Abstract
The carcinogenicity of radon has been convincingly documented through epidemiological studies of underground miners. The risk of lung cancer from radon exposure is due to the continuous radioactive decay of this gas and subsequent emission of high-energy alpha decay particles. And the bronchial epithelial cells are the main targets of radon exposure. However, there is a lack of early warning indicators of lung cancer caused by radon in the physical examination of populations involved in occupations with higher exposure to radon. To assess the potential of a molecular-based marker approach for the early detection of human lung cancer induced by radon, human bronchial epithelial cell injury models induced by alpha-particle irradiation were constructed. The results of transwell migration assay, transwell invasion assay, and the expression of the epithelial–mesenchymal transition-related proteins showed that malignant cell transformation could be triggered by alpha irradiation. Potential microRNAs (miRNAs) (hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257) were screened using miRNA chips in cell models. The pathway analyses of miRNAs selected using DIANA-miRPath v3.0 showed that miRNAs involved in malignant cell transformation were associated with cell adhesion molecules, extracellular matrix receptor interaction, and proteoglycans in cancer, among others, which are closely related to the occurrence and development of carcinogenesis. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR) assay showed that five screened miRNAs were up-regulated in five lung cancer tissue samples. In conclusion, the results indicated that hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257 may be potential early markers of the malignant transformation of bronchial epithelial cells induced by alpha-particle irradiation.
Keywords: bronchial epithelial cells, alpha-particle irradiation, microRNA, lung cancer, radon, biomarker
Introduction
Radon gas is ubiquitous in indoor and outdoor air and contaminates many underground mines [1]. As radon decays, it produces radioactive progeny and emits significant levels of alpha radiation, along with lower levels of beta and gamma radiation, leading to biological damage that can be dangerous to human health [2, 3]. Radon is the second most common cause of lung cancer after smoking, cohort studies of underground miners and epidemiological studies in residential settings have established a strong association between radon exposure and increased risk for lung cancer [3–9].
The development of lung cancer is a multistep process involving the accumulation of genetic and epigenetic materials generated by risk factors, such as cigarette smoking and radon exposure. Different causes of cancer act via different biological mechanisms and are reflected in the varied histopathological subtypes. Primary prevention is fundamental in lung cancer, which is a tumour characterized by a long latency period. So, it is necessary to explore early warning molecular biomarkers for lung cancer induced by radon.
MicroRNAs (miRNAs), a small group of non-coding RNAs, post-transcriptionally modulate gene expression by specific disruption and degradation of target messenger RNAs (mRNAs) [10]. These molecules play a wide variety of regulatory roles in numerous processes, including cell proliferation and differentiation, development, and programmed cell death [11]. MicroRNAs regulate gene expression by binding to the 3′-untranslated region of target mRNAs, resulting in gene translation repression or RNA degradation [12]. A single miRNA may simultaneously regulate a whole set of genes, thereby controlling multiple signalling pathways.
In recent years, there has been great excitement in the use of miRNAs as biomarkers of diseases, such as myocardial infarction, diabetes, and cancer [13–19]. Recent data have indicated that miRNAs are engaged in the regulation of cellular processes induced by radiation, and, consequently, miRNAs can potentially be used as biomarkers to assess the degree of exposure to radiation in humans [20, 21].
Human bronchial epithelial cells are the target cells of radon exposure. The high linear energy transfer alpha particles emitted by radon and radon daughters were found to malignantly transform human bronchial epithelial cells, leading to cell damage or carcinogenesis [22]. Based on these considerations, the human bronchial epithelial cell line (BEAS-2B) was selected to assess the lung injury induced by alpha radiation. In this study, malignant transformation models of BEAS-2B cells induced by alpha radiation were constructed. These models were validated with migration and invasion assays and epithelial–mesenchymal transition (EMT) representative proteins. Then, the potential biomarkers of malignant cellular transformation were screened based on differentially expressed miRNAs induced by alpha irradiation.
Materials and Methods
Cell culture
The human bronchial epithelial cell line BEAS-2B (ATCC number CRL-9609) was obtained from American Type Culture Collections (Manassas, VA, USA) and maintained in our laboratory. The cells were cultured in Bronchial Epithelial Cell Growth Medium (BEGM) [a mixture of the additives dissolved in Bronchial Epithelial Cell Basal Medium (BEBM)] (Lonza/Clonetics Corporation) at 37°C in 5% CO2. The medium was changed every 2 days, and cells were subcultured for 3 days.
Cell irradiation
Overnight after plating, BEAS-2B cells growing on coverslips were irradiated using alpha irradiation equipment (School of Radiation Medicine and Protection, Soochow University) [23]. The alpha irradiation equipment is composed of a 241Am source, a rotatable irradiation source holder, a sample holder, and other necessary parts. The irradiation source holder and the sample holder are 10 mm away and parallel to each other. The 241Am source is put face down on the irradiation source holder and emits alpha particles at a calculated dose rate of 0.138 Gy/min. Coverslips with cells were put on the sample holder and the 241Am source was rotated to the top of the coverslip to irradiate the cells. Cells were irradiated by fractional irradiation. Cells were irradiated cumulatively four times, with subculture for 10 generations after each irradiation, giving 40 total subculture generations. Each irradiation dose was 0.1, 0.5, and 2 Gy. Finally, cell models with different irradiation doses, number of exposures, and subculture times were obtained. The notation ‘2B-xGy(y)-z’ was used to define different cell models, in which 2B represented BEAS-2B cells, x represented radiation dose per exposure, y represented the number of irradiation exposures, and z represented the cell generations. 2B-0Gy(y)-z represented control cells. Thus, the following cell models were collected for subsequent experiments: 2B-0Gy(2)-20, 2B-0.1Gy(2)-20, 2B-0.5Gy(2)-20, 2B-2.0Gy(2)-20, 2B-0Gy(4)-40, 2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40.
Transwell migration and invasion experiments
Cell migration was measured using Transwell chambers (Corning Inc., USA) with polycarbonate filters having 8-μm pores. A total of 50 000 cells suspended in 200 μl of additive-free BEBM were added to the upper insert. The lower wells were filled with 500 μl BEGM containing the additives as chemoattractants. After incubation for 24 h at 37°C, the cells remaining at the upper surface of the membrane were removed with cotton swabs, and the cells on the lower surface of the membrane were the migrated cells. After fixing with 4% paraformaldehyde and staining with 0.1% crystal violet solution, the cells that passed through the filter were photographed and counted under a microscope.
The transwell invasion assay was carried out as described above, except that 50 μl of 1:8 BEBM-diluted Matrigel (BD, USA) was added to each well at 37°C for 6 h before the cells were seeded onto the membrane, followed by incubation for 24 h.
Detection of EMT-related proteins with western blotting
Western blotting was performed to determine the expression of EMT-related proteins, such as E-cadherin, vimentin, Vascular Endothelial Growth Factor A (VEGFA), and Transforming growth factor beta 1 (TGF-β1), after alpha irradiation. The cell models 2B-0Gy(4)-40, 2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40 were washed with ice-cold phosphate-buffered saline and lysed with Radioimmunoprecipitation assay (RIPA) lysis buffer [50 mM Tris HCl, 150 mM NaCl, 1% Triton, and 0.1% sodium dodecyl sulphate (SDS)] containing protease inhibitors (Sigma-Aldrich, USA). The extracted proteins were separated with SDS-polyacrylamide gel electrophoresis and were transferred electrophoretically onto a polyvinylidene difluoride membrane (Millipore, USA). The membranes were blocked with 5% bovine serum albumin (Beyotime Institute of Biotechnology, China) in Tris-buffered saline containing 0.1% Tween-20 and incubated with rabbit anti-E-Cadherin (1:1000), anti-Vimentin (1:10 000, Sangon Biotech, China), anti-VEGFA (1:2000, Sangon Biotech), anti-TGF-β1 (1:800, Sangon Biotech), and anti-Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000, Sangon Biotech) overnight at 4°C, followed by incubation for 40 min with horseradish peroxidase-conjugated secondary antibodies (1:5000, Sangon Biotech). After extensive washing in Phosphate buffered solution (PBST), the expression levels of the proteins were detected by Quantity One software (Bio-Rad Laboratories, USA) using the Enhanced chemiluminescence (ECL)-Chemiluminescent Kit (Sangon Biotech).
Total RNA isolation and miRNA microarray assay
Total RNA including small RNA populations were extracted from cell models using a combination of TRIzol (Invitrogen, USA) and a Qiagen RNeasy Mini Kit (Qiagen, Germany), according to the manufacturer’s protocol. The quality of the extracted RNA was assessed using both NanoDrop ND-2000 and, to acquire an RNA Integrity Number (RIN), the Agilent Bioanalyzer 2100, using an Agilent RNA 6000 Nano Kit (Agilent Technologies, CA, USA). RNA samples with an RIN value ≥8, an A260/280 of ~2, and an A260/230 of ~2 were selected for downstream assessment.
Global expression patterns of miRNAs were examined using an Agilent miRNA Chip. The sample labelling, microarray hybridization, and washing were performed based on the manufacturer’s standard protocols. Briefly, total RNA was dephosphorylated, denatured, and then labelled with Cyanine-3-CTP. After purification, the labelled RNAs were hybridized onto the microarray. After washing, the arrays were scanned with the Agilent Scanner G2505C (Agilent Technologies).
Feature Extraction software (version10.7.1.1, Agilent Technologies) was used to analyse array images to get raw data. Next, Genespring software (version 13.1, Agilent Technologies) was employed to finish the basic analysis with the raw data. To begin with, the raw data were normalized with the quantile algorithm. The probes for which 100% of samples in any one condition have ‘Detected’ flags were chosen for further data analysis. Differentially expressed miRNAs were then identified through fold change. The threshold for up-and down-regulated genes was an absolute fold change ≥2.0, compared with the control.
Pathway analysis
Pathway analyses based on significant miRNA changes were performed using the open-access web server DIANA-miRPath v3.0. A conservative Fisher’s exact test and the false discovery rate method were used to calculate the significantly targeted Kyoto Encyclopedia of Genes and Genome (KEGG) pathways and Gene Ontology (GO) terms, and the P-value threshold was set at <0.05.
Detection of miRNA altered expression in formalin-fixed and paraffin-embedded lung cancer tissue samples
Five cases of lung cancer tissue samples were obtained from untreated patients who underwent tumour resection at Shanxi Provincial Cancer Hospital. Tissue samples were fixed in 4% buffered formalin overnight and then embedded in paraffin. MicroRNA was isolated from the formalin-fixed and paraffin-embedded (FFPE) tumour tissues using miRNAprep Pure FFPE Kits (TIANGEN, China), according to the manufacturer’s instruction. miRcute Plus miRNA First-Strand cDNA Kits (TIANGEN) were used to convert RNA into complementary DNA. Quantitative reverse transcription polymerase chain reaction experiments were performed with miRcute Plus miRNA qPCR Kits (TIANGEN) and miRNA primers (FulenGen, China). The comparative CT method was used to calculate the miRNA expression, with the universally expressed small nuclear RNA U6 used as the endogenous control.
Results
Influence of alpha radiation on cell migration and invasion
Increased migration and invasion are properties of malignant cell transformation. The effects of alpha radiation on cell migration and invasion were evaluated using a transwell migration assay and a transwell Matrigel invasion assay. The results showed that the alpha-irradiated cells displayed higher migration and invasive abilities compared with the control cells (Figs 1 and 2).
Figure 1.
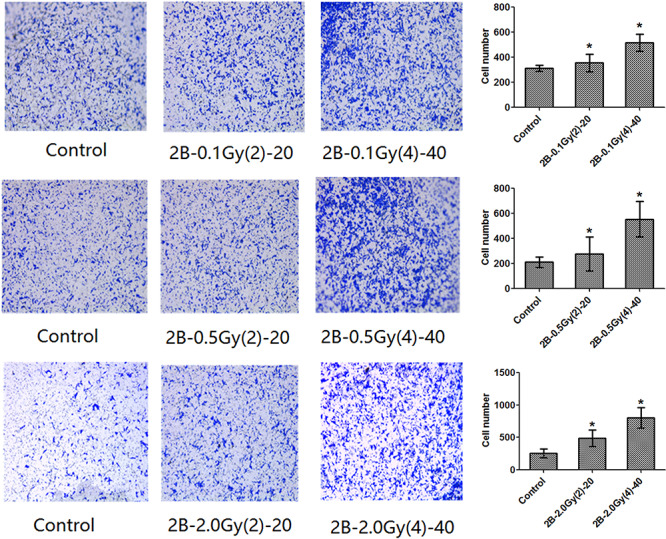
Migration ability of cells as evaluated by transwell assay.
Figure 2.
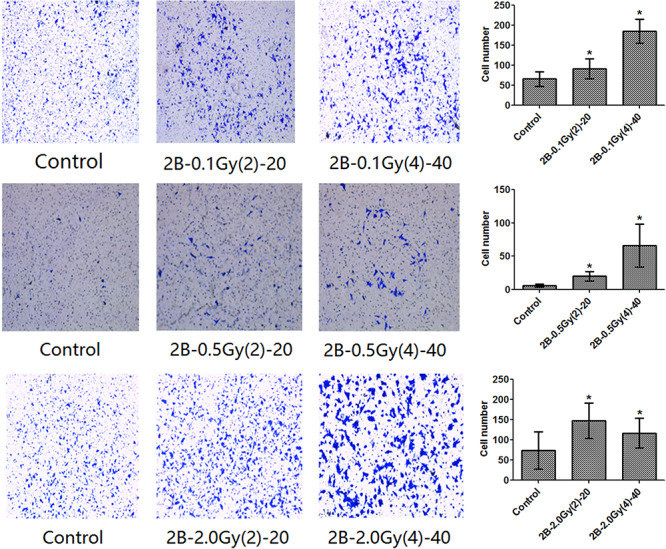
Invasive ability as detected by Matrigel-coated transwell assays.
Changes in expression level of EMT representative proteins in BEAS-2B cells after alpha irradiation
EMT is an important process inducing cancer migration and invasion. Thus, the present study examined whether EMT-associated protein expression levels changed in cells after alpha irradiation. As presented in Fig. 3, the epithelial marker, E-cadherin, was down-regulated in alpha-irradiated cells compared with the control group, whereas the mesenchymal marker, vimentin, was consistently up-regulated in the irradiated cells. Meanwhile, EMT-associated proteins, VEGFA and TGF-β1, were up-regulated in the treated cells. Therefore, it was suggested that alpha irradiation increased the migration and invasion abilities in BEAS-2B cells by promoting EMT.
Figure 3.
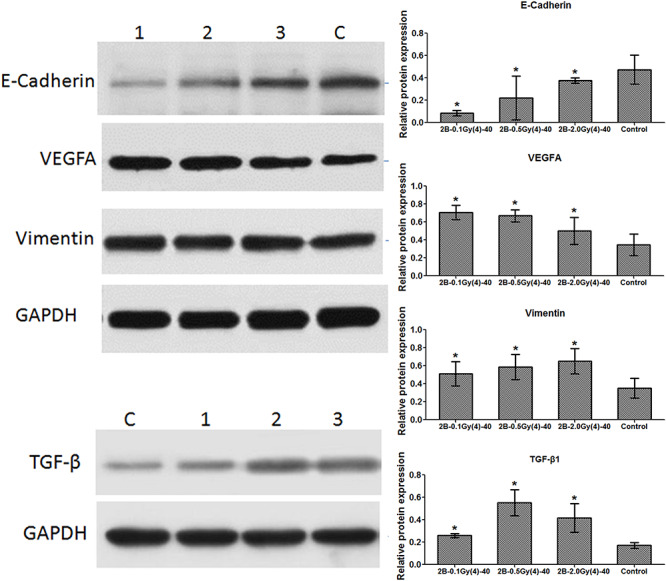
Expression levels of EMT representative proteins.
Differential regulation of miRNA expression in cell models
RNAs from cell models were tested using Agilent miRNA Chips. Relative to 2B-0Gy(2)-20 control cells, 2B-0.1Gy(2)-20, 2B-0.5Gy(2)-20, and 2B-2.0Gy(2)-20 cells had 33, 95, and 55 down-regulated miRNAs, respectively. Relative to 2B-0Gy(2)-20 control cells, 2B-0.1Gy(2)-20, 2B-0.5Gy(2)-20, and 2B-2.0Gy(2)-20 cells had 44, 37, and 79 up-regulated miRNAs, respectively. A total of 24 co-differentially expressed miRNAs in three experimental groups [2B-0.1Gy(2)-20, 2B-0.5Gy(2)-20, and 2B-2.0Gy(2)-20] were screened (Table 1). Relative to 2B-0Gy(4)-40 control cells, 2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40 cells had 38, 110, and 91 down-regulated miRNAs, respectively. Relative to 2B-0Gy(4)-40 control cells, 2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40 cells had 43, 86, and 50 up-regulated miRNAs, respectively. A total of 41 co-differentially expressed miRNAs in three experimental groups [2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40] were screened (Table 2). Furthermore, five differentially expressed miRNAs from the six experimental cell groups, including hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257, were screened (Table 3).
Table 1.
Co-differentially expressed miRNAs in three experimental groups [2B-0.1Gy(2)-20, 2B-0.5Gy(2)-20, and 2B-2.0Gy(2)-20]
| Name of miRNA | Fold change | ||
|---|---|---|---|
| 2B-0.1Gy(2)-20 | 2B-0.5Gy(2)-20 | 2B-2.0Gy(2)-20 | |
| hsa-miR-4730 | -28.69 | -27.00 | -29.59 |
| hsa-miR-4257 | -10.16 | -9.56 | 2.56 |
| hsa-miR-3188 | -9.16 | -8.62 | 4.80 |
| hsa-miR-4788 | -9.00 | -8.46 | 4.13 |
| hsa-miR-30c-2-3p | -8.79 | -8.28 | -4.21 |
| hsa-miR-6872-3p | -8.44 | -7.94 | -8.70 |
| hsa-miR-4298 | -8.05 | -7.58 | 4.14 |
| hsa-miR-3198 | -8.04 | -7.57 | 4.99 |
| hsa-miR-6717-5p | -7.64 | -7.19 | 5.85 |
| hsa-miR-28-5p | -2.47 | -21.23 | 2.04 |
| hsa-miR-148a-3p | -2.35 | -9.16 | 4.70 |
| hsa-miR-6132 | -2.11 | -8.18 | -4.02 |
| hsa-miR-6798-3p | 2.03 | 2.61 | -5.13 |
| hsa-miR-3907 | 2.09 | -10.01 | 2.15 |
| hsa-miR-582-5p | 2.75 | -8.33 | 4.49 |
| hsa-miR-634 | 2.93 | 2.99 | -3.93 |
| hsa-miR-642b-3p | 3.24 | 2.52 | 2.91 |
| hsa-miR-5001-5p | 3.28 | -8.86 | 3.12 |
| hsa-miR-7845-5p | 3.68 | -8.97 | 4.25 |
| hsa-miR-636 | 3.76 | 2.67 | -3.85 |
| hsa-miR-1470 | 9.01 | 21.61 | 3.83 |
| hsa-miR-98-3p | 9.47 | 43.49 | 3.87 |
Table 2.
Co-differentially expressed miRNAs in three experimental groups [2B-0.1Gy(4)-40, 2B-0.5Gy(4)-40, and 2B-2.0Gy(4)-40)]
| Name of miRNA | Fold change | ||
|---|---|---|---|
| 2B-0.1Gy(4)-40 | 2B-0.5Gy(4)-40 | 2B-2.0Gy(4)-40 | |
| hsa-miR-550b-2-5p | 15.48 | 15.18 | 15.12 |
| hsa-miR-3907 | 13.65 | 4.07 | 13.33 |
| hsa-miR-224-3p | 13.51 | 13.25 | 13.19 |
| hsa-miR-6512-5p | 13.40 | 13.14 | 13.08 |
| hsa-miR-1233-5p | 6.68 | 6.36 | 6.52 |
| hsa-miR-195-5p | 4.55 | 15.76 | 15.69 |
| hsa-miR-652-3p | 3.79 | 12.69 | 12.63 |
| hsa-miR-4750-3p | 3.69 | 4.60 | 4.57 |
| hsa-miR-7152-3p | 3.66 | 3.59 | 3.57 |
| hsa-miR-6732-3p | 3.63 | 3.56 | 3.55 |
| hsa-miR-6820-5p | 3.62 | 19.35 | 3.53 |
| hsa-miR-500a-3p | 3.59 | 3.52 | 3.51 |
| hsa-miR-6516-3p | 3.52 | 12.08 | 12.03 |
| hsa-miR-6808-5p | 3.44 | 17.06 | 3.36 |
| hsa-miR-6890-3p | 3.40 | 3.34 | 3.32 |
| hsa-miR-6756-3p | 3.38 | 3.31 | 3.30 |
| hsa-miR-3150b-5p | 3.36 | 3.30 | 3.28 |
| hsa-miR-182-5p | 3.18 | 3.11 | 3.10 |
| hsa-miR-345-5p | 3.18 | 3.11 | 3.10 |
| hsa-miR-3675-3p | 3.18 | 3.11 | 6.78 |
| hsa-miR-454-5p | 3.18 | 3.11 | 3.10 |
| hsa-miR-132-3p | 2.13 | 13.96 | 13.90 |
| hsa-miR-5100 | 2.06 | 4.80 | 2.91 |
| hsa-miR-6087 | 2.47 | 2.60 | 3.96 |
| hsa-miR-1225-5p | 2.50 | 2.07 | 3.96 |
| hsa-miR-4428 | 2.74 | 2.39 | 2.26 |
| hsa-miR-487b-3p | 3.40 | 14.83 | 3.55 |
| hsa-miR-3656 | 3.62 | 7.24 | 3.27 |
| hsa-miR-4254 | 3.90 | 3.43 | 3.42 |
| hsa-miR-6851-5p | 4.77 | 7.52 | 3.10 |
| hsa-miR-6726-5p | 5.50 | 4.25 | 6.32 |
| hsa-miR-4788 | 5.87 | 3.40 | 3.39 |
| hsa-miR-4314 | 6.19 | 3.35 | 3.34 |
| hsa-miR-5006-5p | 6.92 | 3.55 | 5.08 |
| hsa-miR-3679-5p | 9.18 | 11.48 | 4.69 |
| hsa-miR-4466 | 9.26 | 3.33 | 9.26 |
| hsa-miR-5001-5p | 12.72 | 12.89 | 17.97 |
| hsa-miR-8072 | 12.84 | 19.43 | 16.29 |
| hsa-miR-4257 | 14.79 | 12.93 | 13.65 |
| hsa-miR-5787 | 34.19 | 44.04 | 23.61 |
| hsa-miR-4646-5p | 68.03 | 36.58 | 68.77 |
Table 3.
Co-differentially expressed miRNAs in six experimental cell models
| Name of miRNA | Fold change | |||||
|---|---|---|---|---|---|---|
| 2B-0.1Gy(2)-20 | 2B-0.5Gy(2)-20 | 2B-2.0Gy(2)-20 | 2B-0.1Gy(4)-40 | 2B-0.5Gy(4)-40 | 2B-2.0Gy(4)-40 | |
| hsa-miR-3907 | 2.09 | -10.01 | 2.15 | -13.65 | 4.07 | -13.33 |
| hsa-miR-6732-3p | 27.27 | 39.50 | 8.26 | -3.63 | -3.56 | -3.55 |
| hsa-miR-4788 | -9.00 | -8.46 | 4.13 | 5.87 | -3.40 | -3.39 |
| hsa-miR-5001-5p | 3.28 | -8.87 | 3.12 | 12.72 | 12.89 | 17.97 |
| hsa-miR-4257 | -10.16 | -9.56 | 2.56 | 14.79 | 12.93 | 13.65 |
Biological pathways analysis of differentially expressed miRNAs
Five co-differentially expressed miRNAs (hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257) induced by alpha irradiation in six cell models were further considered for GO analysis and KEGG pathway enrichment analysis using DIANA-miRPath v3.0 [24].
GO analysis showed that 14 biological process terms were found to be significantly enriched (Table 4 and Fig. 4). Among the five miRNAs, hsa-miR-6732-3p and hsa-miR-5001-5p were involved in many biological processes, such as ‘biosynthetic process’, ‘cellular nitrogen compound metabolic process’, and ‘cellular protein modification process’. In addition, hsa-miR-4788 was highly associated with ‘cellular protein metabolic process’ and ‘blood coagulation’; and hsa-miR-4257 was involved in ‘cellular nitrogen compound metabolic process’.
Table 4.
DIANA-miRPath GO category enrichment results
| GO category | P-value | miRNAs | Number of target genes |
|---|---|---|---|
| Biosynthetic process | 2.05(E-12) | hsa-miR-6732-3p hsa-miR-5001-5p | 219 |
| Cellular nitrogen compound metabolic process | 3.01(E-11) | hsa-miR-6732-3p hsa-miR-5001-5p hsa-miR-4257 | 298 |
| Cellular protein modification process | 1.15(E-06) | hsa-miR-6732-3p hsa-miR-5001-5p | 130 |
| Gene expression | 8.07(E-05) | hsa-miR-6732-3p hsa-miR-5001-5p | 42 |
| Viral process | 1.20(E-03) | hsa-miR-6732-3p hsa-miR-5001-5p | 36 |
| Symbiosis, encompassing mutualism through parasitism | 1.52(E-03) | hsa-miR-6732-3p hsa-miR-5001-5p | 38 |
| Epidermal growth factor receptor signalling pathway | 4.38(E-03) | hsa-miR-6732-3p | 10 |
| Transcription, DNA templated | 5.24(E-03) | hsa-miR-6732-3p hsa-miR-5001-5p | 139 |
| Neurotrophin TRK receptor signalling pathway | 5.87(E-03) | hsa-miR-6732-3p | 10 |
| Catabolic process | 7.60(E-03) | hsa-miR-5001-5p | 70 |
| Fc-epsilon receptor signalling pathway | 9.99(E-03) | hsa-miR-6732-3p | 8 |
| Cellular protein metabolic process | 1.77(E-02) | hsa-miR-6732-3p hsa-miR-4788 | 14 |
| Blood coagulation | 3.11(E-02) | hsa-miR-4788 hsa-miR-5001-5p | 21 |
| Small molecule metabolic process | 3.14(E-02) | hsa-miR-5001-5p | 75 |
Figure 4.
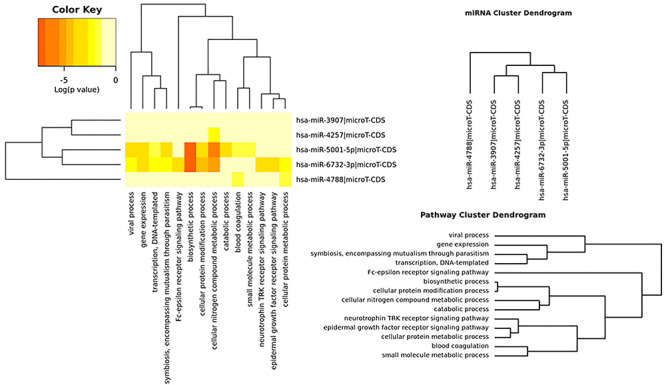
Heatmap of the enriched biological processes related to the predicted miRNA targets.
KEGG analysis showed that five KEGG pathways were significantly enriched (Table 5 and Fig. 5); hsa-miR-5001-5p was highly associated with three KEGG pathways: cell adhesion molecules (CAMs), proteoglycans (PGs) in cancer, and allograft rejection; and hsa-miR-4257 was highly associated with extracellular matrix (ECM)–receptor interaction and hsa-miR-6732-3p was highly associated with glioma.
Table 5.
DIANA-miRPath KEGG pathway enrichment results
| KEGG pathway | P-value | MicroRNAs | Genes |
|---|---|---|---|
| Cell adhesion molecules (CAMs) | 2.56(E-05) | has-miR-5001-5p | L1CAM,SDC3,SDC1,CLDN19,VTCN1,CLDN18,LRRC48,HLA-DRA,PTPRF,HLA-DOB,CADM3,HLA-F,CNTNAP1,CLDN10,ITGB2,HLA-DQA1 |
| ECM–receptor interaction | 2.26 (E-03) | has-miR-4257 | COL2A1,COL4A4,SV2C,CD44 |
| Proteoglycans in cancer | 6.87 (E-03) | has-miR-5001-5p | CAMK2G,SDC1,PTCH1,PIK3R5,WNT2B,RPS6KB2,FLNB,FLNA,AKT3,MAPK1 |
| Allograft rejection | 2.68(E-02) | has-miR-5001-5p | HLA-DRA,HLA-DOB,HLA-F,HLA-DQA1 |
| Glioma | 3.74 (E-02) | has-miR-6732-3p | PRKCA,PDGFB,PIK3CA,PTEN,MDM2 |
Figure 5.
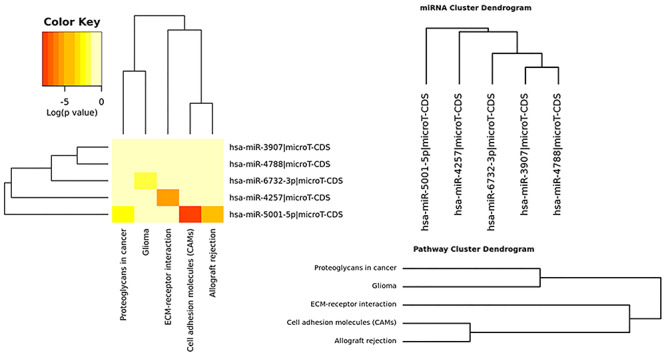
Heatmap of the enriched KEGG pathways related to the predicted miRNA targets.
MicroRNA expression profile in lung cancer tissues
Expression levels of miR-3907, miR-6732-3p, miR-4788, miR-5001-5p, and miR-4257 were evaluated in five paired samples (cancerous and non-malignant adjacent tissues) by RT-qPCR. The relative quantification of miRNA expression was performed by the 2−△△Ct method and expression levels of non-malignant adjacent tissues were used as a calibrator. As shown in Table 6, the expression levels of five miRNAs were up-regulated in all five lung cancer tissue samples.
Table 6.
Expression of miRNAs in lung cancer tissues
| Sample | miR-3907 | miR-4257 | miR-4788 | miR-5001-5p | miR-6732-3p |
|---|---|---|---|---|---|
| 001 | 3.69 ± 0.23 | 1.97 ± 0.22 | 2.27 ± 0.39 | 2.08 ± 0.09 | 1.62 ± 0.35 |
| 002 | 1.00 ± 0.24 | 1.35 ± 0.16 | 3.50 ± 0.18 | 2.09 ± 0.99 | 1.16 ± 0.67 |
| 003 | 1.52 ± 0.03 | 1.77 ± 0.32 | 1.65 ± 0.11 | 1.00 ± 0.15 | 1.87 ± 0.20 |
| 004 | 4.57 ± 0.78 | 2.53 ± 0.39 | 3.14 ± 0.35 | 2.83 ± 0.45 | 1.98 ± 0.38 |
| 005 | 1.78 ± 0.18 | 2.31 ± 0.52 | 2.19 ± 0.27 | 2.33 ± 0.71 | 1.52 ± 0.41 |
Discussion
Radon has already been defined as a lung carcinogen by the International Agency for Research on Cancer based on the radon-exposed miners cohort from 1988 [25]. Damage to epithelial cells of the lung occurs when radiation interacts either directly with DNA in the cell nucleus or indirectly through the effect of free radicals [26]. Mortality from this disease could be reduced greatly through the identification of molecular markers present in individuals at the earliest stages of lung cancer, in which curative resection is feasible [26]. A number of studies have identified aberrant miRNA expression in different types of cancer, including lung cancer [27, 28]. Since miRNAs are involved in the development and progression of lung cancer, there is a particular interest in miRNAs not only as novel biomarkers but also as potential targets for treatment [20, 29].
To assess the potential of a molecular-based marker approach for early detection of human lung cancer induced by radon, alpha irradiation-induced malignant transformation models of human bronchial epithelial cells were constructed. In order to simulate the long-term harm of radon on the human body, the cell model in this study was constructed by means of multiple cumulative irradiation events with alpha particles, which reflect the different stages and degrees of cell damage induced by alpha irradiation. Specifically, after each irradiation event, the cells were subcultured for 10 generations. The cells in each group were irradiated four times and cultured for 40 generations. Cells were treated with one of three doses (0.1, 0.5, and 2 Gy) to reflect the different exposure hazards of different doses of alpha particles. Ultimately, cell models with different irradiation doses, different numbers of irradiation events, and different numbers of generations were obtained. The index properties of malignant cell transformation, i.e. migration and invasion, were measured. The results showed that malignant cell transformation could be significantly triggered by alpha irradiation. EMT is an important process in inducing cancer migration and invasion, which is often associated with a loss or decrease of E-cadherin and an increase of vimentin, TGF-β, and VEGFA [30–33]. Western blot analysis of EMT-associated proteins indicated that alpha irradiation can induce EMT, which in turn leads to the enhancement of cell migration and invasion, and finally leads to the occurrence of malignant transformation of cells.
Expression profiles of miRNAs were measured in cell models irradiated twice and subcultured for 20 generations and cell models irradiated four times and subcultured for 40 generations. Potential miRNAs of bronchial epithelial cells damaged by alpha radiation (hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257) were identified by comparing the differentially expressed miRNAs of different cell models. Literature searches on the functions of hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257 revealed few reports on these five miRNAs. Though no detailed studies have discussed the specific roles of these miRNAs, researchers have shown hsa-miR-4257 may bind to genes involved in the mitochondrial apoptosis pathway [34], and hsa-miR-3907 may be related to the pathophysiological progress of autosomal dominant polycystic kidney disease [35].
In order to further reveal the possible function of miRNAs in BEAS-2B cells damaged by alpha irradiation, DIANA-miRPath was used to predict the target genes of miRNAs. Results showed that five KEGG pathways related to hsa-miR-5001-5p, hsa-miR-4257, and hsa-miR-6732-3p were enriched, including CAMs, ECM–receptor interaction, PGs in cancer, allograft rejection, and glioma. Most of these pathways are closely related to the occurrence and development of carcinogenesis. Tumour angiogenesis, tumour–stroma interactions, and interactions with immune cells, as well as with the ECM and cancer stem cell niches, allow for malignant cell survival and the promotion of metastasis [36–38]. PGs and glycosaminoglycans, comprising structurally diverse constituents of the ECM and cell surfaces, have emerged as novel biomarkers and molecular players both within tumour cells and their microenvironment [37–39]. PGs in the tumour microenvironment have been shown to be key macromolecules that contribute to biology of various types of cancer proliferation, adhesion, angiogenesis, and metastasis, which affects tumour progression. The expression of PGs can be regulated by miRNAs [38]. CAMs are (glyco) proteins expressed on the cell surface and play a critical role in a wide array of biological processes that include haemostasis, immune response, inflammation, embryogenesis, and development of neuronal tissue. Adhesive interactions mediated by CAMs allow cells to sort into groups and form complex structures and as such are of critical importance in organogenesis and tissue reconstruction. In a variety of human malignancies, tumour progression has been associated with changes in CAM expression [40, 41]. Literature reports and our bioinformatics analysis indicate that hsa-miR-3907, hsa-miR-6732-3p, hsa-miR-4788, hsa-miR-5001-5p, and hsa-miR-4257 may affect the biological processes of bronchial epithelial cells by regulating target genes and participate in damage and malignant transformation induced by alpha irradiation. The differential expression of these miRNA molecules in lung cancer tissues also reveals that there is a certain correlation between these molecules and lung cancer, which seems to be consistent with our bioinformatics analysis.
In this study, a series of human bronchial epithelial cell injury models induced by alpha-particle irradiation were constructed. And five miRNAs screened were expected to be potential early markers of damage and malignant transformation of bronchial epithelial cells induced by alpha irradiation. Further research of five miRNAs will be improved in populations living in radon-polluted areas or working in professions with higher exposure, which were expected to be used for early screening of lung cancer caused by radon.
Conflicts of interest
The authors declare no conflict of interest.
Contributor Information
Xuhong Dang, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Haipeng Lin, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Youchen Li, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Xiuli Guo, Department of Pathology, Shanxi Provincial Cancer Hospital, Taiyuan 030013, China.
Yayi Yuan, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Ruifeng Zhang, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Xiaozhen Li, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Dongliang Chai, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
Yahui Zuo, Division of Radiology and Environmental Medicine, China Institute for Radiation Protection, Taiyuan 030006, China.
References
- 1. Sethi TK, El-Ghamry MN, Kloecker GH. Radon and lung cancer. Clin Adv Hematol Oncol 2012;10:157–64. [PubMed] [Google Scholar]
- 2. Robertson A, Allen J, Laney R, Curnow A. The cellular and molecular carcinogenic effects of radon exposure: a review. Int J Mol Sci 2013;14:14024–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization WHO Handbook on Indoor Radon: A Public Health Perspective. Geneva, Switzerland: World Health Organization, 2009. [PubMed] [Google Scholar]
- 4. Kreuzer M, Fenske N, Schnelzer M, Walsh L. Lung cancer risk at low radon exposure rates in German uranium miners. Br J Cancer 2015;113:1367–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nie JH, Chen ZH, Shao CL et al. Analysis of the miRNA-mRNA networks in malignant transformation BEAS-2B cells induced by alpha-particles. J Toxicol Environ Health A 2016;79:427–35. [DOI] [PubMed] [Google Scholar]
- 6. Lino Ada R, Abrahao CM, Amarante MP, Sousa Cruz MR. The role of the implementation of policies for the prevention of exposure to radon in Brazil-a strategy for controlling the risk of developing lung cancer. Ecancermedicalscience 2015;9:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Archer VE, Coons T, Saccomanno G, Hong DY. Latency and the lung cancer epidemic among United States uranium miners. Health Phys 2004;87:480–9. [DOI] [PubMed] [Google Scholar]
- 8. Gilliland FD, Hunt WC, Archer VE, Saccomanno G. Radon progeny exposure and lung cancer risk among non-smoking uranium miners. Health Phys 2000;79:365–72. [DOI] [PubMed] [Google Scholar]
- 9. Darby S, Hill D, Auvinen A et al. Radon in homes and risk of lung cancer: collaborative analysis of individual data from 13 European case-control studies. BMJ 2005;330:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cristino AS, Williams SM, Hawi Z et al. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol Psychiatry 2014;19:294–301. [DOI] [PubMed] [Google Scholar]
- 11. Greco S, Zaccagnini G, Fuschi P et al. Increased BACE1-AS long noncoding RNA and beta-amyloid levels in heart failure. Cardiovasc Res 2017;113:453–63. [DOI] [PubMed] [Google Scholar]
- 12. Martin HC, Wani S, Steptoe AL et al. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol 2014;15:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ai J, Zhang R, Li Y et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun 2010;391:73–7. [DOI] [PubMed] [Google Scholar]
- 14. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 2015;5:1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X, Ba Y, Ma L et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 16. Ogata-Kawata H, Izumiya M, Kurioka D et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang GK, Zhu JQ, Zhang JT et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010;31:659–66. [DOI] [PubMed] [Google Scholar]
- 18. Yang Z, Chen H, Si H et al. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol 2014;51:823–31. [DOI] [PubMed] [Google Scholar]
- 19. Tanaka Y, Kamohara H, Kinoshita K et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159–67. [DOI] [PubMed] [Google Scholar]
- 20. Bulgakova O, Zhabayeva D, Kussainova A et al. miR-19 in blood plasma reflects lung cancer occurrence but is not specifically associated with radon exposure. Oncol Lett 2018;15:8816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaudhry MA. Radiation-induced microRNA: discovery, functional analysis, and cancer radiotherapy. J Cell Biochem 2014;115:436–49. [DOI] [PubMed] [Google Scholar]
- 22. Liu W, Xiao L, Dong C et al. Long-term low-dose alpha-particle enhanced the potential of malignant transformation in human bronchial epithelial cells through MAPK/Akt pathway. Biochem Biophys Res Commun 2014;447:388–93. [DOI] [PubMed] [Google Scholar]
- 23. Tian W, Yin X, Wang L et al. The key role of miR-21-regulated SOD2 in the medium-mediated bystander responses in human fibroblasts induced by alpha-irradiated keratinocytes. Mutat Res 2015;780:77–85. [DOI] [PubMed] [Google Scholar]
- 24. Vlachos IS, Zagganas K, Paraskevopoulou MD et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu Q, Fang L, Chen B et al. Radon induced mitochondrial dysfunction in human bronchial epithelial cells and epithelial-mesenchymal transition with long-term exposure. Toxicol Res (Camb) 2019;8:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su S, Jin Y, Zhang W et al. Aberrant promoter methylation of p16(INK4a) and O(6)-methylguanine-DNA methyltransferase genes in workers at a Chinese uranium mine. J Occup Health 2006;48:261–6. [DOI] [PubMed] [Google Scholar]
- 27. Inamura K. Major tumor suppressor and oncogenic non-coding RNAs: clinical relevance in lung cancer. Cell 2017;6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inamura K. Diagnostic and therapeutic potential of microRNAs in lung cancer. Cancers (Basel) 2017;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inamura K, Ishikawa Y. MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J Clin Med 2016;5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442–54. [DOI] [PubMed] [Google Scholar]
- 31. Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol 2003;4:657–65. [DOI] [PubMed] [Google Scholar]
- 32. Ungefroren H, Witte D, Lehnert H. The role of small GTPases of the Rho/Rac family in TGF-beta-induced EMT and cell motility in cancer. Dev Dyn 2018;247:451–61. [DOI] [PubMed] [Google Scholar]
- 33. Kim M, Jang K, Miller P et al. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene 2017;36:5199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu X, Wang X, Fu B et al. Differentially expressed genes and microRNAs in bladder carcinoma cell line 5637 and T24 detected by RNA sequencing. Int J Clin Exp Pathol 2015;8:12678–87. [PMC free article] [PubMed] [Google Scholar]
- 35. Kocyigit I, Taheri S, Sener EF et al. Serum micro-RNA profiles in patients with autosomal dominant polycystic kidney disease according to hypertension and renal function. BMC Nephrol 2017;18:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 37. Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J 2010;277:3904–23. [DOI] [PubMed] [Google Scholar]
- 38. Ibrahim SA, Hassan H, Gotte M. MicroRNA regulation of proteoglycan function in cancer. FEBS J 2014;281:5009–22. [DOI] [PubMed] [Google Scholar]
- 39. Hassan H, Greve B, Pavao MS et al. Syndecan-1 modulates beta-integrin-dependent and interleukin-6-dependent functions in breast cancer cell adhesion, migration, and resistance to irradiation. FEBS J 2013;280:2216–27. [DOI] [PubMed] [Google Scholar]
- 40. Edelman GM. Cell adhesion molecules in the regulation of animal form and tissue pattern. Annu Rev Cell Biol 1986;2:81–116. [DOI] [PubMed] [Google Scholar]
- 41. Johnson JP. Cell adhesion molecules of the immunoglobulin supergene family and their role in malignant transformation and progression to metastatic disease. Cancer Metastasis Rev 1991;10:11–22. [DOI] [PubMed] [Google Scholar]


