, liver sections were stained with hematoxylin-eosin for histologic analysis or saturated picric acid containing 0.1% Sirius Red and 0.1% Fast Green for collagen deposition.
For immunohistochemistry, liver sections were incubated with specific antibodies overnight at 4°C. The immunodetection was performed on the second day using EnVision immunodetection system (K5007; Dako, Copenhagen, Denmark), according to the manufacturer's instructions.
For immunofluorescence, liver sections or cells were fixed with ice-cold methanol and stained with specific antibodies. And the immunodetection was performed by the species-matched secondary antibodies labeled with Alexa Fluor 647, 555, or 488, according to the manufacturer's instructions. Nuclei were counterstained with DAPI. A Zeiss LSM 800 confocal microscope (Carl Zeiss, Oberkochen, Germany) was used for the immunofluorescence.
Cell Proliferation Assay
Proliferation was determined by seeding 2500 LX-2 cells or 5000 primary mouse HSCs per well in 96-well plates. The cells were treated with indicated chemotherapeutic agents or transfected with siRNA for the indicated time. Cell viability was measured using the Cell Counting Assay Kit-8 (Dojindo, Kumamoto, Japan), according to the manufacturer's instructions.
Cell Cycle and Apoptosis Analysis
Cell cycle and apoptosis of the indicated cells were respectively determined using the cell cycle detection kit (BD Biosciences, San Jose, CA) or Annexin V Apoptosis Detection Kit (BD Biosciences), according to the manufacturer's instructions, by a BD LSRFortessa X-20 (BD, Biosciences). The data were analyzed using FlowJo software version 10.4 (Tree Star, Inc., San Carlos, CA).
Wound-Healing Assays
For the wound-healing assay, LX-2 cells were seeded into a 6-well plate; straight lines were delineated at the bottom of the 6-well plate after incubation for 12 hours. Then, the width was respectively measured after 0 and 12 hours.
Transwell Assay
For the cell migration assay, LX-2 cells were plated in the upper compartment of a polycarbonate Transwell insert in serum-free Dulbecco’s modified Eagle’s medium. Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum as chemoattractant was added in the lower compartment. After 12 hours of incubation, the cells that go through the compartment were stained using Hoechst 33342 (Thermo Scientific, Waltham, MA). Then, the stained cells were counted as the mean number of cells per field of view.
Real-Time Quantitative PCR Assay
Total mRNA was obtained from fresh tissues or cultured cells with TRIzol Reagent (15596-026; Invitrogen), and 1 μg of mRNA was used for cDNA synthesis with a Transcriptor First Stand cDNA Synthesis Kit (TakaRa Bio Inc., Shiga, Japan), according to the manufacturer's protocol. Real-time quantitative PCR was performed according to the manufacturer's instructions using the SYBR Green (Roche) and LightCycler480II system (Roche). Samples were run in triplicates; the results were normalized to glyceraldehyde-3-phosphate dehydrogenase expression by using the 2−ΔΔ Ct method. Gene-specific primers are listed in Table 1.
Western Blot Analysis
Protein was extracted from the cells using radioimmunoprecipitation assay buffer (Thermo Scientific), separated by SDS-PAGE gel electrophoresis and transferred to polyvinylidene difluoride membranes. Then, the indicated primary antibodies were used for immunodetection with horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence reagents (Pierce, Rockford, IL). The antibodies used for Western blot analysis are shown in Table 2.
Table 2.
Antibodies Used in the Study
| Name | Supplier | Catalog no. | Clone no. | RRID |
|---|---|---|---|---|
| PKM2 | Cell Signaling (Danvers, MA) | 4053S | D78A4 | AB_1904096 |
| Phosphorylated PKM2 | Cell Signaling | 3827S | NA | AB_1950369 |
| PKM1 | Cell Signaling | 7067S | D30G6 | AB_2715534 |
| CD31 (PECAM-1) | Cell Signaling | 3528S | 89C2 | AB_2160882 |
| GAPDH | Cell Signaling | 5174S | D16H11 | AB_10622025 |
| PARP | Cell Signaling | 9532S | 46D11 | AB_659884 |
| c-Myc | Cell Signaling | 5605S | D84C12 | AB_1903938 |
| Cyclin D1 | Cell Signaling | 2978S | 92G2 | AB_2259616 |
| Rabbit IgG | Cell Signaling | 7077S | NA | AB_10694715 |
| Anti-mouse IgG (HRP) | Cell Signaling | 7076S | NA | AB_330924 |
| Anti-rabbit IgG (HRP) | Cell Signaling | 7074S | NA | AB_2099233 |
| Histone H3 | Abcam (Cambridge, MA) | ab195277 | NA | NA |
| Histone H3 (acetyl K9) | Abcam | ab12179 | NA | AB_298910 |
| Histone H3 (phosphorylated T11) | Abcam | Ab5168 | NA | AB_10858648 |
| α-SMA | Abcam | ab5694 | NA | AB_2223021 |
| α-SMA (Alexa Fluor 488) | Abcam | ab197240 | E184 | NA |
| PDGFRβ | Abcam | ab32570 | NA | AB_777165 |
| PAI1 | Abcam | ab182973 | NA | NA |
| CK19 | Abcam | A53-B/A2 | NA | AB_1156465 |
| CD68 | Abcam | ab955 | NA | AB_307338 |
| KAT3B/p300 | Abcam | ab14984 | NA | AB_301550 |
| Ep300 (Ser1834) | Bioss (Beijing, China) | BS-5339R | NA | NA |
| Fibronectin | Santa (Santa Cruz, CA) | sc-69681 | NA | AB_1122900 |
| H3K9me3 | ABclonal (Boston, MA) | A2360 | NA | AB_2721266 |
| HDAC3 | Proteintech (Rosemont, IL) | 10255-1-AP | NA | AB_2279733 |
| CYP2E1 | Proteintech | 19937-1-AP | NA | AB_10646444 |
CK19, cytokeratin 19; CYP2E1, cytochrome P4502E1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC3, histone deacetylase 3; HRP, horseradish peroxidase; NA, not applicable; PAI1, plasminogen activator inhibitor type-1; PARP, poly (ADP-ribose) polymerase; PDGFRβ, platelet-derived growth factor receptor-β; PECAM-1, platelet-endothelial cell adhesion molecule-1; PKM2, pyruvate kinase M2; RRID, Research Resource Identifier; α-SMA, α-smooth muscle actin.
Nuclear and Cytoplasmic Extraction
NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) were used according to manufacturer’s recommendations. The extractive of nuclear and cytoplasmic was further analyzed by Western blotting.
Cross-Linking Study to Determine Tetramers, Dimers, and Monomers of PKM2
In LX-2 cells and primary mouse HSCs, we used 2 mmol/L disuccinimidyl suberate (Thermo Scientific), according to the manufacturer's instructions, to cross-link cells for 30 minutes at room temperature. After the cross-linking, equal numbers of cells were lysed with radioimmunoprecipitation assay buffer, and the lysate was boiled with protein loading buffer (Thermo Scientific) for 5 minutes. Then, the samples were separated by SDS-PAGE gel electrophoresis and transferred to polyvinylidene difluoride membranes. Then, membranes were incubated with 0.4% paraformaldehyde in phosphate-buffered saline for 30 minutes at room temperature before PKM2 antibody was added for detection of tetramers, dimers, and monomers of PKM2.
Immunoprecipitation
Cells for co-immunoprecipitation experiments were lysed in ice-cold IP Lysis Buffer (Thermo Scientific) containing a protease and phosphatase inhibitor cocktail (Thermo Scientific) for 5 minutes, followed by centrifugation at 13,000 × g for 10 minutes. The indicated antibodies (Table 2) were incubated with Dynabeads Protein G beads (Thermo Scientific) for 10 minutes at room temperature. Then, the beads were incubated with the obtained cell lysates for 12 hours at 4°C. The immunocomplexes were washed five to six times with lysis buffer, denaturing eluted, and analyzed by Western blotting, as previously described (Western Blot Analysis).
ChIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed using EZ-Magna ChIP Chromatin Immunoprecipitation kit (Millipore, Billerica, MA), according to manufacturer's instructions. Briefly, cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. Then, cells were harvested, lysed, and sonicated for 30 cycles of 30 seconds on/30 seconds off and high mode with Diagenode Bioruptor (Diagenode, Liège, Belgium) to obtain adequate fragment sizes of DNA. Antibody against ace-H3K9 and rabbit IgG was used for immunoprecipitation. The precipitated DNA was subjected to PCR amplification using the ChIP-eluted DNA as template. The specific primers used in ChIP-PCR assay are listed in Table 1.
Glucose and Lactate Detection
The 24-hour glucose consumption by LX-2 cells and primary mouse HSCs was measured with a Glucose (HK) Assay Kit (Sigma-Aldrich). Cells were seeded in 24-well plates with or without treatment of indicated chemotherapeutic agents. After 24 hours, glucose in the medium was detected according to the manufacturer's instructions. Relative fluorescence units were determined at 340 nm using a Multiskan Sky (Thermo Scientific). The levels of lactate production were examined with an l-lactate assay kit (Abcam). Cells were seeded in a 6-well plate at a density of 1 × 106 cells per well. After incubation with or without treatment of indicated chemotherapeutic agents for 24 hours, the cells were collected, lactate assays were performed according to the manufacturer's protocol, and the optical density was measured at 450 nm using a Multiskan Sky.
Public Data Set
Gene expression profiles based on gene chips of fibrotic liver tissues were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo; accession number GSE84044). Expression profiles of these data sets were reanalyzed using R version 3.5.0 and correlated packages (R Software, Vienna, Austria; http://www.r-project.org).
Statistical Analysis
The t-test or the U-test was used to compare values between subgroups. Data are expressed as means ± SD of at least three biological replicates. The threshold for statistical significance was set at P < 0.05. All analyses were performed using SPSS software version 23.0 (IBM, Armonk, NY).
Results
The Expression of PKM2 Is Enhanced in the Fibrotic Liver, and Mainly Occurs in Activated HSCs
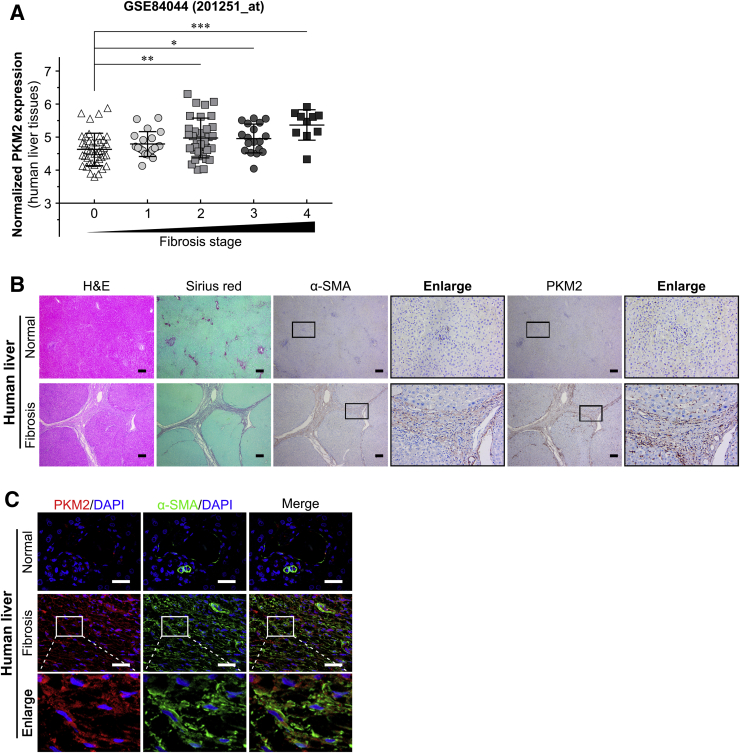
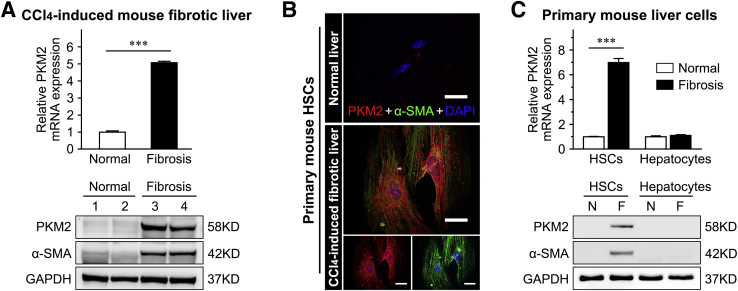
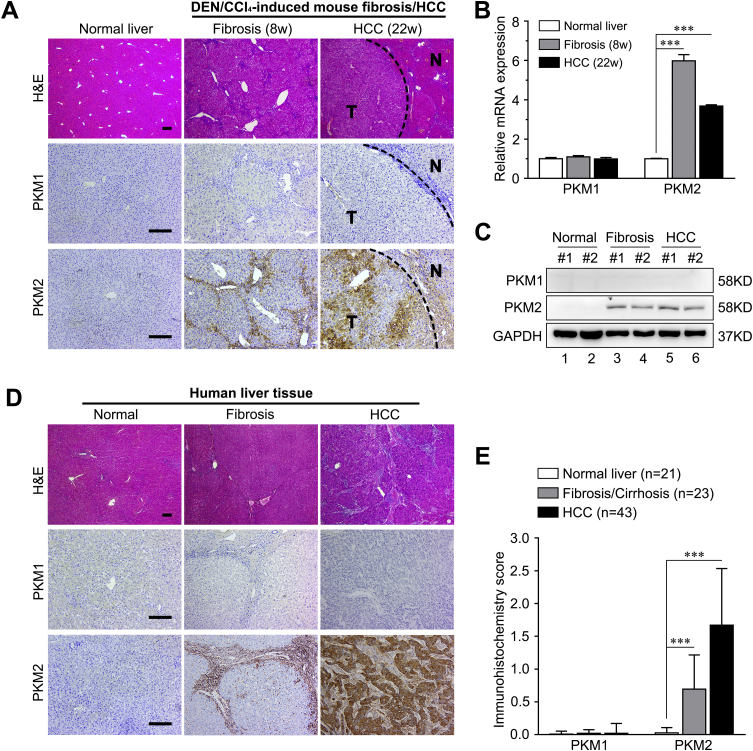
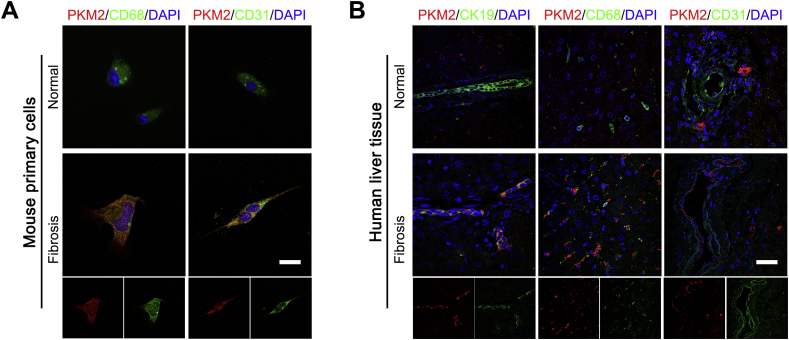
Publicly available data from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE84044) was analyzed to explore the exact role of PKM2 in liver fibrosis/cirrhosis.33 The result indicated that the PKM2 mRNA was markedly increased in human fibrotic livers in association with fibrosis progression (Figure 1A). And the level of PKM2 protein was also significantly enhanced in human fibrotic liver tissues, as assessed by immunostaining (Figure 1, B and C), but PKM2 was absent in normal human or mouse livers (Supplemental Figure S1). Real-time quantitative PCR and Western blotting further confirmed that both mRNA and protein PKM2 expression were dramatically increased in mouse CCl4-induced fibrotic liver tissues (Figure 2A and Supplemental Figure S1). In addition, immunostaining also indicated that, in fibrotic liver tissues, PKM2 expression mainly occurred in mesenchymal cells exhibiting fiber deposition, but not in the hepatocytes (Figure 1B). To further confirm this result and determine the PKM2 localization in fibrotic liver tissues, immunofluorescence experiments were performed by costaining PKM2 with markers of activated HSCs (α-SMA), biliary tract cells (cytokeratin 19), Kupffer cells (CD68), or endothelial cells (CD31). The strongest colocalization was observed between PKM2 and α-SMA, indicating that the increase in PKM2 expression mainly took place in activated HSCs (Figure 1C and Supplemental Figure S2). In addition, PKM2 expression was also up-regulated in Kupffer cells of fibrotic liver tissues, possibly reflecting inflammatory events accompanying liver fibrosis.34 Next, PKM2 expression was compared in primary mouse liver cells isolated from normal or CCl4-induced fibrotic mouse liver. It was found that PKM2 expression was increased in isolated primary activated HSCs rather than quiescent cells from normal mouse liver (Figure 2, B and C), whereas it was absent in primary hepatocytes from both normal and fibrotic mouse liver (Figure 2C). These results indicated that PKM2 expression was enhanced in fibrotic liver, and was mainly contributed by activated HSCs.
Figure 1.
Pyruvate kinase M2 (PKM2, alias PKM) expression is enhanced in human fibrotic liver. A: PKM2 mRNA expression in human fibrotic liver tissues at different Scheuer fibrosis stages (GSE84044). B: Serial sections of human normal or fibrotic liver tissues were subjected to hematoxylin-eosin (H&E) staining, Sirius red staining, and immunohistochemistry [α-smooth muscle actin (α-SMA) and PKM2], and representative images are shown. Boxed areas are shown at higher magnification to the right. C: Representative double immunofluorescence images of α-SMA (green) and PKM2 (red) labeling in human normal and fibrotic liver tissues. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars: 200 μm (B); 50 μm (C).
Figure 2.
Pyruvate kinase M2 (PKM2, alias PKM) expression is enhanced in mouse fibrotic liver and primary activated hepatic stellate cells (HSCs). A: PKM2 mRNA and protein expression in mouse normal or carbon tetrachloride (CCl4)–induced fibrotic livers. B: Representative double immunofluorescence images of α-smooth muscle actin (α-SMA; green) and PKM2 (red) labeling in primary HSCs isolated from mouse normal or CCl4-induced fibrotic livers. C: PKM2 mRNA and protein expression in primary mouse hepatocytes and HSCs isolated from normal or CCl4-induced fibrotic livers. ∗∗∗P < 0.001. Scale bars = 50 μm (B). F, fibrosis; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; N, normal.
The expression of PKM1, the other alternatively spliced product of the PKM gene, was also evaluated in both mouse and human liver samples. PKM1 expression was not detected in either normal or fibrotic liver tissues. In addition, in human or diethylnitrosamine/CCl4–induced mouse hepatocellular carcinoma samples, only the expression of PKM2, not that of PKM1, could be detected and was up-regulated (Supplemental Figure S1).
PKM2 Promotes the Activation and Proliferation of HSCs
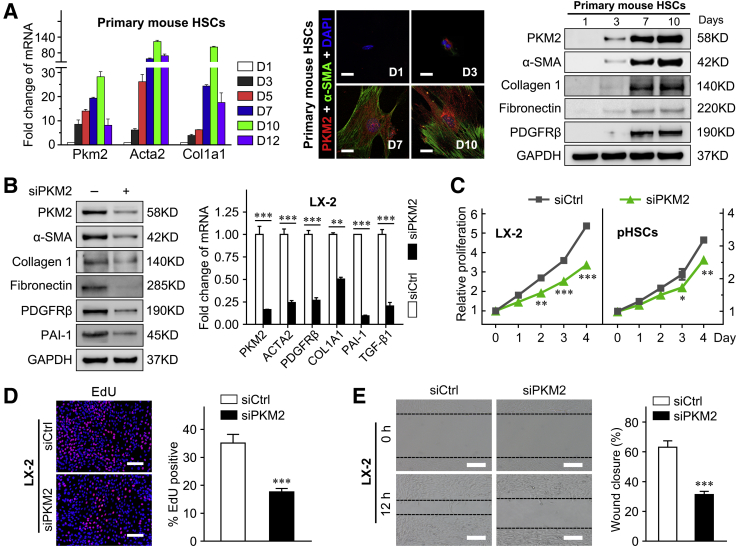
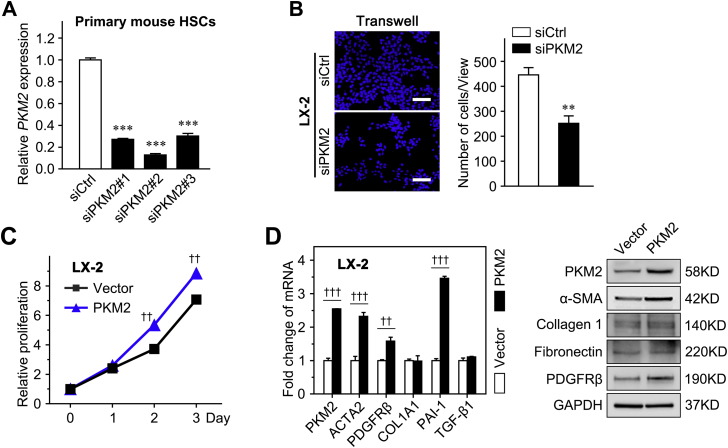
It was already known that primary HSCs plated on plastic gradually activate within 2 weeks.32 Thus, to investigate the exact role of PKM2 in HSC activation, its expression along with that of HSC activation markers (Acta2 and Col1a1) was examined in cultured primary mouse HSCs, at different times of differentiation (from day 1 to 12). It was found the mRNA level of PKM2 gradually increased during HSC activation, along with the up-regulation of Acta2 and Col1a1 (Figure 3A). The PKM2 protein was detected by immunofluorescence and Western blot at an early stage of HSC activation, confirming that PKM2 expression increased in parallel with that of the HSC activation and fibrosis markers [α-SMA, collagen 1, fibronectin, and platelet-derived growth factor receptor-β (PDGFRβ, alias PDGFRB)] (Figure 3A). Then, siRNA was used to interfere with PKM2 expression in the human activated HSC line, LX-2, and showed that PKM2 knockdown dramatically diminished the expression of various profibrogenic genes (Figure 3B). In addition, PKM2 knockdown inhibited the proliferation of both LX-2 cells and primary mouse HSCs (Figure 3, C and D, and Supplemental Figure S3A), and attenuated the migration capability of LX-2 cells (Figure 3E and Supplemental Figure S3B). In contrast, PKM2 overexpression enhanced the expression of HSC activation markers and promoted the proliferation of LX-2 cells (Supplemental Figure S3, C and D).
Figure 3.
Pyruvate kinase M2 (PKM2, alias PKM) promotes the activation and proliferation of hepatic stellate cells (HSCs). A: Left panel: Fold-change of Pkm2, Acta2, and Col1a1 mRNA expression in primary HSCs isolated from mouse normal liver and cultured for the indicated periods. Double immunofluorescence staining (middle panel) and Western blot (right panel) analysis of the indicated genes in cultured HSCs at the indicated days. B: The mRNA and protein level of PKM2 and HSC activation markers in LX-2 cells after transfection with control siRNA (siCtrl) or PKM2 siRNA (siPKM2) for 72 hours. C: LX-2 cells and primary mouse HSCs were transfected with control or PKM2 siRNA, and then subjected to Cell Counting Assay Kit-8 assay. D: The proliferation of LX-2 cells transfected with control siRNA or PKM2 siRNA is measured using the 5-ethynyl-2′-deoxyuridine (EdU) assays. E: The migration capability of LX-2 cells transfected with control siRNA or PKM2 siRNA is measured using the wound-healing assays. Dashed lines indicate the edge of cell migration. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus siCtrl. Scale bars: 50 μm (A, middle panel); 100 μm (D and E). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PAI-1, plasminogen activator inhibitor type-1; PDGFRβ, platelet-derived growth factor receptor-β; pHSC, primary mouse hepatic stellar cell; α-SMA, α-smooth muscle actin.
Collectively, these results indicated that PKM2 promoted HSC activation and proliferation.
Tetramerization of PKM2, Induced by an Allosteric Agent, Inhibits HSC Activation
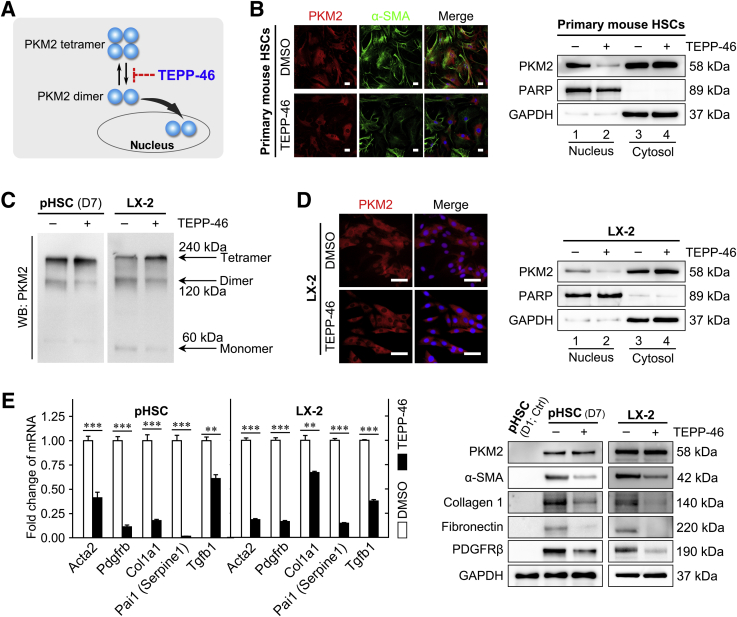
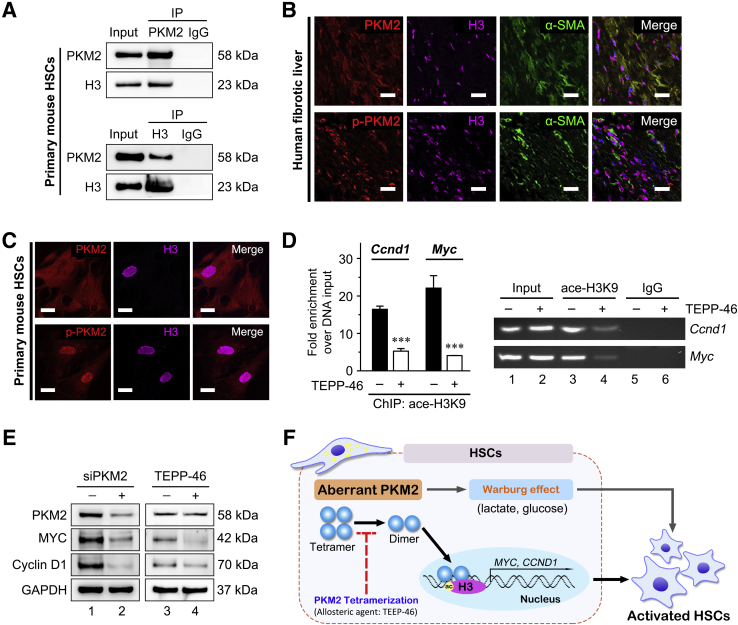
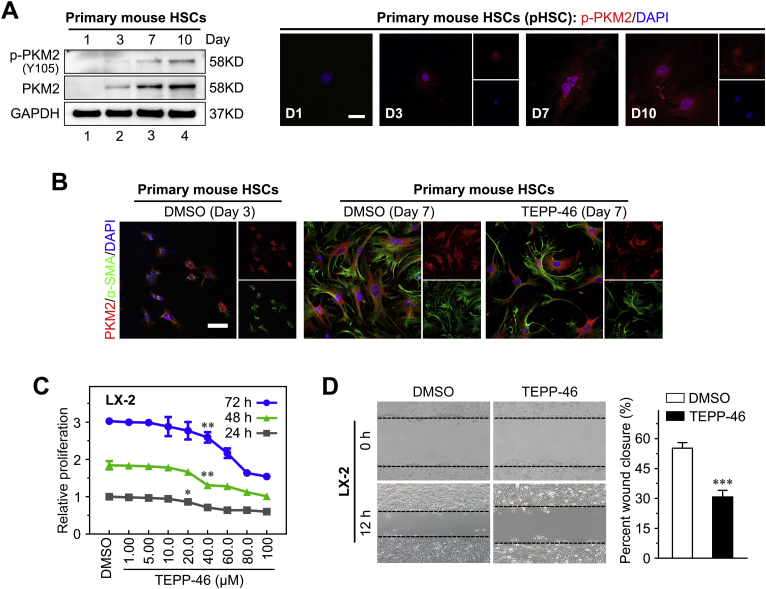
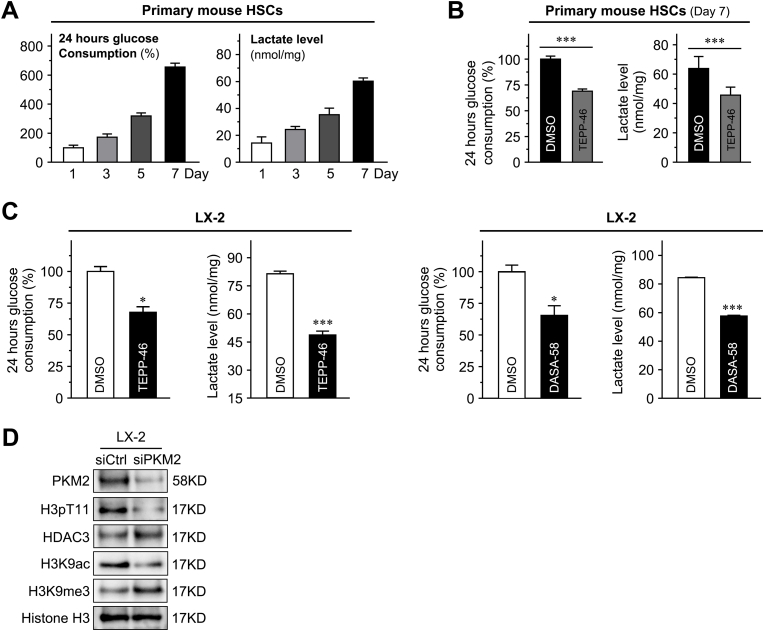
PKM2 can function either as a high-PK activity tetramer or as a low-PK activity dimer. The high-PK activity tetramer localizes in the cytoplasm and drives the tricarboxylic acid cycle, whereas the PKM2 dimer exerts protein kinase activity and translocates to the nucleus, where it regulates the expression of numerous genes.35 In this study, the dimeric PKM2 indicator (ie, phosphorylation of the Y105 residue in PKM2) was gradually up-regulated in cultured primary HSCs during the activation process, and accompanied the increase in the total level of PKM2 (Supplemental Figure S4A). Immunofluorescence performed on cultured primary mouse HSCs at 1, 3, 7, and 10 days after isolation further confirmed the nuclear localization of dimeric PKM2 in activated HSCs (Supplemental Figure S4A). These results indicated that dimeric PKM2 was up-regulated in activated HSCs.
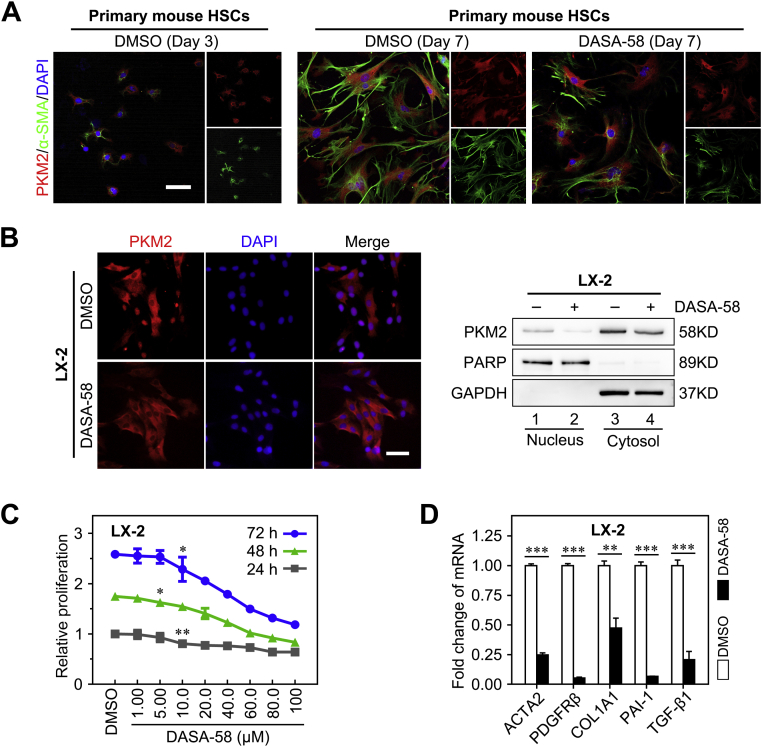
Next, it was investigated whether PKM2 tetramerization could play a role in the inhibition of HSC activation (Figure 4A). To this end, the established tool compound, TEPP-46, a small-molecule PKM2-specific allosteric agent promoting dimer-to-tetramer transformation, was used.36 Immunofluorescence experiments showed that treatment with TEPP-46 resulted in a decreased proportion of nuclear PKM2, which was confirmed by Western blotting on nuclear and cytoplasmic extracts, in both primary mouse HSCs and LX-2 cells (Figure 4, B and D, and Supplemental Figure S4B). In addition, TEPP-46 treatment markedly decreased the dimer/tetramer PKM2 ratio, as assessed by disuccinimidyl suberate cross-linking in both activated primary HSCs and LX-2 cells, suggesting that TEPP-46 promoted the dimer-to-tetramer conversion (Figure 4C). Next, to determine whether TEPP-46–induced PKM2 tetramerization suppressed HSC activation, the expression of activation markers in TEPP-46–treated HSCs was measured. The treatment significantly decreased the expression of HSC activation markers (Figure 4E) and inhibited HSC proliferation and migration (Supplemental Figure S4, C and D). Furthermore, when another PKM2-specific allosteric agent (DASA-58)36 was tested, similar results were obtained in both primary HSCs and LX-2 cells (Supplemental Figure S5).
Figure 4.
Tetramerization of pyruvate kinase M2 (PKM2) induced by an allosteric agent inhibits hepatic stellate cell (HSC) activation. A: Schematic of the TEPP-46–induced PKM2 dimer-tetramer transition. B: Left panel: Immunofluorescence staining for PKM2 and α-smooth muscle actin (α-SMA). Right panel: Western blotting (WB) for PKM2, poly (ADP-ribose) polymerase (PARP), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in nuclear and cytoplasmic extracts of primary HSCs pretreated with dimethyl sulfoxide (DMSO) or TEPP-46 (60 μmol/L) for 24 hours. C: Western blot for PKM2 followed by disuccinimidyl suberate cross-linking in primary HSCs and LX-2 cells treated with TEPP-46 (60 μmol/L) for 24 hours. D: Immunofluorescence staining for PKM2 and Western blotting for PKM2, PARP, and GAPDH of nuclear and cytoplasmic extracts of LX-2 cells pretreated with TEPP-46 (60 μmol/L) for 24 hours. E: The mRNA and protein level of HSC activation markers in primary HSCs and LX-2 cells treated with TEPP-46 (60 μmol/L, 24 hours). ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bar = 50 μm (B and D). ACTA2, actin alpha 2; COL1A1, collagen type I alpha 1; PAI-1, plasminogen activator inhibitor type-1; PDGFRβ, platelet-derived growth factor receptor-β; pHSC, primary mouse hepatic stellar cell; TGF-β1, transforming growth factor-β1.
Taken together, these data suggested that dimeric PKM2 promoted HSC activation, whereas PKM2 tetramerization induced by allosteric agents resulted in antifibrotic effects in vitro.
PKM2 Dimer–Mediated Histone 3 Modification Enhances the Expression of Specific Transcription Factors
Dimeric PKM2 drives aerobic glycolysis (Warburg effect) in tumor cells. Recent evidence indicated that activated HSCs undergo a rapid reprogramming of energy metabolism similar to the Warburg effect, and impairment of this pathway attenuates HSC activation.21 These results confirmed that, in activated primary mouse HSCs, aerobic glycolysis gradually increased, as indicated by high glucose consumption and lactate generation, whereas the PKM2-specific allosteric agent, TEPP-46, reversed the activation of aerobic glycolysis (Supplemental Figure S6, A and B). In addition, both the used PKM2 allosteric agents TEPP-46 and DASA-58 significantly decreased glucose consumption and lactate accumulation in LX-2 cells (Supplemental Figure S6C). On the other hand, the nuclear PKM2 dimer facilitated histone H3K9 acetylation on epidermal growth factor (EGF) receptor activation, and thus up-regulated specific genes, such as MYC and CCND1, in tumor cells.28 Next, immunoprecipitation assays indicated an interaction between PKM2 and histone H3 in activated primary HSCs (Figure 5A). Immunofluorescence confirmed their colocalization in activated HSCs of human fibrotic liver tissues and activated primary mouse HSCs (Figure 5, B and C). Histone deacetylase 3 plays an indispensable role in promoting the H3K9ac/H3K9me3 transition. Further, PKM2 siRNA in LX-2 cells inhibited histone phosphorylation modification on H3T11, which caused the decrease of H3K9Ac expression, and the increase of the H3K9me3 and histone deacetylase 3 (Supplemental Figure S6D). In addition, ChIP assays suggested that TEPP-46 treatment decreased histone H3K9 acetylation at the MYC and CCND1 promoter regions in activated primary HSCs (Figure 5D). MYC and CCDN1 genes are established stimulators of HSC activation, and histone H3K9 acetylation is important for their transcription.28,32 Western blotting verified that TEPP-46 treatment significantly decreased the expression of MYC and cyclin D1 in the activated primary HSCs (Figure 5E). Moreover, siRNA targeting PKM2 also markedly decreased the expression of these gene in LX-2 cells (Figure 5E). It has been reported that PKM2 regulates H3K9 acetylation on EGF receptor activation. EGF is a cytokine that is secreted by activated HSCs (including LX-2 cells) and promotes the proliferation and activation of HSCs via autocrine methods.37,38 EGF stimulation exists during the process of liver fibrosis, which is conformed to the previous reports about the mechanism of PKM2 regulating H3K9 acetylation.28 The further exploration of the mechanism by which PKM2 influences H3K9 acetylation without EGF stimulation is warranted.
Figure 5.
Dimeric pyruvate kinase M2 (PKM2, alias PKM) mediated histone 3 modification enhances the expression of specific transcription factors to promote hepatic stellate cell (HSC) activation. A: Immunoprecipitation (IP) and immunoblotting analyses with the indicated antibodies in activated primary HSCs. B: Three-color fluorescence with the indicated antibodies in human fibrotic liver tissues. C: Immunofluorescence with the indicated antibodies in primary HSCs. D: Primary HSCs were treated with TEPP-46 (60 μmol/L) for 24 hours, then chromatin immunoprecipitation (ChIP) was performed in the indicated whole-cell lysates with anti-histone H3K9 acetylation antibody, and the DNA binding was determined by quantitative PCR. E: PKM2 siRNA (siPKM2) in LX-2 cells and TEPP-46 treatment (60 μmol/L, 24 hours) in primary HSCs reduce MYC and CCND1 expression. F: Schematic of dimeric PKM2 promoting HSC activation. ∗∗∗P < 0.001 versus without TEPP-46. Scale bar = 50 μm (B and C). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin; p-PKM2, phospho-PKM2.
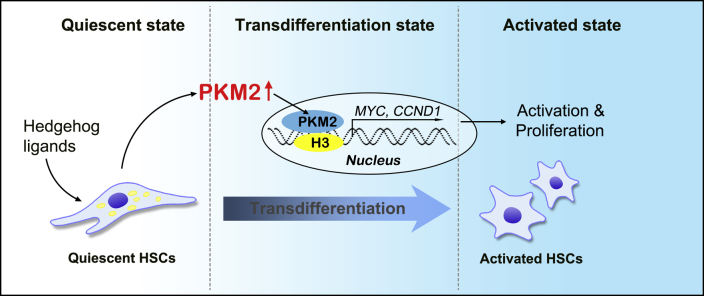
Collectively, these results suggested that the PKM2 dimer induced aerobic glycolysis (Warburg effect), as well as the up-regulation of specific transcription factors via regulation of histone H3 modification, thus promoting HSC activation. PKM2 tetramerization suppressed these events, thus inhibiting HSC activation (Figure 5F and Supplemental Figure S7).
PKM2 Tetramerization Attenuates CCl4-Induced Liver Fibrosis in Vivo
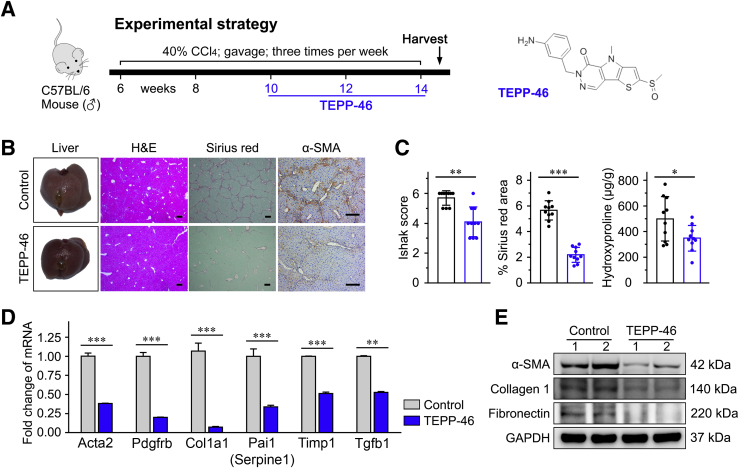
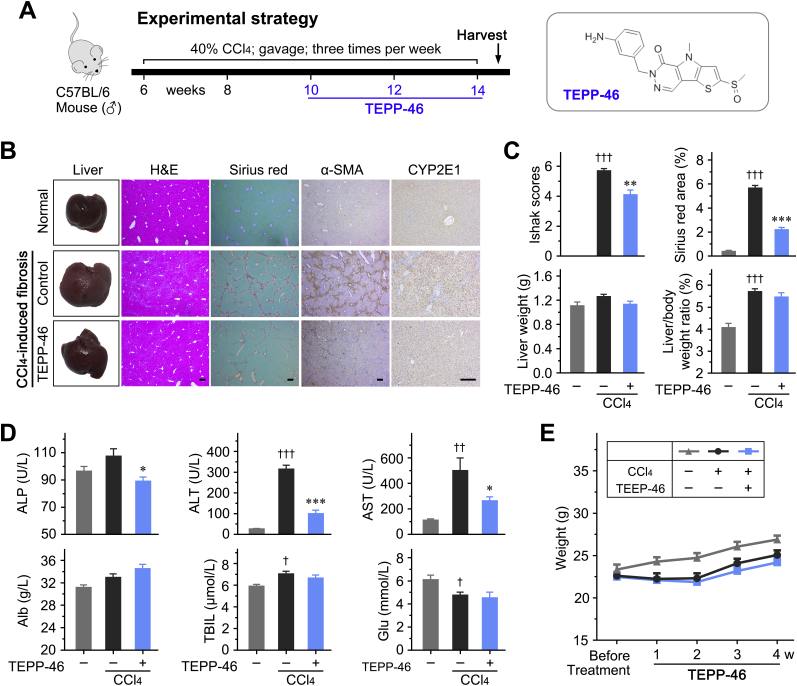
The effect of the PKM2-specific allosteric agent, TEPP-46, promoting PKM2 tetramerization at the expense of the dimer, was first investigated in the CCl4-induced liver fibrosis mouse model (Figure 6A and Supplemental Figure S8A). It was found that TEPP-46 dramatically attenuated liver fibrosis and reduced the Sirius red staining area as well as hepatic hydroxyproline levels in vivo (Figure 6, B and C, and Supplemental Figure S8, B and C). A lower expression of the HSC activation markers a-SMA (Acta2), Pdgfrb, Col1a1, Timp1, Pai-1 (Serpine1), and TGF-β1 (Tgfb1) was observed in liver tissues treated with TEPP-46, compared with control tissues, suggesting that TEPP-46 effectively inhibited HSC activation and attenuated liver fibrosis in vivo (Figure 6, D and E). In addition, TEPP-46 treatment improved hepatic function to some extent, and did not affect body or liver weight in experimental animals (Supplemental Figure S8, C–E). TEPP-46 may inhibit CCl4-induced liver fibrosis. Immunohistochemistry was performed to detect the expression of cytochrome P4502E1 (CYP2E1) in the liver tissues to determine if TEPP-46 affects the expression of CYP2E1, which plays a key role in the metabolism of CCl4. It was found that chronic CCl4 administration increased CYP2E1. However, TEPP-46 did not affect CYP2E1 expression (Supplemental Figure S8B).
Figure 6.
Pyruvate kinase M2 (PKM2) tetramerization attenuates carbon tetrachloride (CCl4)–induced liver fibrosis in vivo. A: The experimental design of TEPP-46 treatment in a CCl4-induced liver fibrosis mouse model. B: Representative images of mouse liver, hematoxylin-eosin (H&E), Sirius red, and α-smooth muscle actin (α-SMA) staining. C: The statistic results of Ishak score, Sirius red–positive area, and hepatic hydroxyproline level in the indicated treatment groups. D and E: TEPP-46 reduces mRNA and protein expression of the indicated hepatic stellate cell (HSC) activation markers in the liver tissues of treated mice. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bar = 200 μm (B). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Shikonin Inhibits HSC Activation and Attenuates Liver Fibrosis
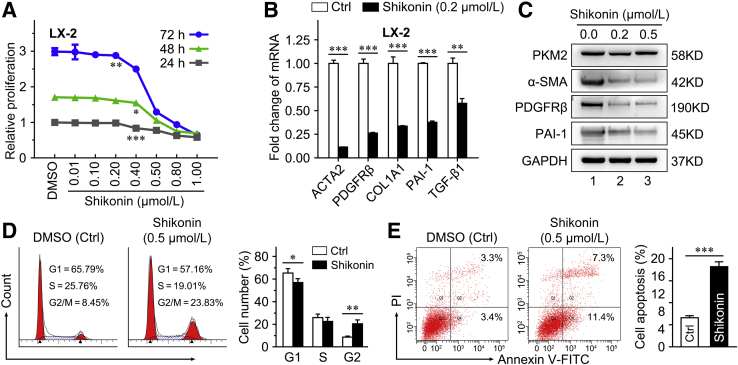
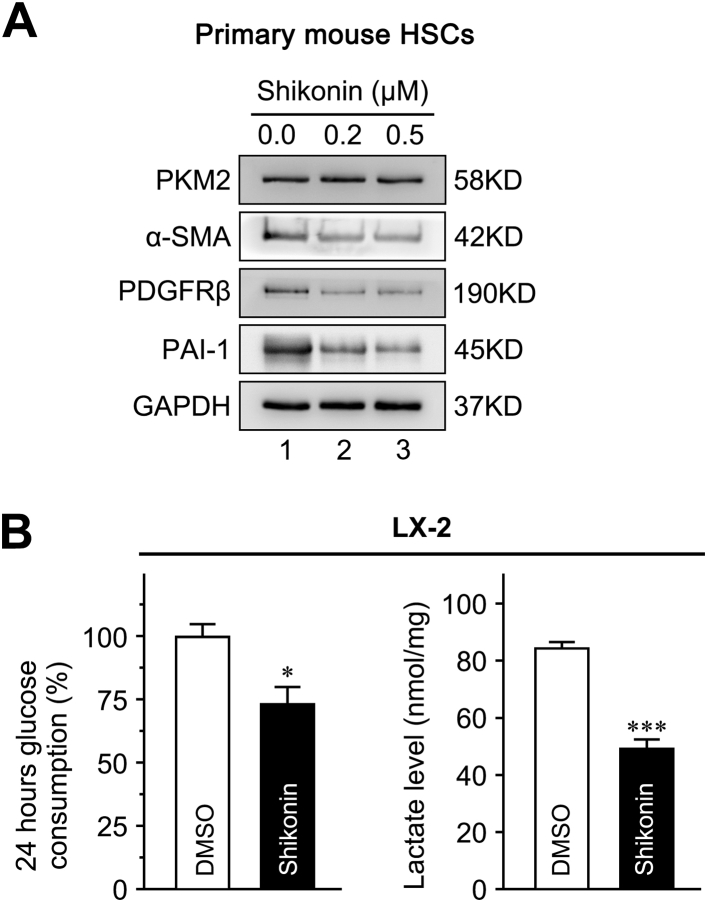
Shikonin, a small-molecule compound isolated from the Chinese herbal medicine, lithospermum, is regarded as an effective PKM2 inhibitor.39,40 Herein, shikonin was used to further investigate the role of PKM2 in HSC activation and liver fibrosis. The treatment with shikonin inhibited LX-2 cell proliferation in a dose- and time-dependent manner (Figure 7A). Our data indicated that 0.2 μmol/L shikonin did not affect LX-2 cell proliferation (Figure 7A); thus, 0.2 and 0.5 μmol/L were selected for further experiments. It was found that 0.2 μmol/L shikonin effectively decreased the expression of the HSC activation markers actin alpha 2 (ACTA2), PDGFRβ, collagen 1A1 (COL1A1), plasminogen activator inhibitor 1 (PAI-1, alias SERPINE1), and transforming growth factor-β1 (TGF-β1, alias TGFB1) in LX-2 cells (Figure 7B). Also, shikonin (0.2 and 0.5 μmol/L) decreased protein expression of α-SMA, PDGFRβ, and PAI-1 in both LX-2 cells (Figure 7C) and primary mouse HSCs (Supplemental Figure S9A). Interestingly, treatment with 0.5 μmol/L shikonin blocked the cell cycle (Figure 7D) and induced apoptosis (Figure 7E) in LX-2 cells.
Figure 7.
Shikonin inhibits hepatic stellate cell (HSC) activation and proliferation in vitro. A: Shikonin inhibits cell proliferation of LX-2 cells in a dose- and time-dependent manner. B: The mRNA level of activated HSC markers in LX-2 cells treated with 0.2 μmol/L shikonin for 24 hours. C: The protein level of activated HSC markers in LX-2 cells treated with shikonin (0.2 and 0.5 μmol/L) for 24 hours. D and E: The cell cycle distribution (D) and apoptotic fraction (E) in LX-2 cells treated with 0.5 μmol/L shikonin for 24 hours is analyzed by flow cytometry. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus control. Ctrl, control; DMSO, dimethyl sulfoxide; FITC, fluorescein isothiocyanate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PAI-1, plasminogen activator inhibitor type-1; PDGFRβ, platelet-derived growth factor receptor-β; PI, propidium iodide; SMA, α-smooth muscle actin; V-FITC, annexin-V fluorescein isothiocyanate.
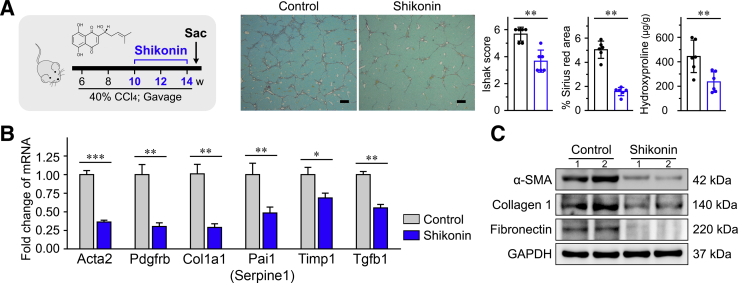
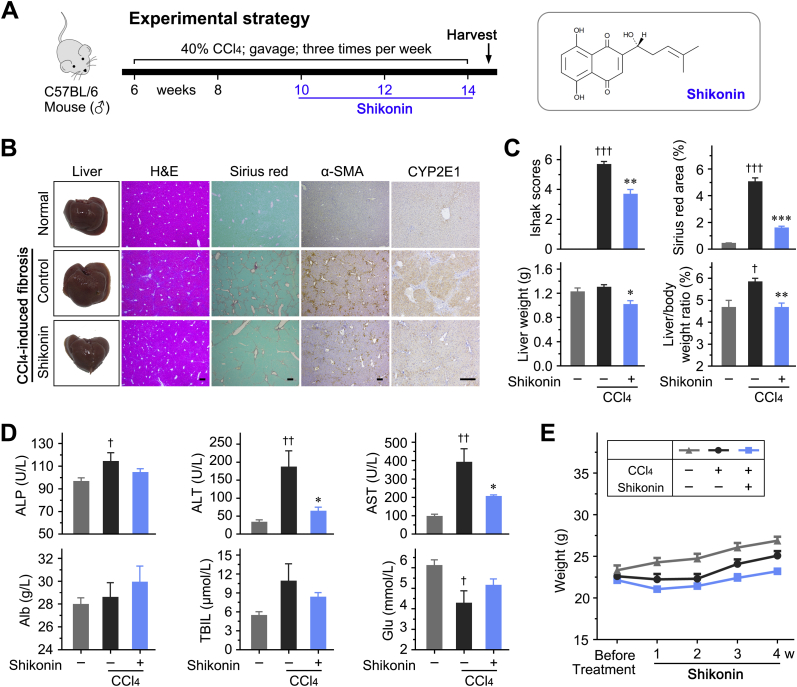
The PKM2 inhibitor, shikonin, blocked the pyruvate kinase activity of tetrameric PKM2 and some function of dimeric PKM2.39,40 Shikonin significantly decreased glucose consumption and lactate accumulation in LX-2 cells (Supplemental Figure S9B). Then, shikonin was used to verify its antifibrosis effect in vivo. Shikonin significantly decreased the expression of HSC activation markers, attenuated liver fibrosis, and improved hepatic function (Figure 8 and Supplemental Figure S10). It was also found that shikonin in CCl4-treated mice decreases CYP2E1 expression in the livers (Supplemental Figure S10).
Figure 8.
Shikonin inhibits hepatic stellate cell (HSC) activation and attenuates liver fibrosis. A: Left panel: The experimental design of shikonin treatment in a carbon tetrachloride (CCl4)–induced liver fibrosis mouse model. Right panel: Representative Sirius red staining images of liver tissues; the statistic results of Ishak score, Sirius red–positive area, and hepatic hydroxyproline level in the indicated treatment groups. B and C: Shikonin reduced mRNA and protein expression of the indicated HSC activation markers in the liver tissues of treated mice. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars = 200 μm (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin.
Taken together, these results demonstrated that PKM2 inhibitor shikonin effectively attenuated HSC activation and liver fibrosis both in vitro and in vivo.
Discussion
Liver fibrosis is a health problem worldwide, for which no effective antifibrosis agents are available.41 Although the activation of HSCs plays an important role in liver fibrosis, its molecular mechanisms are still unclear, which limits therapeutic development. In this study, we found that the expression of PKM2 was markedly increased in activated HSCs, and the nonmetabolic transcription-regulating function of dimeric PKM2 was critical for HSC activation. Furthermore, both PKM2-specific allosteric agents and PKM2 inhibitor hindered HSC activation and attenuated liver fibrosis.
PKM2 is a pyruvate kinase involved in glycometabolism.29 Four pyruvate kinase isoforms are encoded by two genes (PKL and PKR by PKLR; PKM1 and PKM2 by PKM) in mammalian cells, and are expressed in different tissues.42 Unlike other isoforms, PKM2 is mainly expressed in fetal tissues, and is progressively replaced by the other three isozymes after birth. Nevertheless, PKM2 expression is up-regulated in tumor cells, where it activates aerobic glycolysis (Warburg effect).43 PKM2 exists in two different forms (tetramer and dimer) in mammalian cells. The PKM2 tetramer exhibits high PK activity and catalyzes the conversion of phosphoenolpyruvic acid to pyruvate, whereas the PKM2 dimer, with low PK activity, has protein kinase activity and translocates into the nucleus, where it regulates the transcription of specific genes.25 The latter function drives the Warburg effect in tumor cells.24
PKM2 up-regulation is implicated in activated HSCs. The increase of energy requirement and metabolism was important for HSC activation. It might cause the up-regulation of PKM2. During the reprogramming of HSC metabolism, hedgehog and hypoxia-inducible factor-/1α signaling was activated and increased PKM2 expression. Thus, energy metabolism reprogramming and hedgehog/hypoxia-inducible factor-1α signaling may cause up-regulated PKM2 expression in quiescent HSCs during liver injury.21,44,45 However, the exact role of PKM2 in this process and in liver fibrogenesis is unclear. In this study, we found that PKM2 expression was up-regulated in the fibrotic liver and promoted the activation of HSCs. PKM2 silencing significantly inhibited HSC activation in vitro. Interestingly, the current study results demonstrated that dimeric, but not tetrameric, PKM2 could promote HSC activation. Therefore, a well-known PKM2 allosteric agent, TEPP-46, which induces PKM2 tetramerization and simultaneously reduces dimeric PKM2, was utilized to investigate the role of PKM2 tetramerization in liver fibrosis. In vitro experiments suggested that TEPP-46 treatment markedly inhibited the activation and proliferation of HSCs, and in vivo experiments (a CCl4-induced liver fibrosis mouse model) indicated that TEPP-46 significantly attenuated liver fibrosis. These antifibrosis effects depended on the inhibition of dimeric PKM2-induced aerobic glycolysis, and on the down-regulation of specific transcription factors, including MYC and CCND1, which are recognized promoters of HSC activation.46,47 Both MYC and CCND1 are important target effectors of the Wnt/b-catenin signaling pathway to promote the HSC activation and proliferation. Besides, MYC has been identified to promote the transcription of both transforming growth factor-β1 and IL-6, which contribute to activate HSCs via autocrine and paracrine methods. Cyclin D1 is a crucial regulator of cell cycle, and its altered activity is associated with the proliferation of HSCs. Specifically, TEPP-46–induced PKM2 tetramerization inhibited histone H3K9 acetylation at the promoter regions of MYC and CCND1, thereby inhibiting their expression. PKM2 has been demonstrated to modulate H3K4AC, H3K9AC, H3K14AC, and H3K56AC in tumor.48 In this study, it was found that PKM2 promotes HSC activation through H3K9AC as H3K9 acetylation and subsequently increases MYC and cyclin D1 expression. Further studies on PKM2 modulation on other histone modification sites to promote HSC activation are warranted.
In addition, the PKM2 inhibitor, shikonin, a small-molecule compound isolated from the Chinese herbal medicine, lithospermum,49 was used to test PKM2-related antifibrosis effects, in vitro and in vivo. Shikonin has been reported to inhibit multiple functions of dimeric PKM2, including the promotion of the Warburg effect in tumor cells.50,51 Consistently, it was found that shikonin treatment inhibited HSC activation and attenuated liver fibrosis.
Although the antifibrotic effect of TEPP-46 or shikonin on HSCs is not direct, liver fibrosis and HSC activation could also induce liver injury and hepatocyte death. Activated HSCs increase production of transforming growth factor-β and other cytokines to facilitate hepatocyte injury and accelerate disease progression. We therefore hypothesize that inhibiting HSC activation would protect liver injury. The metabolism of CCl4 to its toxic metabolite is also the stimulus for liver fibrosis. However, TEPP-46 did not affect CYP2E1 expression, whereas shikonin in CCl4-treated mice can decrease CYP2E1 expression in the livers. Shikonin might exert its anti–liver fibrosis effect partly by decreasing CYP2E1 expression in CCl4 mouse model. In consideration of the in vitro HSC activation inhibiting effect of TEPP-46 or shikonin, we speculated that inhibiting HSC activation plays a critical role in the TEPP-46– or shikonin-induced liver injury alleviation.
In conclusion, our study indicated that dimeric, but not tetrameric, PKM2 promotes HSC activation and liver fibrogenesis. Both PKM2 tetramerization, induced by allosteric agents, and pharmacologic PKM2 inactivation resulted in potent antifibrosis effects in mouse models. Considering the homology of PKM2 with the other three pyruvate kinase isoforms, it is hard to design highly specific PKM2 agents. Thus, the induction of PKM2 tetramerization to decrease the level of dimeric PKM2 may be a potential therapeutic strategy for liver fibrosis. Although further studies are needed to evaluate the clinical applicability of this approach, our study demonstrated a novel role of PKM2 in liver fibrosis and provided new evidence that PKM2 tetramerization may reverse liver fibrosis.
Footnotes
Supported by National Natural Science Foundation of China grants 81672320 (J.H.), 81871987 (J.H.), 81802423 (C.Q.), and 81772821 (K.H.); Science and Technology Program of Guangzhou, China, grant 201704020128 (J.H.); Municipal University Science and Technology Program of Guangzhou Education Bureau grant 1201410075 (J.H.); Fundamental Research Funds for the Central Universities grant 21620106 (J.H.); Yangcheng Scholar Program grant 1201561579 (J.H.); China Postdoctoral Science Foundation grant 2017M622741 (C.Q.); and Guangdong Natural Science Foundation2017A030313471 (K.H.).
D.Z. and Y.J. contributed equally to this work.
Disclosures: None declared.
Portions of this manuscript were presented as an abstract and oral presentation during the Annual Meeting of the American Association for the Study of Liver Diseases (AASLD), November 8–12, 2019, Boston, MA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2020.08.002.
Author Contributions
D.Z. and C.Q. conceived the study design/experiments and analyzed data; D.Z., C.Q., M.T., J.L., H.C., and L.L. performed molecular experiments; L.H., H.Y., and Z.L. performed histologic analyses, tissue processing, and immunohistochemical staining; D.Z., Y.J., and P.C. performed animal experiments; D.Z. made the figures and tables, and wrote the manuscript; K.H., W.L., J.F., and G.C. critically reviewed the manuscript; all authors had final approval of the submitted version.
Supplemental Data
Supplemental Figure S1.
Pyruvate kinase M2 (PKM2), but not pyruvate kinase M1 (PKM1), is increased in fibrotic liver and hepatocellular carcinoma (HCC). A: Representative images of liver tissue stained with hematoxylin-eosin (H&E), PKM1, and PKM2 for indicated treatment duration. Black dashed lines represent boundaries between tumor (T) and nontumor (N) tissues. B and C: PKM2 (PKM) expression is detected by real-time quantitative PCR (B) and Western blot analysis (C) at the indicated time. D: Representative images of H&E, PKM1, and PKM2 staining in normal liver, fibrotic liver, and HCC tissues. E: Immunohistochemical staining scores of PKM1 and PKM2 in the different tissues. Results are presented as means ± SD (B and E). ∗∗∗P < 0.001. Scale bars = 200 μm (A and D). CCl4, carbon tetrachloride; DEN, diethylnitrosamine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Supplemental Figure S2.
Pyruvate kinase M2 (PKM2) expression in various liver cell types. A: Representative image of PKM2 (red) with CD68 (green) or CD31 (green) in primary cells isolated from normal and carbon tetrachloride–induced fibrotic mouse liver. B: Representative images of immunofluorescence costaining of PKM2 (red) with cytokeratin 19 (CK19; green), CD68 (green), or CD31 (green) in normal and fibrotic liver tissues. Scale bars = 50 μm (A and B).
Supplemental Figure S3.
Pyruvate kinase M2 (PKM2, alias PKM) promotes hepatic stellate cell (HSC) migration. A: Interference efficiency of PKM2 siRNA (siPKM2) in primary mouse HSCs. B: The migration capability of LX-2 cells transfected with control siRNA (siCtrl) or PKM2 siRNA is measured using Transwell assays. C: Cell Counting Assay Kit-8 assays in LX-2 cells infected with Lv-Vector or Lv-PKM2. D: PKM2 and HSC activation marker expression in LX-2 cells that infected with Lv-Vector or Lv-PKM2. ∗∗P < 0.01, ∗∗∗P < 0.001 versus siCtrl; ††P < 0.01, †††P < 0.001 versus vector. Scale bar = 100 μm (B). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PDGFRβ (PDGFRB), platelet-derived growth factor receptor-β; α-SMA, α-smooth muscle actin.
Supplemental Figure S4.
TEPP-46 inhibits hepatic stellate cell (HSC) proliferation and migration. A: The indicated proteins are detected by Western blot analysis or immunofluorescence staining in primary mouse HSCs cultured for the indicated periods. B: Primary HSCs pretreated with TEPP-46 as indicated for 24 hours. Double immunofluorescence staining of pyruvate kinase M2 (PKM2) (red) and α-smooth muscle actin (α-SMA; green) was performed. C: TEPP-46 inhibits cell proliferation in LX-2 cells in a dose- and time-dependent manner. D: The migration capability of LX-2 cells treated with TEPP-46 is measured using the wound-healing assays. Black dashed lines indicate the edge of cell migration. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus DMSO. Scale bars: 50 μm (A); 100 μm (B). DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; p-PKM2, phospho-PKM2; pHSC, primary mouse hepatic stellar cell.
Supplemental Figure S5.
DASA-58 inhibits hepatic stellate cell (HSC) proliferation and migration. A: Double immunofluorescence staining of pyruvate kinase M2 (PKM2) (red) and α-smooth muscle actin (α-SMA; green) was performed in the primary HSCs pretreated with DASA-58 (40 μmol/L) for 24 hours. B: Left panel: Representative immunofluorescence image of PKM2 in LX-2 cells treated with DASA-58. Right panel: Western blot analysis of indicated proteins in the cytosolic and nuclear extractive of LX-2 cells. C: DASA-58 inhibits cell proliferation in LX-2 cells in a dose- and time-dependent manner. D: Expression of HSC activation markers in LX-2 cells treated with DASA-58. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars: 100 μm (A); 50 μm (B). DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PARP, poly (ADP-ribose) polymerase.
Supplemental Figure S6.
Pyruvate kinase M2 (PKM2) promotes glycolysis and regulates histone 3 modification of hepatic stellate cells (HSCs). A: Glucose consumption and lactate accumulation in HSCs isolated from mouse normal liver and cultured for indicated periods. B: TEPP-46 treatment (60 μmol/L, 24 hours) reduces glucose consumption and lactate accumulation in cultured HSCs. C: LX-2 cells were treated with TEPP-46 (left panel) and DASA-58 (right panel) for 24 hours. Glucose in the medium and lactate in the cells were detected to evaluate the aerobic glycolysis. D: Western blotting with PKM2, H3pT11, histone deacetylase 3 (HDAC3), H3K9Ac, H3K9me3, and histone H3 in LX-2 cells transfected with PKM2 siRNA (siPKM2). ∗P < 0.05, ∗∗∗P < 0.001 versus DMSO. DMSO, dimethyl sulfoxide; siCtrl, control siRNA.
Supplemental Figure S7.
The cartoon for the regulation of hepatic stellate cell (HSC) activation by pyruvate kinase M2 (PKM2) expression. Hedgehog signaling enhances PKM2 expression during the HSC transition process. Furthermore, PKM2 up-regulation promotes HSC activation by facilitating histone H3K9 acetylation and increasing the expression of MYC and CCND1, which contributes to HSC activation and proliferation as well as stimulation of collagen synthesis. Dashed lines indicate the boundary between different states of HSCs.
Supplemental Figure S8.
Pyruvate kinase M2 (PKM2) allosteric agent (TEPP-46) treatment reduces carbon tetrachloride (CCl4)–induced mouse liver fibrosis. A: Schematic of the experimental design, and the structural formulas of TEPP-46. B: Representative images of mouse liver, hematoxylin-eosin (H&E), Sirius red, α-smooth muscle actin (α-SMA), and cytochrome P4502E1 (CYP2E1) staining. C: Ishak score, Sirius red–positive area, liver weight, and liver/body weight ratio of the indicated groups. D: Serum alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), albumin (Alb), and glucose (Glu) levels in the indicated groups. E: On-treatment weight change of each group. Results are presented as means ± SD (C–E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus CCl4; †P < 0.05, ††P < 0.01, and †††P < 0.001 versus normal. Scale bars = 200 μm (B).
Supplemental Figure S9.
Shikonin inhibits hepatic stellate cell (HSC) activation and glycolysis. A: Protein expression of pyruvate kinase M2 (PKM2) and HSC activation markers in primary HSCs treated with shikonin. B: LX-2 cells were treated with shikonin for 24 hours. Glucose in the medium and lactate in the cells are detected to evaluate the aerobic glycolysis. ∗P < 0.05, ∗∗∗P < 0.001 versus DMSO. DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PAI-1, plasminogen activator inhibitor type-1; PDGFRβ, platelet-derived growth factor receptor-β; α-SMA, α-smooth muscle actin.
Supplemental Figure S10.
Pyruvate kinase M2 (PKM2) inhibitor (shikonin) treatment attenuates carbon tetrachloride (CCl4)–induced mouse liver fibrosis. A: Schematic of the experimental design, and the structural formulas of shikonin. B: Representative images of mouse liver, hematoxylin-eosin (H&E), Sirius red, α-smooth muscle actin (α-SMA), and cytochrome P4502E1 (CYP2E1) staining. C: Ishak score, Sirius red–positive area, liver weight, and liver/body weight ratio of the indicated groups. D: Serum alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBIL), albumin (Alb), and glucose (Glu) levels in the indicated groups. E: On-treatment weight change of each group. Results are presented as means ± SD (C–E). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus CCl4; †P < 0.05, ††P < 0.01, and †††P < 0.001 versus normal. Scale bars = 200 μm (B).
References
- 1.El-Serag H.B. Current concepts hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Evans H.M., Kefly D.A., McKiernan P.J., Hubscher S. Progressive histological damage in liver allografts following pediatric liver transplantation. Hepatology. 2006;43:1109–1117. doi: 10.1002/hep.21152. [DOI] [PubMed] [Google Scholar]
- 3.Scheenstra R., Peeters P.M.G.J., Verkade H.J., Gouw A.S.H. Graft fibrosis after pediatric liver transplantation: ten years of follow-up. Hepatology. 2009;49:880–886. doi: 10.1002/hep.22686. [DOI] [PubMed] [Google Scholar]
- 4.Schuppan D., Afdhal N.H. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.-P., Schwabe R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J Pathol. 2003;200:500–503. doi: 10.1002/path.1427. [DOI] [PubMed] [Google Scholar]
- 7.Puche J.E., Saiman Y., Friedman S.L. Hepatic stellate cells and liver fibrosis. Compr Physiol. 2013;3:1473–1492. doi: 10.1002/cphy.c120035. [DOI] [PubMed] [Google Scholar]
- 8.Bataller R., Brenner D.A. Liver fibrosis. J Clin Invest. 2005;115:1100. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gressner A.M. Transdifferentiation of hepatic stellate cells (Ito cells) to myofibroblasts: a key event in hepatic fibrogenesis. Kidney Int Suppl. 1996;54:S39–S45. [PubMed] [Google Scholar]
- 10.Mann D.A., Marra F. Fibrogenic signalling in hepatic stellate cells. J Hepatol. 2010;52:949–950. doi: 10.1016/j.jhep.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Higashi T., Friedman S.L., Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C.-Y., Yuan W.-G., He P., Lei J.-H., Wang C.-X. Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 15.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Wong N., De Melo J., Tang D. PKM2, a central point of regulation in cancer metabolism. Int J Cell Biol. 2013;2013:1–11. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altenberg B., Greulich K.O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Fan F., Wu H., Liu Z., Hou X., Chen W., Wang A., Lu Y. Nuclear PKM2 expression, an independent risk factor for ER after curative resection of hepatocellular carcinoma. Biomed Pharmacother. 2016;84:1858–1864. doi: 10.1016/j.biopha.2016.10.108. [DOI] [PubMed] [Google Scholar]
- 19.Fan F.-T., Shen C.-S., Tao L., Tian C., Liu Z.-G., Zhu Z.-J., Liu Y.-P., Pei C.-S., Wu H.-Y., Zhang L., Wang A.-Y., Zheng S.-Z., Huang S.-L., Lu Y. PKM2 regulates hepatocellular carcinoma cell epithelial-mesenchymal transition and migration upon EGFR activation. Asian Pac J Cancer Prev. 2014;15:1961–1970. doi: 10.7314/apjcp.2014.15.5.1961. [DOI] [PubMed] [Google Scholar]
- 20.Wang F.X., Jia Y., Li M.M., Wang L., Shao J.J., Guo Q.L., Tan S.Z., Ding H., Chen A.P., Zhang F., Zheng S.Z. Blockade of glycolysis-dependent contraction by oroxylin a via inhibition of lactate dehydrogenase-a in hepatic stellate cells. Cell Commun Signal. 2019;17:11. doi: 10.1186/s12964-019-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.P., Choi S.S., Michelotti G.A., Chan I.S., Swiderska-Syn M., Karaca G.F., Xie G.H., Moylan C.A., Garibaldi F., Premont R., Suliman H.B., Piantadosi C.A., Diehl A.M. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology. 2012;143:1319. doi: 10.1053/j.gastro.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi T., Inoue H., Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–13812. [PubMed] [Google Scholar]
- 23.Chen M., Zhang J., Manley J.L. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., Cantley L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 25.Tamada M., Suematsu M., Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 26.Iqbal M.A., Gupta V., Gopinath P., Mazurek S., Bamezai R.N. Pyruvate kinase M2 and cancer: an updated assessment. FEBS Lett. 2014;588:2685–2692. doi: 10.1016/j.febslet.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Christofk H.R., Vander Heiden M.G., Wu N., Asara J.M., Cantley L.C. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 28.Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Yung W.K.A., Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Wang D., Li M., Plecita-Hlavata L., D'Alessandro A., Tauber J., Riddle S., Kumar S., Flockton A., McKeon B.A., Frid M.G., Reisz J.A., Caruso P., El Kasmi K.C., Jezek P., Morrell N.W., Hu C.-J., Stenmark K.R. Metabolic and proliferative state of vascular adventitial fibroblasts in pulmonary hypertension is regulated through a MicroRNA-124/PTBP1 (polypyrimidine tract binding protein 1)/pyruvate kinase muscle axis. Circulation. 2017;136:2468. doi: 10.1161/CIRCULATIONAHA.117.028069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mederacke I., Dapito D.H., Affo S., Uchinami H., Schwabe R.F. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc. 2015;10:305–315. doi: 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei Y., Wang D., Jin F., Bian Z., Li L., Liang H., Li M., Shi L., Pan C., Zhu D., Chen X., Hu G., Liu Y., Zhang C.-Y., Zen K. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun. 2017;8:14041. doi: 10.1038/ncomms14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu C., Zheng D.D., Li S., Liu Y.J., Lidofsky A., Holmes J.A., Chen J.N., He L., Wei L., Liao Y.D., Yuan H., Jin Q.M., Lin Z.L., Hu Q.T., Jiang Y.C., Tu M.X., Chen X.J., Li W.M., Lin W.Y., Fuchs B.C., Chung R.T., Hong J. Tyrosine kinase SYK is a potential therapeutic target for liver fibrosis. Hepatology. 2018;68:1125–1139. doi: 10.1002/hep.29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M., Gong Q., Zhang J., Chen L., Zhang Z., Lu L., Yu D., Han Y., Zhang D., Chen P., Zhang X., Yuan Z., Huang J., Zhang X. Characterization of gene expression profiles in HBV-related liver fibrosis patients and identification of ITGBL1 as a key regulator of fibrogenesis. Sci Rep. 2017;7:43446. doi: 10.1038/srep43446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong Q., Li N., Cheng H., Zhang X., Cao X., Qi T., Dai L., Zhang Z., Chen X., Li C., Li Y., Xue B., Fang L., Liu L., Ding Z. HSPA12A is a novel player in nonalcoholic steatohepatitis via promoting nuclear PKM2-mediated M1 macrophage polarization. Diabetes. 2019;68:361–376. doi: 10.2337/db18-0035. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang X.S., Han S.N., Zhang J.Y., Dioletis E., Nemeth B.T., Pacher P., Feng D.C., Bataller R., Cabezas J., Starkel P., Caballeria J., Pongratz R.L., Cai S.Y., Schnabl B., Hoque R., Chen Y.L., Yang W.H., Garcia-Martinez I., Wang F.S., Gao B., Torok N.J., Kibbey R.G., Mehal W.Z. Digoxin suppresses pyruvate kinase M2-promoted HIF-1 alpha transactivation in steatohepatitis. Cell Metab. 2018;27:1156. doi: 10.1016/j.cmet.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Anastasiou D., Yu Y.M., Israelsen W.J., Jiang J.K., Boxer M.B., Hong B.S. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:1008. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma T., Cai X., Wang Z., Huang L., Wang C., Jiang S., Hua Y., Liu Q. miR-200c accelerates hepatic stellate cell-induced liver fibrosis via targeting the FOG2/PI3K pathway. Biomed Res Int. 2017;2017:2670658. doi: 10.1155/2017/2670658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D., Zhang J., Jiang X., Li X., Wang Y., Ma J., Jiang H. Heparin-binding epidermal growth factor-like growth factor: a hepatic stellate cell proliferation inducer via ErbB receptors. J Gastroenterol Hepatol. 2014;29:623–632. doi: 10.1111/jgh.12412. [DOI] [PubMed] [Google Scholar]
- 39.Chen J., Xie J., Jiang Z., Wang B., Wang Y., Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene. 2011;30:4297–4306. doi: 10.1038/onc.2011.137. [DOI] [PubMed] [Google Scholar]
- 40.Xie M., Yu Y., Kang R., Zhu S., Yang L.C., Zeng L., Sun X.F., Yang M.H., Billiar T.R., Wang H.C., Cao L.Z., Jiang J.X., Tang D.L. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat Commun. 2016;7:13280. doi: 10.1038/ncomms13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottcher K., Pinzani M. Pathophysiology of liver fibrosis and the methodological barriers to the development of anti-fibrogenic agents. Adv Drug Deliv Rev. 2017;121:3–8. doi: 10.1016/j.addr.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., Jiang J.K., Israelsen W.J., Keane J., Thomas C., Clish C., Vander Heiden M., Xavier R.J., O'Neill L.A.J. Pyruvate kinase M2 regulates Hif-1 alpha activity and IL-1 beta induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:347. doi: 10.1016/j.cmet.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Mazurek S. Pyruvate kinase type M2: a key regulator within the tumour metabolome and a tool for metabolic profiling of tumours. Ernst Schering Found Symp Proc. 2007;4:99–124. doi: 10.1007/2789_2008_091. [DOI] [PubMed] [Google Scholar]
- 44.Wan L., Xia T., Du Y., Liu J., Xie Y., Zhang Y., Guan F., Wu J., Wang X., Shi C. Exosomes from activated hepatic stellate cells contain GLUT1 and PKM2: a role for exosomes in metabolic switch of liver nonparenchymal cells. FASEB J. 2019;33:8530–8542. doi: 10.1096/fj.201802675R. [DOI] [PubMed] [Google Scholar]
- 45.Hou W., Syn W.-K. Role of metabolism in hepatic stellate cell activation and fibrogenesis. Front Cell Dev Biol. 2018;6:150. doi: 10.3389/fcell.2018.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son G., Hines I.N., Lindquist J., Schrum L.W., Rippe R.A. Inhibition of phosphatidylinositol 3-kinase signaling in hepatic stellate cells blocks the progression of hepatic fibrosis. Hepatology. 2009;50:1512–1523. doi: 10.1002/hep.23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su M., Chao G., Liang M., Song J., Wu K. Anticytoproliferative effect of vitamin C on rat hepatic stellate cell. Am J Transl Res. 2016;8:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., Li Y., You W., Dong Q., Hong T., Yan Z., Jin S., Wang T., Zhao W., Mai H., Huang J., Han X., Ji Q., Song Q., Yang C., Zhao S., Xu X., Ye Q. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 2018;33:368. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Chen X., Yang L., Zhang N., Turpin J.A., Buckheit R.W., Osterling C., Oppenheim J.J., Howard O.M.Z. Shikonin, a component of Chinese herbal medicine, inhibits chemokine receptor function and suppresses human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2003;47:2810–2816. doi: 10.1128/AAC.47.9.2810-2816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L., Xie M., Yang M., Yu Y., Zhu S., Hou W., Kang R., Lotze M.T., Billiar T.R., Wang H., Cao L., Tang D. PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat Commun. 2014;5:4436. doi: 10.1038/ncomms5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X., Zhu Y., Hu J., Jiang L., Li L., Jia S., Zen K. Shikonin inhibits tumor growth in mice by suppressing pyruvate kinase M2-mediated aerobic glycolysis. Sci Rep. 2018;8:14517. doi: 10.1038/s41598-018-31615-y. [DOI] [PMC free article] [PubMed] [Google Scholar]