Graphical abstract
Keywords: Source-separated human urine, Antibiotics, Antibiotic-resistant bacteria, Antibiotic-resistant genes, Short-time storage, Elimination
Highlights
-
•
20 randomly real urine samples were collected and analyzed.
-
•
Totally 30 antibiotics in the fresh real urine were identified.
-
•
Representative antibiotic resistance bacteria and its gene were identified.
-
•
Changes of antibiotics, antibiotic resistance bacteria and gens of urine were monitored.
Abstract
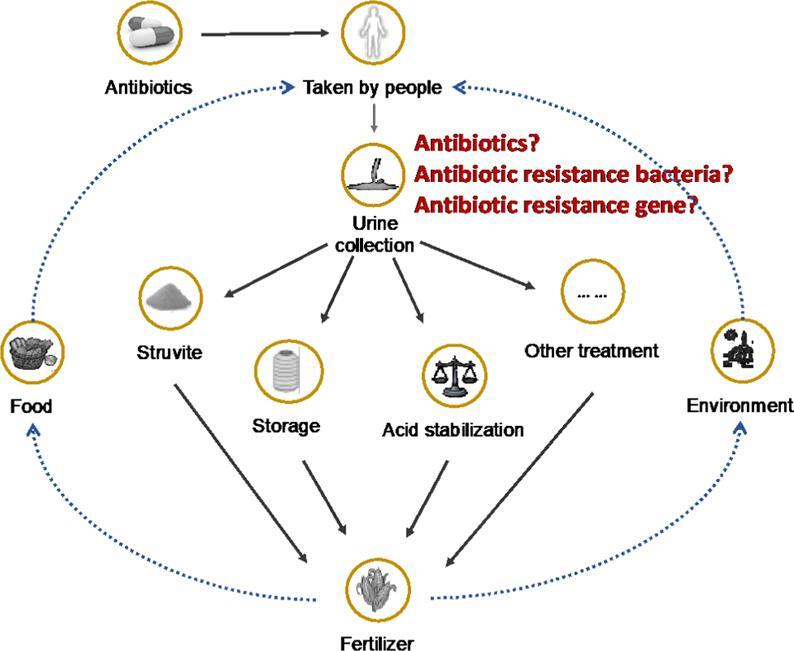
Human urine is a source of fertilizer and, with proper management, it can be reused in agriculture. Determining the contamination issue of antibiotics in source-separated urine is important because the majority of antibiotics are excreted with urine. In this study, source-separated urine samples were randomly collected from a male toilet in a university building and analyzed in terms of 30 typical antibiotics (including 14 sulfonamides, 4 tetracyclines, and 12 fluoroquinolones) and tetracycline-resistant Escherichia coli, as well as its antibiotic-resistant genes to determine the contamination characteristics of antibiotic-related pollution in fresh and stored urine. Results showed that 18 out of 30 typical antibiotics were detected in fresh source-separated human urine. The dominant antibiotic was oxytetracycline with a frequency of 100%, followed by tetracycline, sparfloxacin, enrofloxacin, and ofloxacin, which demonstrated a detection frequency of 55%. Among the detected values, sulfonamides (2 antibiotics), tetracyclines (4 antibiotics), and fluoroquinolones (12 antibiotics) had a concentration range of 0.25–2.94, 0.94–41.2, and 0.06–163.16 ng/mL, respectively. Furthermore, tetracycline-resistant Escherichia coli, which was measured using plate count method, and its related gene, tet M, exhibited a maximum cell density of (200,000 ± 5000) CFU/100 mL and (2.73 ± 0.261) × 107 copies/mL, respectively. When the fresh urine was stored in an ambient environment for 30 days to simulate the real circumstances of urine management, a significant reduction in antibiotics and antibiotic-resistant bacteria was observed, while the change in antibiotic-resistant genes was insignificant. The results of this study suggest that risks associated with antibiotics and their antibiotic-resistant bacteria and genes are retained during collection and storage. Hence, these kinds of microcontaminants must be considered in further urine utilization.
1. Introduction
Human urine may be collected separately in toilets by using sanitation technologies to achieve nutrient recovery because nitrogen, phosphorus, and potassium in urine represent nearly 80%, 50%, and 50% of necessary nutrients in agriculture, respectively (Bischel et al., 2015). Source separation of urine has been increasingly investigated and proposed as a method for efficient fertilizer production to make use of urine and avoid the discharge of large amounts of nutrients in surface water, which can cause eutrophication problems in rivers, lakes, and oceans (Udert et al., 2006). In addition to nutrients, urine also contains micropollutants, such as synthetic hormones and pharmaceuticals and their metabolites, including antibiotics excreted in the urine, which are extensively used by humans in the treatment of various diseases (Jia et al., 2012, Zhou et al., 2013). Antibiotics are only partially digested and absorbed by the human body, and approximately 30%–90% are excreted through urine and feces within 8–24 h after consumption (Frade et al., 2014). For example, the concentration of doxorubicin in a patient’s urine sample collected at 0–4 h ranges from 9.44 mg/L to 17.87 mg/L, which is nearly 95 times the amount of plasma in patients (Gao et al., 2020). Meanwhile, ciprofloxacin is reportedly excreted by 65% in the urine while only 25% is excreted in the feces; thus, urine contains most of excreted antibiotics (Frade et al., 2014). China is the largest producer of antibiotics worldwide with a yearly production of 210,000 tons (Zhang et al., 2015). China is also the largest consumer of antibiotics, and 90% of the produced antibiotics are applied in agriculture (48%) and medicine (42%) (Zhou et al., 2013). However, the indiscriminate use of antibiotics has generated a survival response in microorganisms; this survival response is called antibiotic resistance (Jia et al., 2012), which allows microorganisms to evade the bactericidal action of some agents efficiently and become emerging contaminants that can affect human health and the environmental ecosystem through the direct consumption of contaminated water and/or food (Lamba and Ahammad, 2017, Lind et al., 2001, Zhou et al., 2013). In the US, antibiotic-resistant bacteria and fungi caused at least 2,868,700 infections and 35,900 deaths annually in 2019 (U.S. Centers for Disease Control and Prevention, 2019). According to the World Health Organization, antibiotic resistance has become a critical global public health issue in this century (WHO, 2017). Currently, the majority of studies focus on antibiotic identification in wastewater, especially the fate of antibiotics during the wastewater treatment process, and the health risks associated with antibiotic resistance (Kraemer et al., 2019, Larsson, 2014). Wastewater treatment plants (WWTPs) have been considered reservoirs for antibiotic-resistant genes (Manaia et al., 2018). Human excreta accounts for only 1% of the volume of domestic wastewater but the greatest amount of pollutants. Thus, determining the trend of antibiotic contamination level in source-separated urine when urine is used as fertilizer is important. However, to the best of our knowledge, studies on such topics are limited.
Although source separation has been promoted since the 1990s, previous studies focused on nutrients in urine, and limited studies have been conducted in understanding antibiotics, antibiotic-resistant bacteria and genes. A case study on the behavior of pharmaceuticals related to the treatment of human immunodeficiency virus (HIV) investigated the liquid phase of source-separated urine during storage; however, artificial compounds rather than the real condition of human urine were used in the study (Jaatinen et al., 2016). A study in eThekwini, South Africa, investigated the municipality-scale production of agricultural fertilizers from human urine collected from urine-diverting dry toilets (Bischel et al., 2015). In this case, the presence of sulfamethoxazole was detected in 95% of samples, with a maximum concentration of 6800 ug/L. Recently, research in Lusaka, Zambia, demonstrated that sulfamethoxazole in source-separated urine had a concentration of 7740 µg/L, which was several orders of magnitude higher than that in wastewater (Ngumba et al., 2020). The application of fertilizer by excreta introduced antibiotic-resistant genes to crops and subsequently developed antibiotic resistance in the food chain from wastewater, thereby raising the issue of antibiotic resistance in organic fertilizer (Chen et al., 2017, European Commission DG Environment News Alert Service, 2018). Urine and feces from users may contain considerable amounts of active residues. A study found that antibiotics and antibiotic-resistant genes exist in manure-fertilized soil and a nearby watershed (Camotti Bastos et al., 2018). Thus, if urine is applied as fertilizer, then the occurrence and concentrations of antibiotics must be investigated for the safe usage of urine in agriculture.
On the basis of the above-mentioned points, this study aims to identify the presence of typical antibiotics in source-separated human urine and evaluate their contamination level and detection frequency to address the existing research gap. Tetracycline-resistant Escherichia coli (E. coli) was selected as representative antibiotic-resistant bacteria to determine the potential microbial risk caused by antibiotics. The urine was monitored with antibiotics, tetracycline-resistant E. coli and its genes change for 30 days to understand the behavior of antibiotics and its potential health effect during storage. The results can provide useful information for controlling potential health risks when source-separated urine is intended for agricultural use.
2. Materials and methods
2.1. Urine samples
Urine samples were collected from a male toilet in an office building of the University of Science and Technology Beijing. The building has 12 floors, and each floor has a male toilet with two urinals; the two urinals in the toilet of the 12th floor were transformed into waterless urinals. Thus, the male toilet of the 12th floor was selected as the sampling point for the collection of non-flushed source-separated urine. During the sample collection period, a sterile plastic barrel was directly connected with the urinal at the bottom. The toilet users are mainly young researchers, including master’s and PhD candidates, numbering a total of about 30 volunteers. The sampling was conducted during early summer in Beijing under ambient temperature from 8:00 a.m. to 11:00 a.m., and the collected volume was 3–5 L each time. Samples were collected once each day, and the total sampling time was 20 days; sampling was conducted randomly, not consecutively (May 21st to June 13th, 2019). All the urine samples were transported to the laboratory and immediately prepared for further analysis once sampling was finished.
The sampling conditions of each time are listed in Table 1. The average temperature of source-separated urine in 20 sampling days was (26.2 ± 1.5) °C, which ranged from 22.7 °C to 28.9 °C, and the mean pH was (9.11 ± 0.046). The range of ammonia during sampling was 4593.2–6627.2 mg/L NH4–N with an average of 5158 mg/L NH4–N. Urea in the fresh urine is easily hydrolyzed into ammonia, resulting in increased pH and high ammonia concentration. Hence, the collected urine samples were observed immediately after hydrolysis.
Table 1.
Basic chemical parameters of collected urine samples.
| N° Samples | Temperature (°C) | pH | Ammonia, mg/L NH4- N |
|---|---|---|---|
| 1 | 22.7 | 9.15 | 4631.6 |
| 2 | 26 | 9.09 | 6254.9 |
| 3 | 25.4 | 9.12 | 6100.0 |
| 4 | 25.8 | 9.15 | 6031.5 |
| 5 | 26.8 | 9.11 | 6627.2 |
| 6 | 25.9 | 9.14 | 6143.2 |
| 7 | 24.6 | 9.14 | 6031.5 |
| 8 | 25.7 | 9.12 | 4593.2 |
| 9 | 25.6 | 9.12 | 4657.0 |
| 10 | 23.9 | 9.11 | 4650.0 |
| 11 | 25.7 | 9.08 | 4695.3 |
| 12 | 26.2 | 9.03 | 4600.0 |
| 13 | 26.4 | 8.97 | 4784.6 |
| 14 | 26.6 | 9.17 | 4720.8 |
| 15 | 27.7 | 9.13 | 4657.6 |
| 16 | 28.9 | 9.13 | 4784.6 |
| 17 | 26.6 | 9.13 | 4800.0 |
| 18 | 26.7 | 9.13 | 4790.0 |
| 19 | 28.1 | 9.16 | 4810.0 |
| 20 | 28.9 | 9.11 | 4799.0 |
In practice, urine-diverting toilets collect urine over time. The collected urine is then stored for weeks or months for further treatment or utilization. Hence, the change in the substrate concentrations of urine sample was investigated. Urine samples were collected three times within three days from the same toilet mentioned above. Thereafter, the urine samples were mixed to make 6 L and distributed into three glass bottles as parallels for 30 days of storage under ambient temperature of 27.1 °C ± 0.8 °C. The urine samples were divided into three bottles to avoid cross contamination during sampling. Table 2 presents the pH and ammonia concentrations of the fresh samples and after mixture, and results showed that the urine samples hydrolyzed after mixing. Samples were taken every 5 days to analyze the pH, ammonia, antibiotics, and antibiotic-resistant bacteria and genes. In addition, tetracycline-resistant E. coli was analyzed every 6 h during the first 36 h of storage due to rapid elimination.
Table 2.
pH and ammonia concentration before and after mixture.
| N° Samples | pH | Ammonia, mg/L NH4- N |
|---|---|---|
| Sample 1 | 8.57 | 1693 |
| Sample 2 | 8.49 | 2907.2 |
| Sample 3 | 8.55 | 1953.7 |
| Mixture | 9.1 | 4631.6 |
2.2. Chemical and microbial analysis
Table 3 lists 30 typical antibiotics for the analysis of source-separated urine; these antibiotics are frequently used in China and detected in wastewater surface water, rivers, and sediments (Lu et al., 2018, Peng et al., 2011, Qiao et al., 2018, Zhang et al., 2015, Zhou et al., 2013). The chemical analysis of the antibiotics proceeded with modifications; refer to the literature for details (Yin et al., 2018). Liquid chromatography–mass spectrometry was used to analyze the extracted antibiotics; the device is an Agilent 1200 Series HPLC system (Agilent, Palo Alto, CA, USA) coupled to Applied Biosystems API4000 triple quadrupole mass spectrometer with electrospray ionization interface (Foster City, CA, USA) in positive mode. Nitrogen gas (99.9% purity) was used as the drying and collision gas. Agilent Eclipse XDB-C18 (150 mm × 2.1 mm, 3.5 μm) analytical column (Agilent Technologies, Atlanta, GA) operated at 35 °C was used for chromatographic separation, and the injection volume of the samples was 5 μL. A total of 0.1% formic acid in water (mobile phase A) with 0.1% formic acid in methonal (mobile phase B) were used as mobile phase gradient, which was run at a flow rate of 0.3 mL min−1, and the operation condition was 95% A for 0–1 min, 95%–65% A for 1–2 min, 65–50% A for 2–4.5 min, 50%–35% A for 4.5–6 min, 35% A for 6–8 min, 35%–5% A for 8–10 min, 5% A for 10–12 min, and 5%–95% A for 12–12.1 min, then continued for 4 min. Mass spectrometer was adjusted by using an optimizer (Agilent, Palo Alto, CA, USA) in terms of decluttering potential, collision energy, and multiple reaction monitoring mode independently. The operation condition of the ion source was an ion spray voltage of 5000 V; ion source at a temperature of 500 °C; curtain gas at 30 psi; atomization air pressure of 60 psi; auxiliary gas of 60 psi; entrance potential at an voltage of 10 V; collision cell exit potential at a voltage of 12 V; and dwell time of 30 ms for each transition. Analyte 1.5.2 (Applied Biosystems) was used for data acquisition and instrument control. More details on standards and stock solution, reagents and solvents, and sample preparation are listed in the Supplementary Information.
Table 3.
Identified antibiotics in source-separated urine.
Sulfonamides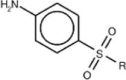
|
Tetracyclines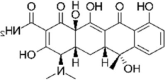
|
Fluoroquinolones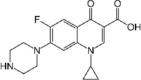
|
|---|---|---|
| Sulfadiazine | Tetracycline | Norfloxacin |
| Sulfadimethylpyrimidine | Oxytetracycline | Fleroxacin |
| Sulfachlorpyridazine | Chlortetracycline/ | Sparfloxacin |
| Sulfamethoxazole | Doxycycline | Orbifloxacin |
| Sulfamethoxine | Enrofloxacin | |
| Sulfathiazole | Pefloxacin | |
| Sulfamethazine | Difloxacin | |
| Sulfaguanidine | Ciprofloxacin | |
| Sulfamonomethoxine | Sarafloxacin | |
| Sulfaphenazole | Lomefloxacin | |
| Sodium sulfacetamide | Ofloxacin | |
| Oxolinic acid | Danofloxacin | |
| Sulfametoxydiazine | ||
| Sulfamethoxypyridiazine |
To detect tetracycline-resistant E. coli from the urine samples, a certain volume of urine sample was extracted by using a sterile filter membrane (0.45 μm). The resistant bacteria were selectively separated by placing tetracycline with concentrations of 4, 16, and 30 µg/ml in Endo agar and then by culturing at 37 °C for 24 h. Thereafter, the membrane was transferred to NA-MUG agar medium in 37 °C for another 4 h of incubation. Subsequently, E. coli was counted under ultraviolet light (6 W at 366 nm) with colony edge or blue fluorescence generation. Antibiotic-resistant genes (ARGs) were identified from the presence of tet A, tet Q, and tet M via PCR. The reaction system is specified in Table 4 (Chen and Zhang, 2013). Data analysis was performed using Origin 8.0.
Table 4.
Primer sequences used for PCR in antibiotic-resistant genes.
| N | Primer name | Primer sequence | Product length/bp | Tm/°C |
|---|---|---|---|---|
| 1 | Tet A-F tet A-R |
GCTACATCCTGCTTGCCTTC CATAGATCGCCGTGAAGAGG |
210 | 60 |
| 2 | tet M-F tet M-R |
ACAGAAAGCTTATTATATAAC TGGCGTGTCTATGATGTTCAC |
171 | 55 |
| 3 | tet Q-F tet Q-R |
AGAATCTGCTGTTTGCCAGTG CGGAGTGTCAATGATATTGCA |
169 | 63 |
3. Results and discussion
3.1. Occurrence of antibiotics in fresh source-separated urine
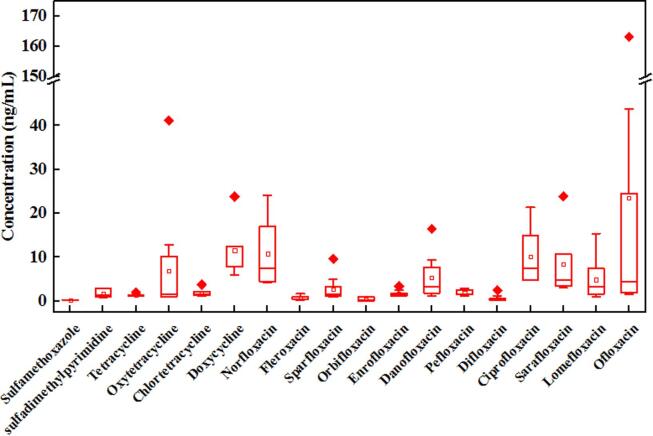
This study detected 18 out of 30 target antibiotics, including 2 sulfonamides and 4 tetracyclines; the remainder included fluoroquinolones, with concentration ranges of 0.25–2.94, 0.94–23.79, and 0.06–163.16 ng/mL. Only sulfamethoxazole and sulfadimethylpyrimidine were identified in the samples from the 14 antibiotics listed in the sulfonamide group (Fig. 1). Sulfamethoxazole was detected in 45% of the samples (0.84–2.94 ng/mL), while sulfadimethylpyrimidine was detected in 2 out of the 20 samples with concentrations of 0.25 and 0.29 ng/mL. In the tetracycline group, oxytetracycline was detected in 100% of the samples with a concentration range of 0.94–41.2 ng/ml. Tetracycline, chlortetracycline, and doxycycline were detected in 55%, 35%, and 25% of the samples, with concentration ranges of 1.16–2.09, 1.2–3.76, and 5.98–23.79 ng/mL, respectively. Twelve antibiotics in the fluoroquinolone group were identified in the samples, with predominant compounds of ofloxacin, norfloxacin, sarafloxacin, ciprofloxacin, danofloxacin, and lomefloxacin, which were detected in concentration ranges of 1.60–163.16, 4.24–24.03, 3.02–23.90, 4.75–21.38, 1.13–16.45, and 1.05–15.42 ng/mL, respectively, indicating a frequency of 55%, 30%, 55%, 30%, 40%, 35%, and accordingly. Low quantities of fleroxacin, sparfloxacin, orbifloxacin, enrofloxacin, pefloxacin, and difloxacin were detected with concentration ranges of 0.24–1.71, 0.99–9.66, 0.06–1, 1.13–3.41, 1.27–2.88, and 0.22–2.51 ng/mL, respectively. Obviously, the majority of antibiotics detected in this study belong to the fluoroquinolone group, which is also frequently detected at high concentrations in the environment, including wastewater, surface water, groundwater, and even drinking water (Jia et al., 2012, Lu et al., 2018, Peng et al., 2011).
Fig. 1.
Box chart of the summary statistics of antibiotic concentration ranges of sulfonamides, tetracyclines, and fluoroquinolones detected in male source-separated urine in 20 sampling days. Note: Only some of the antibiotics described were detected in the collected samples. (Legend: boxes: 25%–75%, whiskers: 5%–95%, diamond: data, dots: maximum and minimum values, crosses: 99% and 1% values, and square: mean value).
Table 5 presents the antibiotic concentration findings in this study and those reported in different studies. Overall concentrations of detected antibiotics in urine samples were higher than the findings reported in other water samples, such as raw wastewater, treated wastewater and surface water. Sulfamethoxazole was detected in WWTPs with concentration ranges of 0.083–0.455 ng/mL (average of 0.303 ng/mL) in the influent and 0.037–0.316 ng/mL (average of 0.157 ng/mL) in the final effluent (Peng et al., 2011); these values are lower than 10% of the concentration values detected in this study. For the tetracycline group, a particular case study in Wisconsin detected oxytetracycline and chlortetracycline in effluents of WWTPs with concentrations of 4.2 and 0.42 ng/mL, respectively (Karthikeyan and Meyer, 2006), which were significantly lower than the value in the current findings. However, another case demonstrated that oxytetracycline is a frequently detected antibiotic in China with a maximum concentration of 72.9 ng/mL (Suzuki and Hoa, 2012). Doxycycline was detected in WWTP with a concentration of 6.7 ng/ml (Borghi et al., 2015), which is 70% less than our findings. For the fluoroquinolone group, norfloxacin was detected with a concentration range of 0.027–0.489 ng/mL in wastewater (Liu et al., 2012), which is approximately one-fiftieth of the finding in the current study. Enrofloxacin was detected in WWTP in Wisconsin with a concentration of 0.27 ng/mL (Karthikeyan and Meyer, 2006), which is even lower than 7% of the value in the current study. Ciprofloxacin was detected in hospital effluents at concentrations of 0.7 and 124.5 ng/mL (Kummerer, 2003), which is six times higher than those in the current study. Meanwhile, ciprofloxacin was detected with concentration ranges of 60–90 and 20–80 ng/L in two typical WWTPs in south China, respectively (Zhou et al., 2013). These values are much lower than the concentration detected in this study. Indian cities registered a concentration range of 0.212–160 ng/mL (Singh et al., 2019), which is similar to the findings in the current study. Notably, the urine samples used in this study were undiluted, being 100 times concentrated compared with the urine that enters wastewater through a large amount of flushing water and graywater, even with rainwater in some cases. From Table 5, we can conclude that the antibiotics in source-separated urine are overall 10 times higher than those in raw wastewater, indicating that source separation can not only concentrate on nutrients at the source but can also be beneficial for antibiotic removal. Besides, the concentrations of the detected antibiotic in the urine samples were even higher than the value reported in rivers in China. For example, the maximum concentration of sulfamethoxazole in Ravi River was 2700 ng/L, and Norfloxacin was reported with concentrations of 6800 ng/L and 1100 ng/L in Hai River system and Pearl River (Singh et al., 2019).
Table 5.
Comparison of antibiotic concentrations between the current study and other water samples.
| Antibiotic | Range (ng/mL) | Other water samples(ng/mL) | References | |
|---|---|---|---|---|
| Sulfonamide | Sulfadimethylpyrimidine | 0.25–0.29 | NR | |
| Sulfamethoxazole | 0.84–2.94 | 0.083–0.455 0.037–0.316 |
Peng et al., 2011 | |
| Tetracyclines | Tetracycline | 1.16–2.09 | 0.0103 | Suzuki and Hoa, 2012 |
| Oxytetracycline | 0.94–41.2 | 4.2,72.9 | Karthikeyan and Meyer, 2006, Suzuki and Hoa, 2012 | |
| Chlortetracycline | 1.2–3.76 | 0.42 | ||
| Doxycycline | 5.98–23.79 | 6.7 | Borghi et al., 2015 | |
| Fluoroquinolones | Norfloxacin | 4.24–24.03 | 0.027–0.489 | Liu et al., 2012 |
| Fleroxacin | 0.24–1.71 | 0.014 | Jia et al.,2012 | |
| Sparfloxacin | 0.99–9.66 | 0.004 | Jia et al.,2012 | |
| Orbifloxacin | 0.06–1 | NR | ||
| Enrofloxacin | 1.13–3.41 | 0.27 | Karthikeyan and Meyer, 2006 | |
| Danofloxacin | 1.13–16.45 | 0.07 | Borghi et al., 2015 | |
| Pefloxacin | 1.27–2.88 | NR | ||
| Difloxacin | 0.22–2.51 | NR | ||
| Ciprofloxacin | 4.75–21.38 | 0.7–160 | Kummerer, 2003, Zhou et al., 2013, Singh et al., 2019 | |
| Sarafloxacin | 3.02–23.9 | 3.02–23.9 | Kummerer, 2003 | |
| Lomefloxacin | 1.05–15.42 | 0.162 | Jia et al.,2012 | |
| Ofloxacin | 1.6–163.16 | 0.212–160 | Singh et al., 2019 |
NR: No reference.
Moreover, the concertation of antibiotics in this study was lower than the reported value in other countries. As mentioned previously, sulfamethoxazole was detected in source-separated urine in eThekwini, South Africa, with high concentrations of approximately 6800 ng/ml (Bischel et al., 2015), which is 2312 times greater than the findings in the current study mainly because sulfamethoxazole is commonly used by HIV-positive patients. Thus, comparing this value with the data in this study is unnecessary. Moreover, the concentration of sulfamethoxazole in source-separated urine sampled in Lusaka, Zambia, was 7740 µg/L, which is thousands of orders of magnitude higher than that in this study (Ngumba et al., 2020). On the basis of the above-mentioned comparisons, antibiotic pollution varies considerably in terms of species and concentrations, which is highly dependent on the individual consumption of antibiotics; more attention should be paid to developing countries and rural regions in this regard. In this study, the fluoroquinolone group was detected with high frequency, suggesting that fluoroquinolones were commonly used among the toilet users. The urine samples collected in this study were mainly from young people. Theoretically, these individuals have strong immune systems; thus, the concentration of such antibiotics should be potentially low.
Notably, sarafloxacin was detected in the concentration range of 3.02–23.9 ng/mL in this study, but this antibiotic was approved only for the prevention of poultry diseases (Kummerer, 2003). Table 6 lists the antibiotics in the current study and their different usages in human and animal infections. To the best of our knowledge, rivers increase the susceptibility of humans and animals to illness because wastewater from hospitals and livestock feedlots as well as effluent and sewage sludge from municipal sewage treatment plants that end up in rivers contain antibiotics to some extent (Jia et al., 2012). Thus, antibiotics transformed within the whole circle, thereby persisting in the environment. Emphasizing the antibiotic contamination of source-separated urine may be an efficient way to control health risks, especially when urine is utilized as fertilizer for eatable crops.
Table 6.
Usage of different antibiotics for humans and animals.
| Antibiotics | antibiotic usages for anti-infections |
References | |||
|---|---|---|---|---|---|
| Human | Animal |
||||
| Pig | Poultry | Cattle | |||
| Sulfamethoxazole Sulfadimethylpyrimidine | Zhou et al., 2013 | ||||
| Norfloxacin, Fleroxacin | Suzuki and Hoa, 2012 | ||||
| Enrofloxacin | √ | Zhang et al, 2015 | |||
| Danofloxacin | √ | √ | √ | Schneider et al,1993 | |
| Pefloxacin, Ciprofloxacin | √ | √ | √ | Suzuki and Hoa, 2012 | |
| Sarafloxacin | √ | Kummerer, 2003 | |||
| Ofloxacin | Zhang et al., 2015, Singh et al., 2019 | ||||
| Danofloxacin | Kibret and Abera (2011) | ||||
| Tetracycline, Oxytetracycline, Chlortetracycline, Doxycycline | Zhang et al, 2015, | ||||
3.2. Occurrence of antibiotic-resistant bacteria and genes in fresh source-separated urine
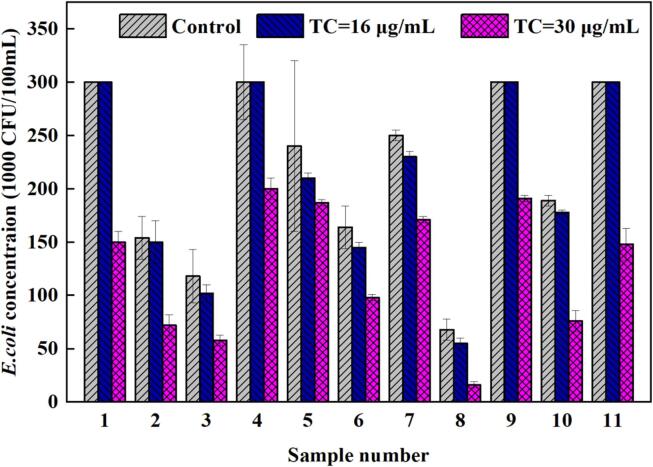
E. coli was incubated at 4, 16, and 30 µg/mL of tetracyclines in the media to assess the potential concentration of antibiotic resistance in fresh urine. The culture media remained unchanged at a tetracycline dosage of 4 µg/mL and without antibiotic. Eleven out of 20 samples were tested with live E. coli at concentrations of 16 and 30 µg/mL (Fig. 2). Hence, the detection frequency of antibiotic-resistant bacteria was 55%. The range of density of colonies in control plates was 68,000–300,000 CFU/100 mL, and the number of colonies was approximately 55,000–300,000 and 16,000–200,000 CFU/100 mL with 16 and 30 µg/mL of tetracyclines, respectively.
Fig. 2.
E. coli resistant at 16 and 30 µg/mL of tetracyclines.
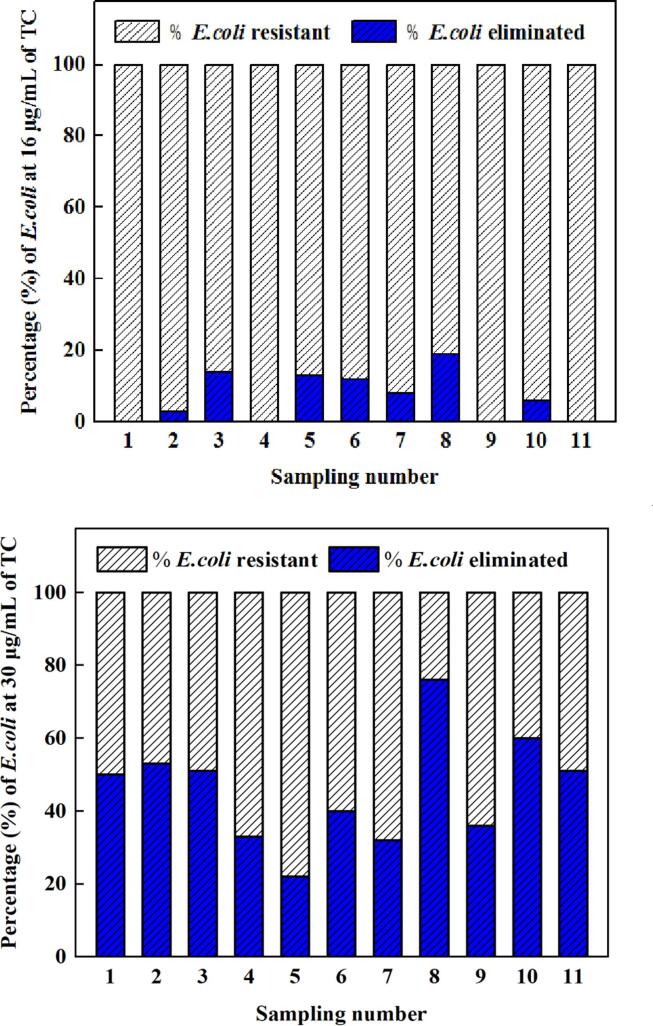
The elimination percentage when E. coli in 11 samples is exposed to 16 and 30 µg/mL of tetracyclines were calculated and shown in Fig. 3. With respect to the control samples, the maximum percentage of elimination was 19% at 16 µg/mL and 76% at 30 µg/mL of tetracyclines. Moreover, 55% of detected E. coli in 11 samples demonstrated elimination percentages of ≥50% under the condition of 30 µg/mL of tetracyclines.
Fig. 3.
Percentage of E. coli eliminated at (a) 16 and (b) 30 µg/mL of tetracyclines.
Further tests involving the three tetracycline-resistant genes (tet A, tet Q, and tet M) showed that the tet A and tet Q genes were not found, and they showed the presence of tet M with a concentration of (2.73 ± 0.261) × 107 copies/mL, indicating that the antibiotics in the urine may motivate antibiotic resistance. However, antibiotic-resistant bacteria may also come from the environment, and their pathway remains unclear.
3.3. Changes in antibiotics and antibiotic-resistant bacteria and genes in stored source-separated urine
Urine-diverting toilets collect urine over time, and the samples are temporarily stored for weeks or months before further treatment or final utilization. We place the collected urine in storage for 30 days in an ambient environment to determine the effect of aging on the micropollutant changes (Table 7). The pH was 9.1–9.17 with an average of (9.14 ± 0.02) and the concentration of ammonia ranged between 4631.6–5389.8 mg/L with an average of (4951.4 ± 167) mg/L after 30 days of storage in the mixed samples. Three antibiotics remained detectable after 30 days of storage; fleroxacin decreased from 9.05 ng/mL to 0.344 ng/mL, enrofloxacin decreased from 11.44 ng/mL to 0.367 ng/mL, and pefloxacin decreased from 6.36 ng/mL to 4.66 ng/mL with a removal percentage of 96%, 97%, and 27%, respectively. Sulfamethoxazole, norfloxacin, ofloxacin, oxytetracycline, chlortetracycline, and doxycycline were below the detection limit after 5 days of storage. After 10 days of storage, sparfloxacin was reduced to under the detection limit from an initial concentration of 26.49 ng/mL. After 15 days of storage, the concentrations of orbifloxacin, sarafloxacin, and lomefloxacin became undetectable in the samples. The concentrations of danofloxacin and ciprofloxacin decreased from 64.42 ng/mL to 1.14 ng/mL and from 85.52 ng/mL to 2.25 ng/mL, respectively, after 20 days of storage but were reduced to under the detection limit after 25 days of storage. Difloxacin and tetracycline were undetected after 30 days of storage, and their last concentrations registered at 0.211 and 0.193 ng/mL, respectively, after 25 days of storage.
Table 7.
Changes in antibiotic concentration during storage.
| Antibiotic Group | Antibiotics | days |
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 15 | 20 | 25 | 30 | ||
| ng/mL | ||||||||
| Sulfonamides | Sulfamethoxazole | 2.54 | ND | ND | ND | ND | ND | ND |
| Fluoroquinolones | Norfloxacin | 89.9 | ND | ND | ND | ND | ND | ND |
| Fleroxacin | 9.05 | 2.92 | 1.41 | 1.25 | 0.69 | 0.638 | 0.344 | |
| Sparfloxacin | 26.49 | 6.5 | ND | ND | ND | ND | ND | |
| Orbifloxacin | 4.97 | 2.72 | 1.41 | ND | ND | ND | ND | |
| Enrofloxacin | 11.44 | 2.44 | 1.17 | 1.1 | 0.695 | 0.366 | 0.367 | |
| Dafloxacin | 62.42 | 1.68 | 1.78 | 1.17 | 1.14 | ND | ND | |
| Pefloxacin | 6.36 | 4.03 | 3.445 | 2.84 | 1.753 | 4.062 | 4.66 | |
| Difloxacin | 10.65 | 1.74 | 0.72 | 0.435 | 0.384 | 0.211 | ND | |
| Ciprofloxacin | 85.52 | 4.87 | 2.07 | 1.995 | 2.25 | ND | ND | |
| Sarafloxacin | 94.41 | 3.96 | 1.44 | ND | ND | ND | ND | |
| Lomefloxacin | 68.95 | 1.92 | 0.96 | ND | ND | ND | ND | |
| Ofloxacin | 12.16 | ND | ND | ND | ND | ND | ND | |
| Tetracyclines | Tetracycline | 2.69 | 0.656 | 0.39 | 0.254 | 0.322 | 0.193 | ND |
| Oxytetracycline | 2.09 | ND | ND | ND | ND | ND | ND | |
| Chlortetracycline | 6.52 | ND | ND | ND | ND | ND | ND | |
| Doxycycline | 7.69 | ND | ND | ND | ND | ND | ND | |
ND: not detected.
Drugs have the following possible fates in the environment: (1) biodegradable drugs may be mineralized into carbon dioxide and water, (2) drugs may undergo specific metabolic processes or be partially degraded, or (3) drugs may be persistent (Frade et al., 2014). In this study, urine samples remained untreated during storage, and hydrolysis was the main chemical process that could occur. Hydrolyzed urine may promote a biological and chemical process for antibiotic degradation. However, Jaatinen et al.(2016) obtained 1%–99% antibiotic removal with or without amendments throughout a urine storage period of 6 months and showed that bacterial activity and urea hydrolysis slightly influence the antibiotic removal efficiency. Thus, abiotic self-degradation may be the primary cause of antibiotic removal. Furthermore, risks associated with antibiotics may reduce when biodegradation and sorption occur in the soil after urine fertilization (Winker et al., 2008). However, this finding requires further evaluation because more than 100 antibiotics have been detected.
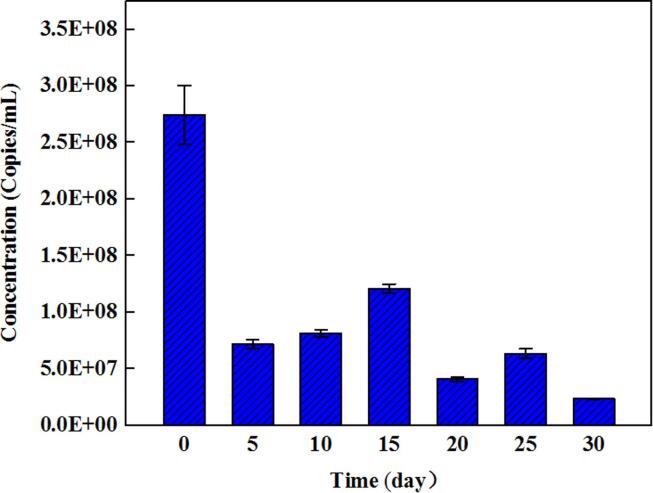
The cell density of samples that are resistant to 30 µg/mL of tetracycline decreased from 120,000 UFC/100 mL to 5000 UFC/100 mL after 30 h of storage, which equals a reduction of 1.38 log, and decreased to be undetectable at 36 h. The concentration range of tet M was (2.73–0.261) × 108 to (2.35 ± 0.0463) × 107 copies/mL with a reduction of less than 1 log after 30 days of storage (Fig. 4), and the concentration remained high even after storage. A previous study demonstrated that ampicillin- and tetracycline-resistant genes carried by extracellular DNA released into aged urine demonstrate a low potential to spread antibiotic-resistant genes to bacteria when released to the environment (Goetsch et al., 2020). Meanwhile, a study demonstrated that the ARG in manure fertilized soil would transfer into the non-manured soils, and ARGs abundance below 1.62 × 109 copies/g was suggested to be the typical level of non-manured soil. In regards to the tet M concentration found in the urine in this study, it seems that the concentration is safe to soil (Pu et al.,2020). However, this is only one strain, thus, the ecological risk should not be ignored and the gene transfer risk must be considered because the concentration remained high during storage. Overall, statistical changes in terms of log reduction were absent in antibiotic-resistant genes after 30 days of storage.
Fig. 4.
Concentration of tet M genes during storage.
4. Conclusion
Antibiotics and antibiotic-resistant bacteria and genes were investigated in this study to further understand the risks of antibiotic-related issues in terms of urine utilization. The variation in antibiotics and representative antibiotic-resistant bacteria and genes during the temporary storage of urine was also investigated. Results showed that 18 antibiotics belonging to three groups (84% fluoroquinolones, 14% tetracyclines, and 2% sulfonamides) were detected in fresh source-separated urine at different concentrations and frequencies. Tetracycline-resistant E. coli was detected at a frequency of 55% with maximum cell densities of (200,000 ± 5000) CFU/100 mL, and tet M was detected with a frequency of 36% in the samples. Furthermore, during 30 days of storage, aging urine showed significant elimination of antibiotics and antibiotic-resistant bacteria but only a slight reduction in antibiotic-resistant genes. Therefore, such kinds of pollutants should be emphasized during urine treatment for the purpose of utilization because urine may act as a transport medium of antibiotics, antibiotic-resistant bacteria and genes.
CRediT authorship contribution statement
Xiaoqin Zhou: Conceptualization, Writing - review & editing. Gabriela Jacqueline Perez Cuasquer: Investigation, Writing - original draft. Zifu Li: Supervision. Heinz Peter Mang: Supervision. Yaping Lv: Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to acknowledge the financial support of the National Key Research and Development Program of China (2017YFC0403401) and the Reinvent the Toilet Challenge–China Regional Program (OPP1051913) of the Bill and Melinda Gates Foundation. The authors would also like to thank the Beijing Natural Science Foundation (grant no. 8194069), the Fundamental Research Funds for the Central Universities (FRF-IC-19-001), the Central Public-interest Scientific Institution Basal Research Fund (BSRF201803), and the National Environment and Energy International Cooperation Base for their support. The authors would like to express their sincere thanks to Dr. Yin Fubin from the Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agriculture Science, for the assistance with the analysis of 30 antibiotics.
Handling Editor: Thanh Nguyen
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2020.106280.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Bischel H.N., Özel Duygan B.D., Strande L., McArdell C.S., Udert K.M., Kohn T. Pathogens and pharmaceuticals in source-separated urine in eThekwini, South Africa. Water Res. 2015;85:57–65. doi: 10.1016/j.watres.2015.08.022. [DOI] [PubMed] [Google Scholar]
- Borghi A.A., Silva M.F., Al Arni S., Converti A., Palma M.S.A. Doxycycline degradation by the oxidative Fenton process. J. Chem. 2015;2015 doi: 10.1155/2015/492030. [DOI] [Google Scholar]
- Camotti Bastos M., Rheinheimer dos Santos D., Aubertheau É., de Castro Lima J.A.M., Le Guet T., Caner L., Mondamert L., Labanowski J. Antibiotics and microbial resistance in Brazilian soils under manure application. L. Degrad. Dev. 2018;29:2472–2484. doi: 10.1002/ldr.2964. [DOI] [Google Scholar]
- Chen H., Zhang M. Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ. Sci. Technol. 2013 doi: 10.1021/es401091y. [DOI] [PubMed] [Google Scholar]
- Chen Q.L., An X.L., Zhu Y.G., Su J.Q., Gillings M.R., Ye Z.L., Cui L. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 2017;51:8149–8157. doi: 10.1021/acs.est.7b01420. [DOI] [PubMed] [Google Scholar]
- European Commission DG Environment News Alert Service Antibiotic resistance in struvite fertiliser from waste water could enter the food chain. Sci. Environ. Policy. 2018 [Google Scholar]
- Frade V.M.F., Dias M., Teixeira A.C.S.C., Palma M.S.A. Environmental contamination by fluoroquinolones, Brazilian. J. Pharm. Sci. 2014 doi: 10.1590/s1984-82502011000100004. [DOI] [Google Scholar]
- Gao L., He Q., Xing J., Ge Z. Removal of doxorubicin by magnetic copper phosphate nanoflowers for individual urine source separation. Chemosphere. 2020;238:124690. doi: 10.1016/j.chemosphere.2019.124690. [DOI] [PubMed] [Google Scholar]
- Goetsch H.E., Love N.G., Wigginton K.R. Fate of extracellular DNA in the production of fertilizers from source-separated urine. Environ. Sci. Technol. 2020;54:1808–1815. doi: 10.1021/acs.est.9b04263. [DOI] [PubMed] [Google Scholar]
- Jaatinen S.T., Palmroth M.R.T., Rintala J.A., Tuhkanen T.A. The effect of urine storage on antiviral and antibiotic compounds in the liquid phase of source-separated urine. Environ. Technol. (United Kingdom) 2016;37:2189–2198. doi: 10.1080/09593330.2016.1144799. [DOI] [PubMed] [Google Scholar]
- Jia A., Wan Y., Xiao Y., Hu J. Occurrence and fate of quinolone and fluoroquinolone antibiotics in a municipal sewage treatment plant. Water Res. 2012;46:387–394. doi: 10.1016/j.watres.2011.10.055. [DOI] [PubMed] [Google Scholar]
- Karthikeyan K.G., Meyer M.T. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 2006;361:196–207. doi: 10.1016/j.scitotenv.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Kraemer S.A., Ramachandran A., Perron G.G. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7:1–24. doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer K. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 2003;52:5–7. doi: 10.1093/jac/dkg293. [DOI] [PubMed] [Google Scholar]
- Kibret M., Abera B. Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr. Health Sci. 2011;11 doi: 10.4314/ahs.v11i3.70069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba M., Ahammad S.Z. Performance comparison of secondary and tertiary treatment systems for treating antibiotic resistance. Water Res. 2017;127:172–182. doi: 10.1016/j.watres.2017.10.025. [DOI] [PubMed] [Google Scholar]
- Larsson D.G.J. Antibiotics in the environment. Ups. J. Med. Sci. 2014;119:108–112. doi: 10.3109/03009734.2014.896438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind B.B., Ban Z., Bydén S. Volume reduction and concentration of nutrients in human urine. Ecol. Eng. 2001 doi: 10.1016/S0925-8574(00)00107-5. [DOI] [Google Scholar]
- Liu C., Nanaboina V., Korshin G.V., Jiang W. Spectroscopic study of degradation products of ciprofloxacin, norfloxacin and lomefloxacin formed in ozonated wastewater. Water Res. 2012;46:5235–5246. doi: 10.1016/j.watres.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Lu L., Liu J., Li Z., Liu Z., Guo J., Xiao Y., Yang J. Occurrence and distribution of tetracycline antibiotics and resistance genes in longshore sediments of the three gorges reservoir, China. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaia C.M., Rocha J., Scaccia N., Marano R., Radu E., Biancullo F., Cerqueira F., Fortunato G., Iakovides I.C., Zammit I., Kampouris I., Vaz-Moreira I., Nunes O.C. Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 2018;115:312–324. doi: 10.1016/j.envint.2018.03.044. [DOI] [PubMed] [Google Scholar]
- Ngumba E., Gachanja A., Nyirenda J., Maldonado J., Tuhkanen T. Occurrence of antibiotics and antiretroviral drugs in source-separated urine, groundwater, surface water and wastewater in the peri-urban area of chunga in lusaka, Zambia. Water SA. 2020;46:278–284. doi: 10.17159/wsa/2020.v46.i2.8243. [DOI] [Google Scholar]
- Peng X., Zhang K., Tang C., Huang Q., Yu Y., Cui J. Distribution pattern, behavior, and fate of antibacterials in urban aquatic environments in South China. J. Environ. Monit. 2011;13:446–454. doi: 10.1039/c0em00394h. [DOI] [PubMed] [Google Scholar]
- Pu Q., Zhao L.X., Li Y.T., Su J.Q. Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China. J. Hazard. Mater. 2020;391:122267. doi: 10.1016/j.jhazmat.2020.122267. [DOI] [PubMed] [Google Scholar]
- Qiao M., Ying G.G., Singer A.C., Zhu Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Schneider R.P., Ericson J.F., Lynch M.J., Fouda H.G. Confirmation of danofloxacin residues in chicken and cattle liver by microbore high-performance liquid chromatography electrospray ionization tandem mass spectrometry. Biol. Mass Spectrom. 1993;22:595–599. doi: 10.1002/bms.1200221007. [DOI] [PubMed] [Google Scholar]
- Singh V., Pandey B., Suthar S. Phytotoxicity and degradation of antibiotic ofloxacin in duckweed (Spirodela polyrhiza) system. Ecotoxicol. Environ. Saf. 2019;179:88–95. doi: 10.1016/j.ecoenv.2019.04.018. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Hoa P.T.P. Distribution of quinolones, sulfonamides, tetracyclines in aquatic environment and antibiotic resistance in Indochina. Front. Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udert K.M., Larsen T.A., Gujer W. Fate of major compounds in source-separated urine. Water Sci. Technol. 2006:413–420. doi: 10.2166/wst.2006.921. [DOI] [PubMed] [Google Scholar]
- US. Centers for Disease Control and Prevention, 2019. Antibiotic resistance threats in the United States, Centers for Disease Control and Prevention. https://doi.org/CS239559-B.
- WHO, 2017. Global priority list of antibiotic-resistant batceria to guide research, discovery, and development of new antibiotics. Who 7.
- Winker, M., Gulyas, H., Otterpohl, R., Clemens, J., 2008. Fertilization of rye grass with carbamazepine, ibuprofen , and 17 α – ethinylestradiol spiked urine – a greenhouse experiment.
- Yin F., Dong H., Zhang W., Zhu Z., Shang B. Antibiotic degradation and microbial community structures during acidification and methanogenesis of swine manure containing chlortetracycline or oxytetracycline. Bioresour. Technol. 2018;250:247–255. doi: 10.1016/j.biortech.2017.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang Q.Q., Ying G.G., Pan C.G., Liu Y.S., Zhao J.L. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015;49:6772–6782. doi: 10.1021/acs.est.5b00729. [DOI] [PubMed] [Google Scholar]
- Zhou L.J., Ying G.G., Liu S., Zhao J.L., Yang B., Chen Z.F., Lai H.J. Occurrence and fate of eleven classes of antibiotics in two typical wastewater treatment plants in South China. Sci. Total Environ. 2013;452–453:365–376. doi: 10.1016/j.scitotenv.2013.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.