Abstract
Double homeobox genes are unique to eutherian mammals. It has been proposed that the DUXC clade of the double homeobox gene family, which is present in multicopy long tandem arrays, plays an essential role in zygotic genome activation (ZGA). We generated a deletion of the tandem array encoding the DUXC gene of mouse, Double homeobox (Dux), and found it surprisingly to be homozygous viable and fertile. We characterize the embryonic development and ZGA profile of knockout (KO) embryos, finding that zygotic genome activation still occurs, with only modest alterations in 2-cell embryo gene expression, no defect in in vivo preimplantation development, but an increased likelihood of post-implantation developmental failure, leading to correspondingly smaller litter sizes in the KO strain. While all known 2-cell specific Dux target genes are still expressed in the KO, a subset is expressed at lower levels. These include numerous genes involved in methylation, blastocyst development, and trophectoderm/placental development. We propose that rather than driving ZGA, which is a process common throughout the animal kingdom, DUXC genes facilitate a process unique to eutherian mammals, namely the post-implantation development enabled by an invasive placenta.
Keywords: double homeobox, DUXC, Dux, zygotic genome activation, post-implantation development
Summary sentence: Mouse Dux is not absolutely required for viability or fertility, nor for zygotic genome activation, however the subset of Dux knockout embryos, post-implantation development fails.
Introduction
At the beginning of each generation, in the germ cells, the activity state of the genome is reset, allowing pluripotency to be reestablished. After fusion of egg and sperm, however, the reset genome of the zygote is essentially inactive, with maternal (oocyte) RNAs governing cellular processes. Although a minor wave of transcription can be detected after fusion, the major wave of transcription first occurs one or more cell divisions later (at the 2-cell stage in mice, the 4/8 cell stage in humans) [1], an event referred to as zygotic genome activation (ZGA). Recently, the double homeobox gene Dux (DUX4 in humans) has been implicated as an activator of many genes expressed during the major wave of ZGA [2, 3], with one study suggesting that Dux is in fact the key inducer of ZGA [4].
The double homeobox (DUX) family of DNA-binding proteins is unique to eutherian mammals—in no other species do two homeodomains exist in tandem in the same protein [5]. Eutherian mammalian genomes actually harbor a clade of 3 DUX genes, categorized as DUXA, B and C. While all 3 DUX genes encode proteins with an N-terminal double homeodomain, only the DUXC branch harbors a conserved C-terminal activation domain [6], which allows DUXC representatives, such as DUX4 and mouse Dux, to induce expression of downstream targets [7, 8]. DUX4 is a pioneer factor, with the capacity to bind within inaccessible chromatin, where it recruits the histone acetyltransferases p300 and cAMP-response element binding protein (CBP) to remodel chromatin enabling transcription of its target genes [9]. A further interesting feature of the DUXC genes vis-à-vis the rest of the clade is that DUXC genes exist in large arrays of tandem head-to-tail repeats. In humans, DUX4 is present in two such large arrays, both subtelomeric, at 4qter in on average ~30 copies and at 10 qter in on average ~20 copies [10]. This means that DUXC genes are present at enormous copy number. However, in spite of their massive copy number, the proteins encoded by the DUXC genes are not expressed, because repeat-induced silencing, orchestrated by structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) [11], and mediated by excessive DNA methylation [12–14] locks the arrays into inactive heterochromatin. This silencing is weaker in lower copy number arrays, and mutations reducing copy number lead to the genetic myopathy, facioscapulohumeral muscular dystrophy (FSHD) [15], presumably by allowing very low levels of DUX4 protein expression to leak from the locus in muscle cells [16].
After fusion of egg and sperm, zygotic pronuclei undergo massive demethylation, particularly the male pronucleus [17], coincident with their reprogramming, meaning that for a brief moment in development, hypermethylation of the DUXC gene arrays is lost. Since DNA methylation is necessary for silencing of the array [12], DUXC proteins and downstream target genes should be briefly detected during preimplantation development, and this is indeed the case in both human and mouse for DUX4 and Dux, respectively [2].
Whether this blip of expression has important developmental consequences requires loss-of-function analysis. A study using CRISPR injection into zygotes to cleave at either end of the mouse Dux array suggested that this approach led to the absence of Dux expression and catastrophic early developmental failure [4]. However, since cutting at two sites followed by NHEJ is predicted to render the excision as a large DNA circle carrying the Dux gene array, which would only be lost after multiple cell divisions by dilution, this strategy seems less reliable than a true knockout (KO) for evaluating 2-cell embryos. In addition, injection of zygotes and in vitro culture both introduce significant nonphysiological stress. A recent study has generated a Dux KO mouse, and found that the KO was viable and fertile, although KOs produced smaller litter sizes [18]. Although numerous Dux target genes were affected at the 2-cell stage in the KOs, the embryos did not arrest at this stage. Later embryonic development was not specifically addressed in that study. Here, we analyze the in vivo development of an independent genomic Dux KO and find that in the absence of mouse Dux, catastrophic failure of development does not occur. Subtle differences in developmental success, particularly after in vitro culture, suggest that early cleavage-stage mammalian embryonic development has adapted to the presence of DUXC proteins, but not to the point of making them essential to the process. Instead, we find an increased failure rate of post-implantation development of Dux KO embryos.
Materials and methods
Ethics
Animals were maintained under protocol 1708-35046A approved by the University of Minnesota IACUC. Animals were euthanized by cervical dislocation.
Genetic engineering
C57BL/6 embryonic stem cells (ESCs) were engineered by homologous recombination in which 5′ and 3′ homology arms (7.0 kb and ~6.9 kb, respectively) representing unique sequence immediately outside of the Dux array were amplified by high-fidelity polymerase chain reaction (PCR) and cloned pCR4.0 vector. A single Dux repeat (5.3 kb) was amplified by high-fidelity PCR into pLoxFtNwCD, next to an Frt-flanked neo gene, and subcloned into the targeting vector. Amplification primers comprised the following sequences, where capital lettering denotes mouse genomic sequence: 5PH-F: 5-gactcactaggcggccgcGACACACAGTTCATCCTTCCATCCAC-3′; 5PH-R: 5′-attgcggcctttggtaccatatgAAGACAACCCACCCATAAAAATAAAGAA-3′; 3PH-F: 5′- ctggccaatcggcgcgccCATGGTCTCTTCCTCTGTTCTTTCTCCTT-3′; 3PH-R: 5′- gcgagagctcggcgcgCCTAAATCCAATCCAGAACGCATAACA-3′; DUX-F: 5′-gtcgatcgagaccgtacgTGCTGGGTTTGGTTTGGTTCTGTG-3′; DUX-R: 5′- aagctcgatgcatcgtacgATCTACAGATTGATTTAGGCATGGATGGA-3′. The recombination arms and inserted Dux unit were fully sequenced, and the recombination plasmid was linearized and electroporated into C57BL/6 mouse ES cells. Recombinant G418-resistant colonies were picked, expanded, and evaluated for replacement of the Dux array with the floxed Dux unit by Southern blot to generate Dux1UnitFL-neo mouse ES cells. The neo selection gene was eliminated by Frt recombination to generate Dux1UnitFL mouse ES cells.
Mice
Mice were maintained, and in vivo experiments were conducted, under a protocol approved by the University of Minnesota IACUC. Dux1UnitFL mice, bearing a single floxed copy of the Dux repeat, were derived from DuxFL engineered C57BL/6 mouse ES cells. The DuxFL allele was converted into a deletion allele by crossing to Protamine-cre mice [19] and identifying offspring heterozygous for the DuxΔ allele. Heterozygotes were bred to derive a DuxΔ/Δ stock, and the stock maintained by sibling mating. Dux1UnitFL and DuxΔ/Δ mice were genotyped with the following primers: mDUX-LoxP F (floxed) 5′-CAG GCC AGT CTT TTA ATA TCT ACT ACC CAT AGG-3′; mDUX-LoxP R (floxed) 5′-CAC AGT ACA CAA ACA CAC AAA CAA ATT TTG TC-3′; mDUX-KO R 5′-GTT AGT ACT GCT TCC TTG AGA GAC ATT GCC-3′.
Embryo collection, culturing, and preparation for transcriptional analysis
Two-cell stage embryos were collected from female mice (4 weeks old) that were superovulated by intraperitoneal injections of PMSG (5 IU/0.1 mL, Accurate Chemical and Scientific Corporation) followed, 48 hours later, by hCG (5 IU/0.1 mL, Sigma-Aldrich). Following the second hormonal treatment, the females were mated with either C57BL/6 or KO males. 48 h after the last hormone injection, 2-cell stage embryos were collected. Blastocyst embryos were collected from animals that were normally mated, considering the morning of detection of vaginal plug to be 0.5 days post-fertilization. Oviducts were dissected and the embryos were flushed out and washed from debris with M2 medium as previously described [20]. For transcriptional analyses, embryos from each female were pooled together (6–9 embryos) and libraries were prepared using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara) and Nextera XT DNA Library Preparation Kit (Illumina) following the manufacturer’s instructions. In total, 50 base single-end sequencing was performed on an Illumina HiSeq instrument at the University of Minnesota Genomics Center. For the embryo culture experiments, 2-cell stage embryos were cultured in modified human tubal fluid medium [21]. For evaluating number and morphology of blastocysts in vivo, mice were naturally mated and at 3.5 days post-coitum (d.p.c) uteri were dissected and flushed with M2 media. E3.5 embryos were photographed at ×20 on a Zeiss Axiovert 40C microscope.
Bioinformatics analysis
Single-end 50 bp Illumina reads were trimmed with cutadapt (v 1.18) and Trim Galore (v0.6.0) and mapped to the GRCm38 genome with STAR (v2.6.1a) using the optional parameter —outFilterMultimapNmax 100 to report reads that map to endogenous retroviruses. Gene abundance was estimated by enumerating the number of reads overlapping gene annotations in Gencode M22 using RSubread (1.28.1). For gene abundance estimates, multimapping reads with quality scores less than 55 were excluded. The abundances of repeat elements in the transcriptome were enumerated using the repeat masker table from UCSC including the multimapping reads without a quality filter threshold. Differential expression was determined with DESeq2 (c1.24.0) and the Benjamini–Hochberg procedure was applied for multiple test correction. Pathway analysis was performed using gene ontology (GO) data from the Mouse Genome Informatics database (https://informatis.jax.org) and hypergeometric tests were performed with the goseq package (v1.40.0) taking into account the transcript length bias. Enriched categories were consolidated using REViGO (https://revigo.irb.hr). Figures were generated using ggplot2 (v3.2.0) and ComplexHeatmap (v2.0.0) with custom scripts available at https://github.com/micahgearhart/zga. Raw fastq files from GSE45719, GSE121746 and GSE85627 were downloaded and mapped as above. Gene abundance estimates from GSE45719, GSE121746 and data from this study were combined into a single matrix to compute principal components. Batch effects corrections were not applied as the first two components were due to developmental timing rather than batch. Data from GSE85627 was used to define Dux overexpression targets as genes that had a log2 fold change > 1 or < −1, Benjamini–Hochberg adjusted P-value < 0.05 and mean counts > 10. The Gene Expression Omnibus accession number is GSE142043.
Statistics
GraphPad Prism software was used for statistical analyses. Differences between groups were evaluated by one-way analysis of variance followed by Tukey’s post hoc tests. Data are presented as mean ± standard error of the mean (SEM). Differences were considered significant at P-values ≤ 0.05.
Results
Because of the possibility that Dux genes could be essential, instead of deleting the locus in its entirety, we replaced the Dux array with a single floxed copy of one 5 kb unit of the array, on one allele in C57BL/6 ES cells (Figure 1A). Mice homozygous for the Dux1unitFL allele were viable and fertile; therefore, we deleted the remaining single copy with a germ-line cre [19] and crossed heterozygotes to generate homozygous KOs, which were also viable and fertile. Quantitative PCR (qPCR) on genomic DNA allowed us quantify the number of copies of Dux in C57BL/6, revealing the Dux array to be 22 units in size (Figure 1B). Anecdotally, we noted that litters from DuxΔ/Δ mice were often small (Figure 1C); therefore we set up a number of crosses testing litter sizes with different combinations of parental genotypes. Genotyping almost 2000 pups over 338 litters revealed a statistically significant difference between litter size from DuxΔ/Δ parents (4.4 pups) vs. wild-type (WT) B6 parents (6.2 pups) (Figure 1C). Other parental combinations were not statistically different from WT × WT, although male +/Δ × female Δ/Δ trended (P = 0.1) to slightly smaller litters (5.4 pups). To determine whether litter size reduction was due to loss of animals bearing the Δ/Δ genotype, we evaluated genotypes of progeny from different types of crosses. The Δ/Δ genotype was indeed underrepresented in both classes of heterozygous backcross (male KO or female KO, Figure 1D).
Figure 1.
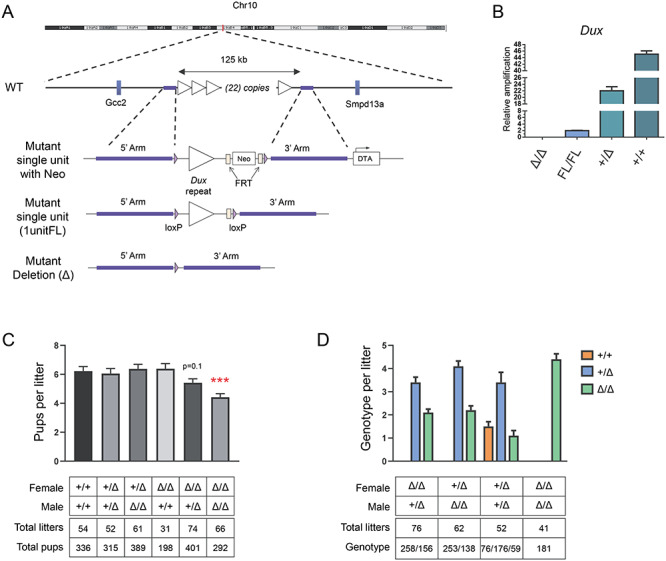
Dux KO generation, inheritance and effect on litter size. A. C57BL/6 ES cells were modified with a recombination vector that deleted the entire Dux array, and replaced it with a floxed single copy of the repeat. After removal of the selectable marker, mutant mice carrying the single floxed unit on one allele were generated by blastocyst injection. This single copy was then deleted with a germ-line cre to generate the deletion allele. B. qPCR for Dux on genomic DNA of mice of different genotypes, normalized to Dux copy number in the 1unitFL allele. The B6 WT allele is estimated to carry 22 copies of the Dux gene. C. Litter sizes of crosses between different genotypes. Data represents mean ± SEM; ***P < 0.001. D. Genotypes of progeny from different types of crosses. The Δ/Δ genotype is underrepresented in both classes of heterozygous backcrosses.
To determine whether the modest underrepresentation of Δ/Δ genotypes was due to failure of fertilization or failure of embryogenesis, we investigated embryos derived from Δ/Δ females bred to males of different genotypes. Counting all decidual masses without regard to viability revealed similar numbers across all genotypes, suggesting that loss occurred at some point after fertilization (Figure 2A). Evaluating genotypes of live embryos after E8.5 revealed a reduced representation of the KO genotype relative to the heterozygote in heterozygous backcrosses (Figure 2B). As discerning genotypes of dead embryos is unreliable due to resorption and contamination with maternal cells, we evaluated the frequencies of live and dead/resorbed embryos in pregnancies from various parental genotypes (Figure 2C and D). This demonstrated an increased ratio of dead: live embryos when both parents were homozygous Δ/Δ, suggesting that KO embryos have a higher rate of developmental failure than heterozygotes (Figure 2D).
Figure 2.
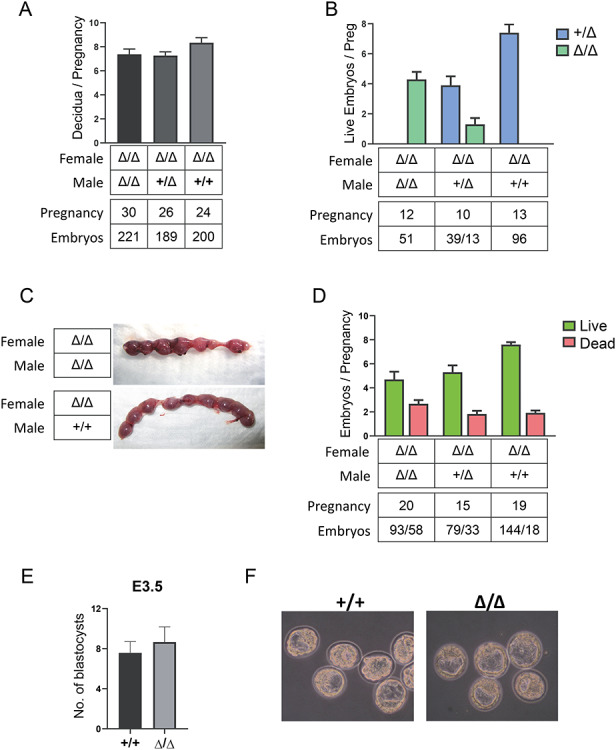
Dux KO effects on embyogenesis. A. Number of implantations per pregnancy (all decidual masses were counted, normal or resorbing). Data is combined from stages between E5.5 and E18.5. B. Genotypes of the live embryos combined from E8.5 to E12.5. C. Representative images of dissected uterus from Δ/Δ females (E13.5) crossed with either Δ/Δ or WT males. Note the evident embryos resorptions in the upper image. D. Number of live and dead embryos per pregnancy from E8.5 to E18.5 in crosses of different genotypes. E. Number of embryos per pregnancy isolated from Dux KOs (Δ/Δ females crossed with Δ/Δ males) and C57BL/6 (+/+) controls at 3.5 d.p.c. (n = 6 KO litters and 6 heterozygote litters). F. Representative example showing morphology of the embryos analyzed in (E) isolated at 3.5 d.p.c.
To clarify whether embryos regress before or after implantation we isolated and quantified E3.5 blastocysts from Δ/Δ and WT B6 mice. A specific defect in ZGA would predict that a subset of embryos would be arrested at the 2-cell stage or thereabouts. However, we did not observe any difference in embryo number (Figure 2E) and KO embryos reached the blastocyst stage as efficiently as WT embryos (Figure 2F). Notably, at 3.5 d.p.c. all of the isolated embryos were at late morula or blastocyst stage and we did not find any embryos arrested at the 2-cell stage or at any cleavage stage.
Because resorptions can indicate developmental failure of post-implantation embryos, and these numbers accounted fairly well for the moderately reduced litter size of the KO strain, this data suggests that in vivo, Dux KO embryos appear to undergo cleavage divisions, blastocyst development, and implantation normally.
The initial report of developmental arrest was made based on in vitro culture of CRISPR-injected embryos [4]; therefore, we also investigated in vitro development of KO and heterozygous embryos harvested at the 2-cell stage (Figure 3A). Surprisingly, under in vitro culture conditions, almost half of the KO embryos failed to develop into blastocysts (Figure 3B). Therefore, although in vivo development proceeds apparently normally through early stages in the KO, the embryo is nevertheless more sensitive to the stress of removal from its normal maternal environment.
Figure 3.
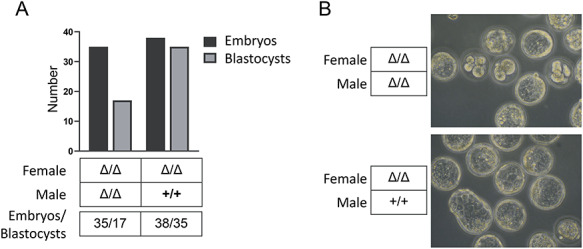
In vitro culture of Dux KO embryos. A. Number of embryos that developed into blastocysts from each genotype. B. Images of embryos harvested at the 2-cell stage, cultured for 5 days in vitro.
Because a large subset of target genes upregulated by DUX4 and Dux in transient overexpression studies is also expressed at the 2-cell stage, it has been proposed that Dux is the key driver of the early cleavage-stage gene expression profile [2, 3]. We therefore investigated the gene expression profile of pooled 2-cell embryos from KOs and compared these to similarly staged WT embryos. Principal component analysis of these profiles in the context of profiles obtained from a study of mouse cleavage stage development [22] placed our embryos in the mid-late 2-cell stage as defined by PC1 (Figure 4A). We also compared to a recent study of 2 cell Dux KO and WT embryos [18] and found very close approximation on both major PC axes. The proximity of the two Dux studies vis-à-vis the cleavage development study is likely enhanced by strain background: both Dux studies were on a C57BL/6 background whereas the other employed an F1 background. Both studies showed a right shift of the KOs, potentially indicating a slight developmental delay.
Figure 4.
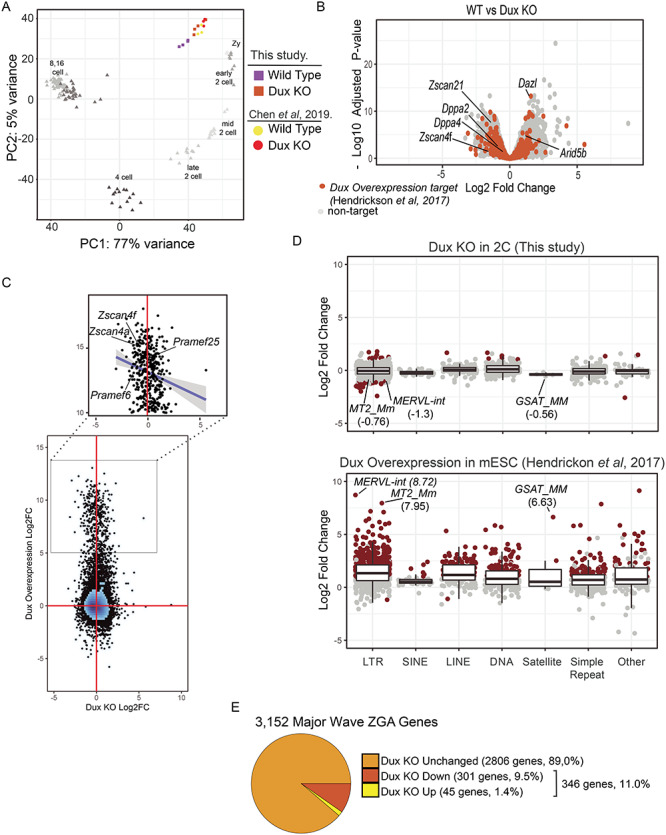
Gene expression changes at the 2-cell stage in Dux KO embryos. A. Principal component analysis of WT (purple) and Dux KO (orange) gene expression profiles at the 2-cell stage. Prior data from the 2-cell stage is shown for WT (yellow) and Dux KO (red) [18]. The principal components were calculated by including developmental time course from zygotes to the 16 cell stage using data from [22]. Principal component 1 captures 77% of the variance across samples reflecting the developmental stage. B. Gene expression changes between WT and Dux KO embryos. The x-axis corresponds to the log2Fold change and the y-axis corresponds to the negative log10 of the Benjamini–Hockberg corrected P-value. Orange points represent genes upregulated >2-fold in Dux overexpression experiments in ESCs [2]. C. Comparison of gene expression between Dux KO in 2-cell stage embryos to Dux overexpression in ESCs. Figure 4. (continued) The x- and y-axis correspond to the log2 fold change in expression for overexpression and KO experiments, respectively. The zoomed region corresponds to genes that are upregulated by more than 32-fold in the overexpression experiment. The trend line represents a linear fit to this subset. D. Expression changes were estimated for Dux KO (upper panel) and Dux overexpression experiments in ESCs (lower panel) [2] and grouped together by repeat class. Points correspond to repeat families and are colored red to indicate statistically significant expression changes (log2 fold change > 1 or < −1, Benjamini–Hochberg adjusted P-value < 0.05 and mean counts > 25). E. Proportions of major wave ZGA genes that are upregulated at the 2-cell stage in the Dux KO. Major wave ZGA genes were defined as genes expressed more highly at mid-2 cell stage relative to zygotic embryos in the data from [22].
Comparison of the WT and KO embryos identified a significant number of differentially expressed genes: 1091 and 731 genes were at least 2-fold changed (log2 fold change > 1 or < −1, Benjamini–Hochberg adjusted P-value < 0.05 and mean counts > 25). Only 109 genes previously identified as upregulated in Dux overexpression studies were differentially expressed at the 2 -cell stage upon loss of Dux (Figure 4B). Of these, 53 were upregulated and 56 were downregulated (orange points, Figure 4B). For example, zinc finger and SCAN domain containing 4F (Zscan4f) was downregulated 2.7 fold in the KO (adjusted P = 1.8e-2) but deleted in azoospermia like (Dazl) was upregulated 3.0-fold (adjusted P = 6.2e-14). We directly compared these datasets by plotting the log2 fold change in each dataset (Figure 4C). While no correlation was observed across the entire transcriptome, genes that were upregulated by more than 32-fold by Dux overexpression showed a slight negative correlation (expanded region Figure 4C, Spearman correlation coefficient = −0.2) as expected if a subset are expressed at lower levels in the KO. Similarly, Dux has been reported to induce the expression of endogenous retroviruses and other repeat elements. We quantified the expression of all major families of repeat elements in the Dux KO, and compared to the same analysis in the Dux overexpression study of Hendrickson et al. [2] (Fig 4D). While 667 repeat families were found to be differentially expressed upon Dux overexpression, only 28 were differentially expressed upon loss of Dux in the 2-cell embryos (log2 fold change > 1 or < −1, Benjamini–Hochberg adjusted P-value < 0.05 and mean counts > 25). However, a few of the key retroviral families, MERVL-int (log2 fold change = −1.34, adjusted P = 3.1e-17), MT2_Mm (log2 fold change = −0.76, adjusted P = 2.2e-4), and GSAT_MM (log2 fold change = −0.56, adjusted P = 0.4), were slightly down regulated in the Dux KO. Together, these results indicate that the genes induced by Dux overexpression in ESCs are not highly overlapping with the genes whose expression requires Dux during ZGA.
To evaluate changes to genes associated with ZGA, we identified the genes differentially expressed between the zygotic and the mid 2-cell stage in the reference dataset [22]. This identified 3152 genes expressed more highly at the mid 2-cell stage and 1428 genes expressed more highly at the zygotic stage (log2 fold change > 1 or < −1, Benjamini–Hochberg adjusted P-value < 0.05 and mean counts > 25). These gene sets were used as proxies for the major wave of ZGA and genes lost to maternal degradation respectively. Next, we evaluated how the Dux KO affected the pattern of ZGA overall. Of the 3152 genes more highly expressed at the mid 2-cell stage, 2806 (89%) were unaffected in the Dux KO. Of the 346 ZGA genes that were differentially expressed in the Dux KO, 301 (87% of 346) were expressed at a lower level, but none of these genes were nonexpressed in the KO. These data are consistent with the notion that Dux is present at this stage, and are responsible for elevated expression of a small portion of the genes up regulated at the 2-cell stage (Figure 4E), but not with the notion that Dux is uniquely responsible for activation of zygotic transcription at any of these loci.
We next performed GO enrichment analysis of the genes that were differentially expressed in the Dux KO. Using the gene lists defined above we found the ontology category ribosome biogenesis to be the most enriched category (not shown). Inspection of the enriched ontologies using a less stringent 1.5 fold cutoff revealed a number of pathways potentially related to the observed phenotypes (Figure 5A). Notably, among the downregulated genes, we found that there are 16 involved in blastocyst formation and 39 genes involved in methylation (Figure 5B). The gene NOP2 nucleolar protein (Nop2) is a member of both of these ontology groups and encodes a 5-methylcytosine RNA methyltransferase that has shown to be required for blastocyst formation and also promotes ribosome biogenesis [23]. Downregulation of Nop2 (log2 fold change = −0.954, adjusted P = 7.3e-7) may therefore be partially responsible for the reduction in the expression of genes involved in ribosome biogenesis observed for the ontology analysis of the downregulated genes at the lower and higher stringency cutoffs. Although ontology terms for trophectoderm specification or placentation were not among the significantly enriched categories, manual interrogation of genes in categories based on these key words did identify some important genes among the down regulated set for these processes (Figure 5C). For example, TEA domain transcription factor 4 (Tead4) (log2 fold change = −0.81, adjusted P = 2.0e-2) is involved in trophectoderm lineage specification [24] and (transcription factor AP-2, gamma) Tfap2c (log2 fold change = −0.76, adjusted P = 2.2e-4) is a key regulator of extraembryonic development [25]. By activating important transcription factors like Tead4 and Tfap2c at the 2-cell stage, Dux expression may help establish a transcriptional profile that is appropriate for later stages of development.
Figure 5.
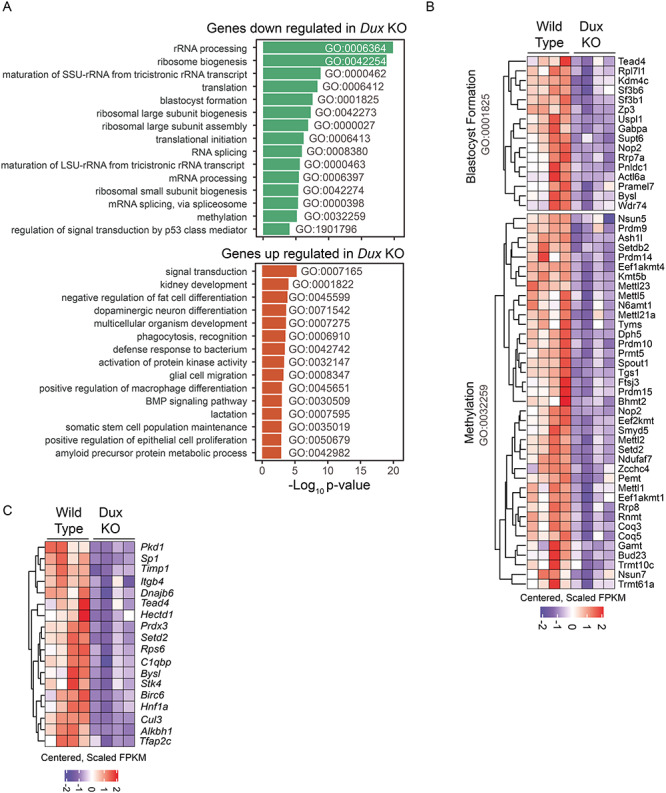
GO enrichment analysis of genes regulated by Dux at the 2-cell stage. A. Enriched GO terms for genes down regulated (upper panel) or up regulated (lower panel) by greater than 1.5-fold in 2C Dux KO embryos. The x-axis corresponds to the negative log10 of the p-value calculated using a hypergeometric test with a transcript length bias correction. B. Heatmap showing scaled and centered FPKM values for down regulated genes by greater than 1.5 fold in 2C Dux KO embryos with ontology associations for blastocyst formation (GO:0001825) and methylation (GO:0032259). C. Heatmap showing scaled and centered FPKM values for down regulated genes by greater than 1.5 fold in 2C Dux KO embryos with ontology associations matching keywords trophoblast, trophectoderm or placenta.
Discussion
In contrast to our initial expectations, these studies reveal only minor disruptions in gene expression at the 2-cell stage in Dux KO embryos. While embryos developed reliably to the blastocyst stage in vivo, a modest effect on viability at later stages of embryogenesis was observed, resulting in a higher risk of post-implantation developmental failure of KO embryos compared to their heterozygous counterparts, resulting in a smaller litter size in the homozygous KO strain. In other respects, the KO strain appeared normal and fertile. In contrast to these modest changes in vivo, KO embryos were much less robust to ex vivo culture, suffering a high failure rate of development to blastocyst when cultured from the 2-cell stage. This is consistent with the originally reported in vitro experiment using CRISPR to delete the Dux locus in zygotes [4], thus potentially explaining the conflicting results between that study and the study of Chen et al. [18]. The elevated ex vivo developmental failure to blastocyst may indicate that the gene expression changes we identify modestly perturb preimplantation development. We speculate that when DUXC first appeared in the eutherian mammal progenitor, early cleavage-stage embryonic development may have adapted to compensate for the short blip of DUXC expression, and the presence of certain of its target genes, but that this compensatory adaptation becomes destabilizing in the absence of DUXC. The modest perturbation in the absence of Dux is not sufficient to cause developmental failure in vivo, but when paired with the shock of removal of the embryo from its normal developmental environment and the stress of culture in an artificial medium under hyperoxic conditions, this greater combined perturbation leads to failure of development to blastocyst in vitro. It would be interesting to test more optimized types of embryo culture medium and hypoxic culture to identify the specific stress that these embryos are sensitized to.
The gene expression profiles that we observed show similarly modest effects compared to those recently described in the similar study by Chen et al. [18]. The KO produced in that study was also reported to be compatible with normal development albeit with a greater rate of embryo loss, however blastocyst development was not specifically investigated, either in vitro or in vivo. More recently, two additional Dux knockouts have documented animals that are viable and fertile, with different degrees of perturbation of the 2 cell transcriptional profile [26, 27]. Interestingly, Chen et al. saw a dramatic decrease in the expression of the DNA-binding factor Arid5b, which is encoded by a locus 10 Mb upstream of Dux, while we see a 2-fold increase in the expression of AT rich interactive domain 5B (Arid5b) (Figure 3B). The proximity of Arid5b and Dux and the differential results in these two KOs suggest the possibility that the Chen and Zhang KO might have made a change in the 3D topology of this region, perhaps by deleting an enhancer element. Such topological differences may account for the differences in the gene expression profiles and lethalities among the models.
The fact that absence of Dux led to only a very subtle phenotype leads to the question of whether activation of its target genes during ZGA is indeed the key function of DUXC in the eutherian mammalian genome. The model proposing that the prime function of DUXC is to initiate ZGA has one very strong feature: it can potentially explain the necessity of the repeated nature of the gene. Since many genes that are kept silent through methylation and perhaps protamine-specific mechanisms can be momentarily derepressed when the genome transitions from being carried in gametes to being zygotic, we might expect an extremely slight blip of expression of many factors at this stage. The fact that the DUXC genes are repeated would amplify this momentary blip of expression, and thus make it possible that DUXC target genes could be transiently induced to a meaningful level. This would in theory allow a single, highly repeated, methylation-sensitive gene to activate a cascade of downstream transcription factors that would ultimately establish the initial genome expression profile, i.e., be the key inducer of ZGA, even while its own expression was being extinguished. In spite of this interesting mechanism, the model suffers from a number of theoretical problems, the first being that ZGA occurs in all animals while DUXC genes occur only in eutherian mammals, therefore it does not account for ZGA and the absence of DUXC genes in the rest of the animal kingdom. Secondly, both DUX4 [28, 29] and Dux [30] are highly cytotoxic, in both somatic cells, as well as pluripotent stem cells [8]. If Dux target genes are responsible for toxicity, the lack of their toxicity at the 2-cells stage would need to be explained.
The transcriptional profile of 2-cell embryos lacking Dux was modestly perturbed, with a subset of ZGA genes expressed at lower levels than in WT 2 cell embryos, including a large number of genes not thought to be direct targets of Dux. We did identify a number of statistically significant down regulated genes related to trophectoderm specification, trophoblast expansion and placenta formation albeit at a less stringent fold change cutoff. This suggests that early Dux expression may help prime these genes for their eventual use post-implantation. Indeed, a recent report has shown that genes transiently induced by DUX4 are expressed at a higher level when activated at later time points than they would have been without the transient DUX4-stimulated activation [31]. Binding by Dux at the 2-cell stage may thus be responsible for an epigenetic change that allows for higher levels of expression of these genes during later trophoblast/placenta development. Furthermore, a recent report suggests that developmental pluripotency associated 2 (Dppa2) and developmental pluripotency associated 4 (Dppa4) upregulate Dux expression during the minor wave of ZGA [32]. In the absence of Dux, we found both Dppa2 and Dppa4 to be downregulated (log2 fold change = −0.96 and −0.61, adjusted P = 2.4e-3 and 2.2e-2, respectively) suggesting that Dux may participate in a feedback loop to activate Dppa2/4, facilitating chromatin decompaction at many loci [33]. This decompaction may be necessary for proper expression of trophectoderm-derived genes upon implantation and therefore may explain the increased number of resorptions observed in the Dux KO.
It has recently been discovered that in addition to inducing specific target genes near sites of DNA binding, DUX4 also drives a genome-wide increase in H3K27ac [34], an effect dependent on the C-terminal domain of DUX4, which interacts with p300 and CBP [9]. While our data clearly shows that activation of the zygotic genome is achieved in the absence of Dux, this global enhancement of histone acetyltransferase activity could potentially explain the large number of genes that are not known to be direct Dux targets but are nevertheless changed in its absence.
Perhaps the most relevant finding is that implanted KO embryos are lost at greater rates than WT embryos. While ZGA is common to all animals, implantation and placental invasion are unique to eutherian mammals. Rather than being the central activator of the zygotic genome, it is possible that DUXC genes, which are unique to eutherian mammals, play a role in placental invasion or function, either by establishing a transcriptional state in the cleavage-stage embryo that later facilitates cytotrophoblast development or function, or by being expressed later, during placental development itself. The loss and resorption of embryos at different stages of post-implantation development is consistent with a placental insufficiency phenotype. Such a function would explain the restriction of the DUXC gene family to eutherian mammals.
Conflict of interest
The authors have no competing financial interests to declare.
Author contributions
D.B. and S.H.C. performed investigation. D.B., M.D.G, and M.K. were involved in analysis. M.D.G. completed bioinformatics analysis. M.K. and D.B. designed and supervised the research. And D.B, M.D.G, and M.K. prepared the manuscript.
Acknowledgments
We thank Elizabeth T. Ener, Ben Mai, Christian Palumbo Johannes Weiblen, and Shenar Dinkha for genotyping embryos and mice. We thank Yun You for assistance harvesting and evaluating 2-cell embryos and blastocysts.
Footnotes
† Grant support: This work was supported by a gift from the Bob and Gene Smith Foundation, and by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR055685). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Darko Bosnakovski, Lillehei Heart Institute, University of Minnesota, Minneapolis, MN, USA; Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA; Faculty of Medical Sciences, University Goce Delcev - Shtip, Shtip, R. North Macedonia.
Micah D Gearhart, Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, MN, USA.
Si Ho Choi, Lillehei Heart Institute, University of Minnesota, Minneapolis, MN, USA; Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA.
Michael Kyba, Lillehei Heart Institute, University of Minnesota, Minneapolis, MN, USA; Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA.
References
- 1. Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013; 500:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 2017; 49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet 2017; 49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 2017; 49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leidenroth A, Hewitt JE. A family history of DUX4: Phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol Biol 2010; 10:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet 2007; 81:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, Tapscott SJ. DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev Cell 2012; 22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppee F, Belayew A, Perlingeiro RR et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 2008; 27:2766–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, Kyba M. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res 2016; 44:5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaap M, Lemmers RJ, Maassen R, van der Vliet PJ, Hoogerheide LF, van Dijk HK, Basturk N, de Knijff P, van der Maarel SM. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations: Evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics 2013; 14:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012; 44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boogaard ML, Lemmers RJLF, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, van der Vliet P, Straasheijm KR, van den Akker R, Kriek M, Laurense-Bik MEY, Raz V et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of Facioscapulohumeral dystrophy. Am J Hum Genet 2016; 98:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Overveld PG, Lemmers RJFL, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJB, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 2003; 35:315–317. [DOI] [PubMed] [Google Scholar]
- 14. Hartweck LM, Anderson LJ, Lemmers RJ, Dandapat A, Toso EA, Dalton JC, Tawil R, Day JW, van der Maarel SM, Kyba M. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology 2013; 80:392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 1992; 2:26–30. [DOI] [PubMed] [Google Scholar]
- 16. Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLoS Genet 2010; 6:e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature 2000; 403:501–502. [DOI] [PubMed] [Google Scholar]
- 18. Chen Z, Zhang Y. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 2019; 51:947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A 1997; 94:14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagy A, Gertsenstein M, Vintersten K, Behringer R. Collecting two-cell- to compacted morula-stage embryos. CSH Protoc 2006; 2006:pdb–rot4359. [DOI] [PubMed] [Google Scholar]
- 21. Kito S, Hayao T, Noguchi-Kawasaki Y, Ohta Y, Hideki U, Tateno S. Improved in vitro fertilization and development by use of modified human tubal fluid and applicability of pronucleate embryos for cryopreservation by rapid freezing in inbred mice. Comp Med 2004; 54:564–570. [PubMed] [Google Scholar]
- 22. Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014; 343:193–196. [DOI] [PubMed] [Google Scholar]
- 23. Cui W, Pizzollo J, Han Z, Marcho C, Zhang K, Mager J. Nop2 is required for mammalian preimplantation development. Mol Reprod Dev 2016; 83:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007; 134:3827–3836. [DOI] [PubMed] [Google Scholar]
- 25. Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 2002; 129:2733–2747. [DOI] [PubMed] [Google Scholar]
- 26. De Iaco A, Verp S, Offner S, Grun D, Trono D. DUX is a non-essential synchronizer of zygotic genome activation. Development 2020; 147:dev177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo M, Zhang Y, Zhou J, Bi Y, Xu J, Xu C, Kou X, Zhao Y, Li Y, Tu Z, Liu K. Precise temporal regulation of Dux is important for embryo development. Cell Res 2019; 29:956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, Nunez NG, Leo O et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 2007; 17:611–623. [DOI] [PubMed] [Google Scholar]
- 29. Bosnakovski D., Xu Z., Liu M., Belayew A., Perlingeiro R.C.R., Kyba M, Conditional Gain of Function Analysis Points to a Key Role for DUX4 in FSHD Pathology, in FSHD Annual Research Symposium. 2006: New Orleans. [Google Scholar]
- 30. Bosnakovski D, Daughters RS, Xu Z, Slack JM, Kyba M. Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One 2009; 4:e7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Resnick R, Wong CJ, Hamm DC, Bennett SR, Skene PJ, Hake SB, Henikoff S, van der Maarel SM, Tapscott SJ. DUX4-induced histone variants H3.X and H3.Y mark DUX4 target genes for expression. Cell Rep 2019; 29:1812, e5–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eckersley-Maslin M, Alda-Catalinas C, Blotenburg M, Kreibich E, Krueger C, Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev 2019; 33:194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hernandez C, Wang Z, Ramazanov B, Tang Y, Mehta S, Dambrot C, Lee YW, Tessema K, Kumar I, Astudillo M, Neubert TA. Dppa2/4 facilitate epigenetic Remodeling during reprogramming to Pluripotency. Cell Stem Cell 2018; 23:396, e8–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bosnakovski D, da Silva MT, Sunny ST, Ener ET, Toso EA, Yuan C, Cui Z, Walters MA, Jadhav A, Kyba M. A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression and cell death. Sci Adv 2019;5:eaaw7781. [DOI] [PMC free article] [PubMed] [Google Scholar]


