Abstract
Recent studies suggested a link between long-term exposure to air-pollution and COVID-19 mortality. However, due to their ecological design based on large spatial units, they neglect the strong localised air-pollution patterns, and potentially lead to inadequate confounding adjustment. We investigated the effect of long-term exposure to NO2 and PM2.5 on COVID-19 mortality in England using high geographical resolution. In this nationwide cross-sectional study in England, we included 38,573 COVID-19 deaths up to June 30, 2020 at the Lower Layer Super Output Area level (n = 32,844 small areas). We retrieved averaged NO2 and PM2.5 concentration during 2014–2018 from the Pollution Climate Mapping. We used Bayesian hierarchical models to quantify the effect of air-pollution while adjusting for a series of confounding and spatial autocorrelation. We find a 0.5% (95% credible interval: −0.2%, 1.2%) and 1.4% (95% CrI: −2.1%, 5.1%) increase in COVID-19 mortality risk for every 1 μg/m3 increase in NO2 and PM2.5 respectively, after adjusting for confounding and spatial autocorrelation. This corresponds to a posterior probability of a positive effect equal to 0.93 and 0.78 respectively. The spatial relative risk at LSOA level revealed strong patterns, similar for the different pollutants. This potentially captures the spread of the disease during the first wave of the epidemic. Our study provides some evidence of an effect of long-term NO2 exposure on COVID-19 mortality, while the effect of PM2.5 remains more uncertain.
Keywords: COVID-19, Mortality, Nitrogen dioxide, Particular matter, Air-pollution, Bayesian spatial models
1. Introduction
As of 30th of June 2020, COVID-19 has caused more than 500,000 deaths globally, with an estimated case fatality of 1–4% (Hauser et al. 2020). The UK is one of the countries most affected, with an estimated 57,300 more deaths in England and Wales than it would be expected from mid-February to end of May 2020 had the pandemic not taken place (Kontis et al. 2020). Established risk factors of COVID-19 mortality include age, sex and ethnicity (Wu et al. 2020). Previous studies have observed a correlation between pre-existing conditions such as stroke, hypertension and diabetes (Williamson et al., 2020, Yang et al., 2020). Long-term exposure to air-pollution has been hypothesised to worsen COVID-19 prognosis: either directly, as it can suppress early immune responses to the infection (E. Conticini et al. 2020), or indirectly, as it can increase the risk of stroke, hypertension and other pre-existing conditions (Giorgini et al., 2016, Scheers et al., 2015).
Previous studies suggested an effect of long-term exposure to air-pollution on COVID-19 mortality (Cole et al., 2020, Liang et al., 2020, Travaglio et al., 2020, Wu et al., 2020), however several methodological shortcomings limit their interpretability. They were based on data aggregated on large spatial units and thus suffer from ecological fallacy (grouped levels association do not reflect individual ones) (Wakefield 2008). Air pollution is characterised by high spatial variability, making the availability of mortality data at the same high spatial resolution crucial (Villeneuve and Goldberg 2020). In addition, a coarse geographical resolution might lead to inadequate adjustment for confounders, when these are available at higher resolution (Villeneuve and Goldberg 2020). Most previous studies assessed cumulative deaths until mid or end of April and thus the generalisability of their results is limited to the early stages of the epidemic (Liang et al., 2020, Travaglio et al., 2020, Wu et al., 2020). One study had data available up to June 5, 2020 (Cole et al. 2020) and another up to June 12, 2020 (Statistics 2020), capturing a proportion COVID-19 deaths attributable to the first wave.
In this nationwide study in England, we investigated the effect of long-term exposure to air pollution on COVID-19 mortality during the entire first wave of the epidemic, after accounting for confounding and spatial autocorrelation. We focused on exposure to NO2 and PM2.5 (atmospheric particulate matter that has a diameter of less than 2.5 µm). We downscaled the LTLA geographical information to the Lower Layer Super Output Area (LSOA) to alleviate the effect of ecological bias and exploit the variability of the exposure at high geographical resolution.
2. Methods
2.1. Study population
We included all COVID-19 deaths as reported to Public Health England (PHE) by June 30, 2020. These include deaths that had a laboratory confirmed report of COVID-19 (including at post-mortem) (EpiCell 2020), as well as suspected COVID-19 deaths, defined as deaths without a positive test but with mention of COVID-19 in the death certificate. These definitions were consistent during the study period and over the study region. The main outcome of this study was laboratory confirmed deaths. We selected COVID-19 deaths up to June the 30th to ensure we captured COVID-19 deaths attributable to the first wave of epidemic that in England and Wales was over by the end of May, when all-cause mortality was no longer elevated (Kontis et al. 2020). Individual data on age, sex, ethnicity, lower tier local authority (LTLA) of the residential address and type of residence type (i.e. nursing homes, prisons, medical facilities etc.) were available. Population at risk in England was available through Office for National Statistics (ONS). Information at the LSOA level about age and sex was available for 2018, whereas about ethnicity for 2011 (the most recent years available at time of analysis).
2.2. Downscaling
There were 317 LTLAs in England in 2019 (Supplemental Material Fig. S1). Such a coarse geographical unit is not expected to capture the strong localised spatial patterns of air-pollution. We thus downscaled the LTLA geographical information to the LSOA level. LSOAs are high resolution geographical units in England (32,844 units in 2011, see Supplemental Material Fig. S2). The median population per LSOA in 2018 was 1617, varying from 591 to 14,696 (min to max) (Supplemental Material Fig. S3), and the median area per LSOA was 0.4, varying from 0.0002 to 68.4(min to max). The LTLA boundaries are revised every year, whereas the LSOA ones at census. Let denote that the l-th LSOA belongs to the m-th LTLA, the number of deaths in the m-th LTLA and the population in the i-th age group (1<, 1–4, 5–9, …, 85–90, >90), j-the sex (male or female), k-th ethnic group (White, Mixed, Asian, Black, Other) and -th LSOA. We sampled individual deaths at the l-th LSOA level from a Multinomial distribution with probabilities:
and repeated the procedure 100 times.
2.3. Exposure
We considered exposure to NO2 and PM2.5 as indicators of air pollution. We selected these pollutants because: 1) they reflect different sources of air-pollution (NO2 reflects traffic related air-pollution, whereas PM2.5 is a combination of traffic and non-traffic sources), 2) they were considered in previous studies (Cole et al., 2020, Liang et al., 2020, Travaglio et al., 2020, Wu et al., 2020), and 3) they are responsible for the highest number of years of life lost compared to other pollutants in Europe (Ortiz 2019). We retrieved NO2 and PM2.5 concentration in England from the Pollution Climate Mapping (PCM; https://uk-air.defra.gov.uk/). The PCM produces annual estimates during 2001–2018 for NO2 and 2002–2018 for PM2.5 at 1x1km resolution for the UK. The PCM model is calibrated using monitoring stations across the nation and has high predictive accuracy, R2 = 0.88 for NO2 and R2 = 0.63 for PM2.5 (Brookes 2017). We defined long-term exposure to these compounds as the mean of the past 5 years for which data was available at the time of analysis, i.e. 2014–2018. An alternative is calculating the median, however the distribution of the air-pollutants using any of these metrics is almost identical, (Supplemental Material Fig. S4). We weighted the exposure using a combination of population estimates available from the fourth version of Gridded Population of the World collection at 1x1km grid as of 2020 (Center for International Earth Science Information Network - CIESIN - Columbia University 2018) and from ONS at LSOA level as of 2018. Let be the pollutant and the population in the intersection of the -th grid cell and -th LSOA. Assuming the is constant (i.e. for all intersections) in the -th grid cell, we define the population weighted version of as:
To calculate , we first compute , where is the area weight per intersection. Then calculate the population per intersection: . We then use the (LSOA populations) and obtain , where is the normalised , ie .
2.4. Confounders
We considered confounders related with meteorology, socio-demographics, disease spread, healthcare provision and health related variables (Table 1 ). As meteorological confounders, we considered temperature and relative humidity and calculated the mean for March-June 2018 as this is the latest year with data available at 1x1km grid retrieved from the MetOffice. We weighted temperature and relative humidity using the population weights calculated for the air-pollution exposure. As socio-demographical confounders we considered age, sex, ethnicity, deprivation, urbanicity, population density and occupation. Information on age (2018), sex (2018), ethnicity (2011), urbanicity (2011) and population density (2018) was available at the LSOA level from ONS (the most recent years available at time of analysis). To adjust for deprivation, we used quintiles of the index of multiple deprivation at LSOA level in 2019 (Ministry of Housing, Communities and Local Government), excluding the dimension related to air quality. We used estimates of occupational exposures to COVID-19, as calculated by ONS, to adjust for high risk exposure to COVID-19, defined as those with a score higher than 80/100 (corresponding to at least >1 per week exposed to someone infected, Supplemental Material Text S1.1 and Table S1). To account for disease progression, we used the number of days since the 1st reported case and the number of positive cases in each LTLA (as of 30th of June 2020, as retrieved from PHE). Adjustment for the latter factors is expected to attenuate geographical differences generated due to regional differences about the timing on the pandemic curve. For healthcare provision, we used the number of intensive care unit beds per population, in February 2020 per NHS trust, as retrieved from NHS. Last, as health-related variables, we considered smoking and obesity prevalence at the GP practice level during 2018–2019, as retrieved from PHE (Supplemental Material Text S1.1).
Table 1.
Data sources used in the analysis.
| Confounders | Source | Spatial Resolution | Temporal Resolution | Type |
|---|---|---|---|---|
| Temperature | MetOffice https://www.metoffice.gov.uk/ |
1 km2 | March-June 2018 | continuous |
| Relative humidity | MetOffice https://www.metoffice.gov.uk/ |
1 km2 | March-June 2018 | continuous |
| Index of Multiple Deprivation | Ministry of Housing, Communities and Local Government https://www.gov.uk/ |
Lower layer super output area | 2019 | rank (quintiles) |
| Urbanicity | Office for National Statistics https://www.ons.gov.uk/ |
Lower layer super output area | 2011 | urban/rural |
| Days since 1st reported case | Public Health England | Lower tier local authority | Until 30th June | continuous |
| Number of positive cases | Public Health England | Lower tier local authority | Until 30th June | discrete (counts) |
| Population density | Office for National Statistics https://www.ons.gov.uk/ |
Lower layer super output area | 2018 | continuous (log transformed) |
| Number of intensive care unit beds | National Health Service https://www.england.nhs.uk/ |
National Health Service trust | February 2020 | continuous (per population) |
| Smoking | Public Health England https://fingertips.phe.org.uk/ |
General practitioner catchment area | 2018–2019 | continuous (prevalence) |
| Obesity | Public Health England https://fingertips.phe.org.uk/ |
General practitioner catchment area | 2018–2019 | continuous (prevalence) |
| High Risk Occupation | Office for National Statistics https://www.ons.gov.uk/ |
Middle layer super output area | 2011 | continuous (prevalence) |
2.5. Statistical methods
We specified Bayesian hierarchical Poisson log-linear models to investigate the association of COVID-19 deaths and NO2 and PM2.5 independently. The LSOA specific standardised mortality ratio is known to be an unstable estimator with high variance when the number of expected deaths is small. To overcome this problem, we used a well-established hierarchical framework, specifying spatially structured and unstructured random effects, so that the model borrows strength from the other areas across the entire study region, as well as from the neighbouring ones (Best et al., 2005, Wakefield et al., 2000, Wakefield, 2006). We model these random effects using a re-parametrisation of the Besag-York-Molliè conditional autoregressive prior distribution (Besag et al., 1991, Simpson et al., 2017). We fitted four models including: 1) each pollutant (model 1), 2) each pollutant and the spatial autocorrelation term (model 2), 3) each pollutant and all confounders (model 3) and 4) each pollutant, the spatial autocorrelation term and all confounders (model 4). All models were adjusted for age, sex and ethnicity using indirect standardisation; we used the English population as the standard population to calculate the rates. We do not report results from the joint analysis including both pollutants since they are highly correlated (Supplemental Material Figure S5).
In order to propagate the uncertainty resulted from the sampling we used for the downscaling, we fitted the models over 100 downscaled samples and then performed Bayesian model averaging to combine the estimates (Gómez-Rubio et al. 2020). We performed a complete case analysis since for only 1.1% of the cases information about age, sex and ethnicity is missing. We report results as posterior median of % increase in mortality risk for every 1 μg/m3 increase in the air-pollutants, 95% credible intervals (CrI) and posterior probability that the estimated effect is positive. We also report posterior median of spatial mortality relative risks (exponential of the spatial autocorrelation term) and posterior probabilities that the spatial relative risks are larger than 1.
The mathematical formulation of the models and prior specifications are given in the Supplemental Material Text S1.2.
All models were fitted in INLA (Rue et al. 2009). Covariate data and code for running the analysis are available at https://github.com/gkonstantinoudis/COVID19AirpollutionEn.
2.6. Sensitivity analyses
We performed a series of sensitivity analyses. First, we repeated the main analyses using data at the LTLA level with all exposures and confounding weighted by population. Second, we examined if there is a differential effect of long-term exposure to air-pollution at the early stages of the epidemic, considering the lockdown (23rd of March 2020) as a landmark. Third, we assessed the correlation between the latent field of the full model (model 4) with that of the model excluding or including only covariates indicating disease spread (i.e. number of tested positive cases and days since first reported cases). Fourth, we categorised pollutants into quintiles to allow more flexible fits. Fifth, we repeated the analysis including the suspected cases to the outcome. Sixth, we repeated the analysis changing the definition of long-term exposure to the mean of the past 3 and 10 years for which data was available at the time of analysis, i.e. 2016–2018 and 2009–2018. Seventh, we fitted a zero-inflated Poisson model to account for the proportion of zeros in the data (36% in the 100 samples – see Supplemental Material Fig. S6).
3. Results
3.1. Study population
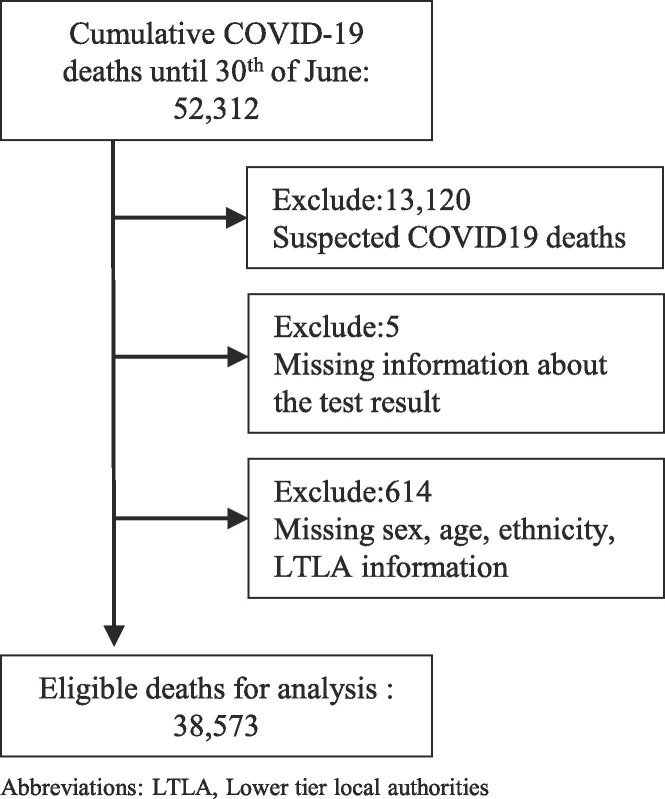
We identified 38,573 COVID-19 deaths with a laboratory confirmed test in England between 2nd March and 30th June (Fig. 1 ). The age, sex and ethnicity distribution of the deaths follows patterns reported previously (Supplemental Material Tables S2-3).
Fig. 1.
Flowchart of the COVID-19 deaths.
3.2. Exposure
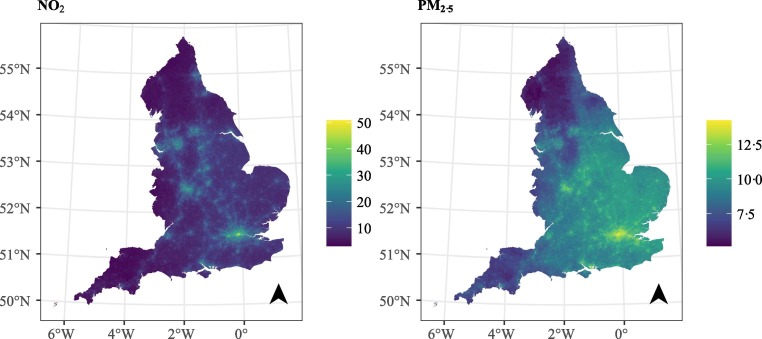
Fig. 2 shows the population weighted air-pollutants at LSOA level in England. We observe that the localised variation of NO2, for instance due to the highways, is adequately captured at the spatial resolution of the LSOAs. The mean of NO2 is 16.17 μg/m3 and it varies from 2.99 μg/m3 in highly rural areas to 50.69 μg/m3 in the big urban centres (Fig. 2). The mean of PM2.5 is 9.84 μg/m3 with a smaller variation, 5.14–14.22 μg/m3 (Fig. 2).
Fig. 2.
Population weighted exposure per LSOA.
3.3. Confounders
Plots and maps of the confounders can be found in Supplemental Material, Fig. S7-17.
3.4. No2
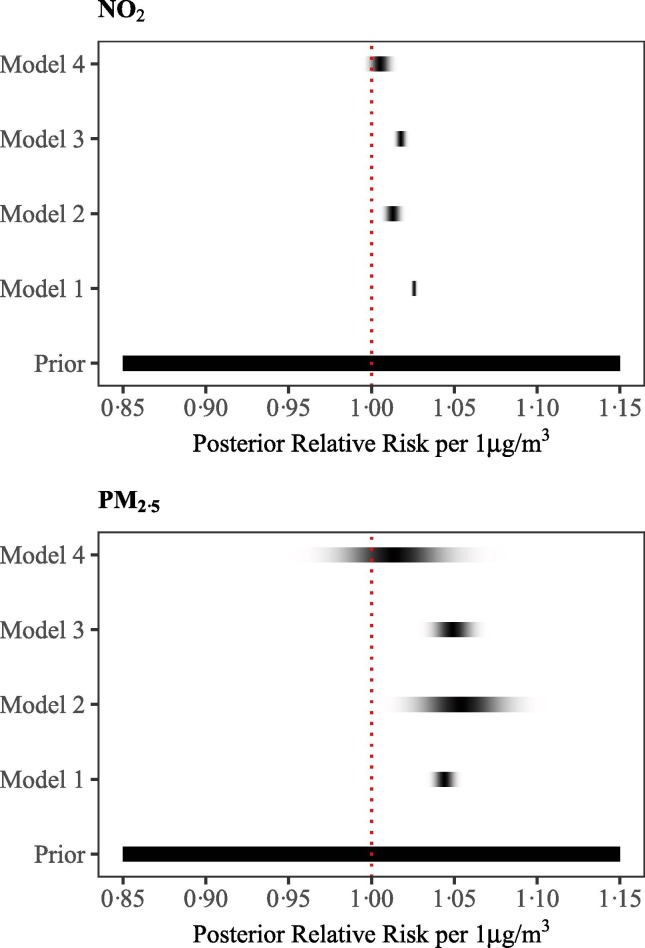
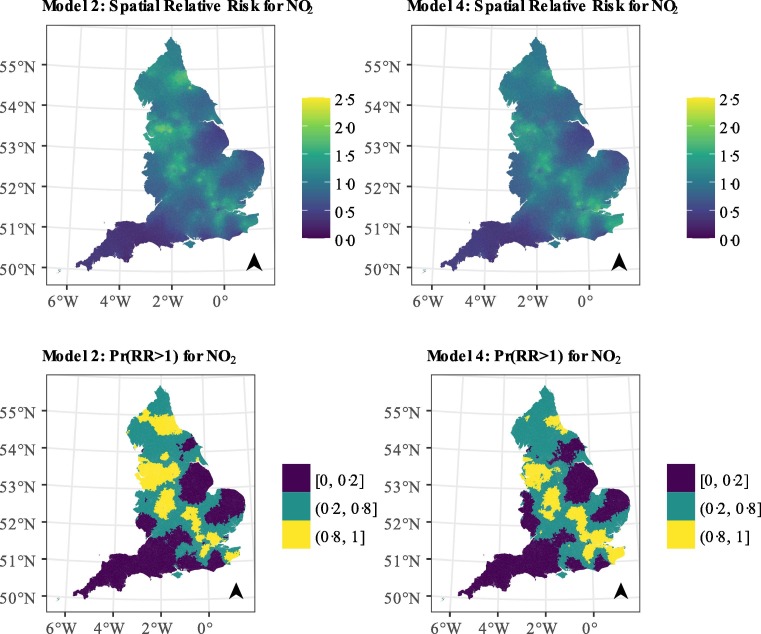
We observe a 2.6% (95%CrI: 2.4%, 2.7%) increase in the COVID-19 mortality risk for every 1 μg/m3 increase in the long-term exposure to NO2, based on model 1 (Fig. 3 & Supplemental Material Table S4). There is still evidence of an effect, albeit smaller, once we adjust for spatial autocorrelation or confounders, with increases in the long-term exposure to NO2 of, respectively, 1.3% (95% CrI: 0.8%, 1.8%), 1.8% (95% CrI: 1.5%, 2.1%) for every 1 μg/m3. When we adjust for both autocorrelation and confounders the evidence is less strong, with estimates of 0.5% (95% CrI: −0.2%, 1.2%) for every 1 μg/m3 (Fig. 3 & Supplemental Material Table S4) and posterior probability of a positive effect reaching 0.93. The spatial relative risk in England varies from 0.24 (95% CrI: 0.08, 0.69) to 2.09 (95% CrI: 1.30, 3.11) in model 2 and from 0.30 (95% CrI: 0.10, 0.84) to 1.87 (95% CrI: 1.18, 2.93) in model 4, implying that the confounders explain very little of the observed variation (Fig. 3). The variation is more pronounced in the cities and suburban areas (with posterior probability higher than 1; Fig. 3).
Fig. 3.
Density strips for the posterior of COVID-19 mortality relative risk with 1 μg/m3 increase in NO2 (top panel) and PM2.5 (bottom panel) averaged long-term exposure.
3.5. Pm2.5
We observe a 4.4% (95% CrI: 3.7%, 5.1%) increase in the mortality risk for every 1 μg/m3 increase in the long-term exposure to PM2.5, based on model 1 (Fig. 3 & Supplemental Material Table S5). When we adjust for spatial autocorrelation the effect increases slightly but the credible intervals are wider, 5.4% (95% CrI: 2.5%, 8.4%), whereas it is similar when we adjust for confounding 4.9% (95% CrI: 3.7%, 6.2%) (Fig. 3 & Supplemental Material Table S5). The effect is weak when we account for confounders and spatial autocorrelation 1.4% (95% CrI: −2.1%, 5.1%) (Fig. 3 & Supplemental Material Table S5). The posterior probability of a positive effect is lower than observed for NO2, and equal to 0.78. The spatial relative risk follows similar patterns as the one reported in the models for NO2, with the posterior median relative risk varying from 0.24 (95% CrI: 0.12, 0.46) to 2.26 (95% CrI: 1.32, 3.85) in model 2 and from 0.30 (95% CrI: 0.15, 0.57) to 1.90 (95% CrI: 1.14, 3.17) in model 4 (Supplemental Material, Fig. S18).
3.6. Sensitivity analyses
When LTLAs are the main geographical unit for analysis, the results are consistent, but higher in magnitude, potentially due to inadequate covariate and spatial autocorrelation adjustment due to the coarse geographical resolution (Supplemental Material Tables S6-7, Fig. S19-20). Restricting the study period to March 23, 2020 (N = 698) also results in similar estimates for both pollutants, however the uncertainty is higher (Supplemental Material Tables S8-9, Fig. S21-22). The latent field of model 4, with NO2 as the pollutant, is similar to the latent fields of the models with and without the disease progression variables, with a correlation coefficient of 0.94 and 0.93 respectively (Supplemental Material Fig. S23). The use of quintiles of the pollutants justifies the linearity assumption (Supplemental Material Fig. S24). The results are consistent, but the evidence weaker, when suspected COVID-19 deaths are included (Supplemental Material Tables S10-11, Fig. S25-26). The results are also similar when we used a 3 or a 10-year mean of the air-pollutants concentration (Supplemental Material Fig. S27). The results are consistent when we fitted a zero-inflated Poisson (Supplemental Material Tables S12-13 and Fig. S28-29).
3.7. Post-hoc analysis
In a post-hoc analysis we investigated if the evidence of an effect of NO2 on COVID-19 mortality can be attributed to pre-existing conditions. We selected hypertension, chronic obstructive pulmonary disease (COPD) and diabetes, because of 1) indications of previous literature that they increase the COVID-19 mortality risk (Williamson et al., 2020, Yang et al., 2020), 2) previous literature that suggest an effect with long-term exposure NO2 (Balti et al., 2014, Cai et al., 2016, Zhang et al., 2018) and 3) data availability. We retrieved prevalence data for these pre-existing conditions from PHE available at the GP practice level during 2018–2019 (https://fingertips.phe.org.uk/), Supplemental Material Fig. S30-32. The effect of NO2 remains similar, 0.6% (95% CrI: −0.1%, 1.3%) with the posterior probability being 0.94 whereas the spatial relative risk highlights the same geographical locations, Supplemental Material Fig. S33.
4. Discussion
4.1. Main findings
This is the first nationwide study in England investigating the effect of long-term exposure to NO2 and PM2.5 during 2014–2018 on COVID-19 mortality at LSOA level. The unadjusted models indicate that for every 1 μg/m3 increase in the long-term exposure to NO2 and PM2.5 the COVID-19 mortality risk increases. After considering the effect of confounding and spatial autocorrelation there is still some evidence of an effect, albeit is less strong, for NO2, while for PM2.5 there is larger uncertainty. The spatial relative risk has strong spatial patterns, identical for the different pollutants, potentially highlighting the effect of disease spread.
4.2. Comparison with previous studies focusing on NO2
Our study is comparable with previous studies in the US, England and the Netherlands assessing the long-term effect of NO2 in COVID-19 mortality. The study in the US focused on deaths reported by April 29, 2020, using 3122 counties. For the exposure, they calculated the mean of daily concentrations during 2010–2016 as modelled by a previously described ensemble machine learning model (R2 = 0.79) (Di et al., 2019a). They reported a 7.1% (95% Confidence Interval: 1.2%, 13.4%) increase in mortality per 4.5 ppb (1 ppb = 1.25 μg/m3) increase in NO2 after adjusting for confounders and spatial autocorrelation(Liang et al. 2020)(that is approximately 1.3% increase per 1 μg/m3). A study in England, with partly overlapping data as in our analysis, also reported a significant association between NO2 and COVID-19 mortality (p < 0.05). For the analysis they focused on COVID-19 deaths reported in England up to April 10, 2020, used 317 LTLAs, and did not account for spatial autocorrelation (Travaglio et al. 2020). The study in the Netherlands using 335 municipalities, mean exposure during 2015–2019 and COVID-19 deaths up to June 5, 2020 reported 0.35 (95% CI: 0.04, 0.66) additional COVID-19 deaths for every 1 μg/m3 increase in NO2 after adjusting for confounders and certain spatial controls, such as transmission beyond the Dutch national borders (Cole et al. 2020). Since the mean number of deaths in their sample is 16.86, the above estimate translates to a 2.0% increase in the COVID-19 mortality for every 1 μg/m3 increase in NO2. An ONS report in England using 175 sampling units, 10-year averaged NO2 exposure (PCM) and COVID-19 deaths up to June 12, 2020 found a 0.6% (95% CI: −0.1%, 2.2%) increase in the COVID-19 mortality for every 1 μg/m3 increase in averaged NO2 exposure (Statistics 2020).
4.3. Comparison with previous studies focusing on PM2.5
Our study is comparable with previous studies assessing the long-term effect of PM2.5 on COVID-19 mortality. The aforementioned study in the US also assessed the effect of PM2.5 on COVID-19 mortality(Liang et al. 2020). Their exposure model was previously validated having an R2 = 0.89 for the annual estimates (Di et al. 2019b). The evidence for PM2.5 was weak, namely 10.8% (95% CI:-1.1%, 24.1%) per 3.4 μg/m3 increase in PM2.5 concentration (that is approximately 3.2% increase per 1 μg/m3) after adjusting for confounding and spatial autocorrelation. The ONS report in England found a 1% (95% CI: −3%, 6%) increase in the COVID-19 mortality for every 1 μg/m3 increase in the 10-year averaged PM2.5 exposure (Statistics 2020). Our study comes in contrast with another study in the US that used deaths reported until April 22nd, 2020 and counties as the geographical unit (Wu et al. 2020). For the exposure, they used previously validated monthly PM2.5 concentrations (R2 = 0.70) (Van Donkelaar et al. 2019) and averaged them during 2000 and 2016. After adjusting for confounding but not for spatial autocorrelation, they found an 11% (95% CI: 6%, 17%) increase in the COVID-19 death rate for an increase of 1 μg/m3 in PM2.5 concentration (Wu et al. 2020). Our study comes also in contrast with the study in the Netherlands that reported 2.3 (95% CI: 1.3, 3.0) additional COVID-19 deaths for an increase of 1 μg/m3 in the averaged long-term PM2.5 concentration (Cole et al. 2020). Having a mean number of deaths equal to 16.86, the above estimate translates to a 13.6% increase in the COVID-19 mortality rate for an increase of 1 μg/m3 in PM2.5 concentration.
4.4. Strengths and limitations
Our study is the first study to examine the association between long-term exposure to NO2 and PM2.5 and COVID-19 mortality at very high geographical precision. The spatial unit of our analysis is LSOAs, for which there are 32,844 in England (~130 000 km2), whereas previous studies have used 317 LTLAs or 175 sampling units in England, counties in the US (3 122 in an area ~9.8 million km2) and municipalities in the Netherlands (334 in an area ~41 500 km2). Such high-resolution allows capturing the localised geographical patterns of the pollutants but also ensures adequate confounding and spatial autocorrelation adjustment. Our study also covers, so far, the largest temporal window of the epidemic (capturing COVID-19 deaths attributable to the first wave, Supplemental Material Fig. S34), while most previous studies focused on the early to mid-stages of the first wave. This ensures better generalisability of the results. In addition, physical distancing and other public health interventions were introduced nationwide in England during the first epidemic, mitigating any distortion between air-pollution and COVID-19 mortality due to potential regional level differences. Our results are also consistent in a sensitivity analysis focusing on the pre-lockdown period, in the absence of public health interventions. Based on the scientific literature, we adjusted for several variables which would act as the confounders of the relationship between air pollution and COVID-19 mortality. Nevertheless, since the aetiology and the factors contributing to COVID-19 mortality are not fully understood yet, we included a spatial random effect to capture unknown spatial confounding. The spatial random effect was found to be a crucial component in the model. Not accounting for spatial autocorrelation, when spatial autocorrelation is present, is expected to give rise to narrower credible intervals and false positive effects (Lee and Sarran 2015).
Our study has also some limitations. The downscaling procedure will likely inflate the reported credible intervals. However, this naturally reflects the uncertainty of the place of residence resulted from the downscaling approach. Although we consider small areas, the study is still an ecological one and thus the reported effects do not reflect individual associations (Wakefield 2008). Case fatality might have been a more appropriate metric for the analysis, since disease spread is accounted for in the denominator. Nevertheless, given the asymptomatic infections and the fact that number of reported infections is not a random sample of the general population, the number of COVID-19 cases per LTLA is not reliable at this stage. For the same reason, using the number of reported cases to adjust for disease progression and clustering of cases and deaths might not adequately capture disease progression and clustering of cases and deaths. However, part of this clustering was captured in the spatial autocorrelation term. We did not account for population mobility during 2014–2018 and assumed constant residence and thus levels of exposure to air-pollution. While this is a limitation, we believe that it would have a minimal impact on the results given that 1) the exposure period is relatively short and 2) almost 93% of the deaths in our dataset occurred in people 60 years or older (Supplemental Material Table S2). This comprises a population less likely to have moved during the past 5 years (Burgess and Quinio 2020). We also could not account for non-residential air-pollution exposure. Spatiotemporal variation in the strains of COVID-19 can introduce bias (Villeneuve and Goldberg 2020), however at the time of publication there was no evidence supporting that strain types can confound the relationship between COVID-19 mortality and air-pollution.
4.5. Interpretation
Compared to the previous studies, our results are the smallest in magnitude, likely because of the high geographical precision that allows more accurate confounding and spatial autocorrelation adjustment. In addition, we report weak evidence of an effect, which could also be due to lack of power and individual exposure data. Nevertheless, as for NO2 we find a high posterior probability of an effect on mortality, we argue that a potential explanation might be the mediation effect of pre-existing conditions. While in our analysis the inclusion of area-level prevalence of hypertension, diabetes and COPD did not change the results, the ecological nature of the pre-existing conditions data does not allow us to account for the mediation effect at the individual level. Our study focuses on the mortality after contracting SARS-CoV-2, however we cannot rule out individual susceptibility to becoming infected as an explanation to the uncertainty in the effect estimates (Villeneuve and Goldberg 2020). Such susceptibility can reflect immunosuppression, leading to later increases in inflammation (Edoardo Conticini et al. 2020) and thus worse prognosis, or even disease spread, as recent studies have suggested that PM2.5 can proliferate COVID-19 transmission (Bianconi et al. 2020).
Our analysis captured strong spatial autocorrelation. The observed pattern could reflect residual variation from a potential inadequate covariate adjustment (including disease spread), spatial variation of pre-existing conditions, other unknown spatial confounders or a combination from all above. In a sensitivity analysis, we observed that the factors associated with disease transmission left the latent field unchanged (Supplemental Material Fig. S21), as did the inclusion of hypertension, diabetes and COPD (Supplemental Material Fig. S33). When we restricted the analysis to the pre-lockdown period, the latent field for both pollutants captured London and Birmingham, i.e. the cities with the first outbreaks. Considering the above, and the fact that COVID-19 is an infectious disease, we believe that large variation of Fig. 4 is likely due to disease spread, which is not adequately captured in the disease progression covariates.
Fig. 4.
Median posterior spatial relative risk (exponential of the spatial autocorrelation term) and posterior probability that the spatial relative risk is larger than 1 for the models with NO2 and a spatial autocorrelation term and the fully adjusted NO2 model.
5. Conclusion
Overall, this study provides some evidence of an association between averaged exposure during 2014–2018 to NO2 and COVID-19 mortality, while the role of PM2.5 remains more uncertain.
CRediT authorship contribution statement
Garyfallos Konstantinoudis: Conceptualization, Methodology, Software, Formal analysis, Writing - original draft, Writing - review & editing, Funding acquisition. Tullia Padellini: Methodology, Software, Formal analysis, Writing - review & editing. James Bennett: Writing - review & editing, Methodology. Bethan Davies: Writing - review & editing. Majid Ezzati: Conceptualization, Project administration, Writing - review & editing, Funding acquisition. Marta Blangiardo: Conceptualization, Methodology, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
GK is supported by an MRC Skills Development Fellowship [MR/T025352/1]. MB and TP are supported by a National Institutes of Health, grant number [R01HD092580-01A1]. JB and ME are supported by the Pathways to Equitable Healthy Cities grant from the Wellcome Trust [209376/Z/17/Z] and by a grant from the US Environmental Protection Agency (EPA), as part of the Centre for Clean Air Climate Solution (assistance agreement no. R835873). This article has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the EPA. The EPA does not endorse any products or commercial services mentioned in this publication. Infrastructure support for this research was provided by the National Institute for Health Research Imperial Biomedical Research Centre (BRC). This work was part supported by the MRC Centre for Environment and Health, which is currently funded by the Medical Research Council (MR/S019669/1, 2019-2024).
Handling Editor: Hanna Boogaard
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2020.106316.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Balti E.V., Echouffo-Tcheugui J.B., Yako Y.Y., Kengne A.P. Air pollution and risk of type 2 diabetes mellitus: A systematic review and meta-analysis. Diabetes Res. Clin. Pract. 2014;106:161–172. doi: 10.1016/j.diabres.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Besag J., York J., Mollié A. Bayesian image restoration, with two applications in spatial statistics. Ann. Inst. Stat. Math. 1991;43:1–20. [Google Scholar]
- Best N., Richardson S., Thomson A. A comparison of Bayesian spatial models for disease mapping. Stat. Methods Med. Res. 2005;14:35–59. doi: 10.1191/0962280205sm388oa. [DOI] [PubMed] [Google Scholar]
- Bianconi V., Bronzo P., Banach M., Sahebkar A., Mannarino M.R., Pirro M. Particulate matter pollution and the COVID-19 outbreak: Results from Italian regions and provinces. Arch. Med. Sci. 2020;16 doi: 10.5114/aoms.2020.95336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, D.S., John; Kent, Andrew; Whiting, Sally; Rose, Rebecca; Williams, Chris. 2017. Technical report on UK supplementary assessment under the air quality directive (2008/50/ec), the air quality framework directive (96/62/ec) and fourth daughter directive (2004/107/ec) for 2015.
- Burgess G., Quinio V. Unpicking the downsizing discourse: Understanding the housing moves made by older people in England. Housing Stud. 2020:1–16. [Google Scholar]
- Cai Y., Zhang B., Ke W., Feng B., Lin H., Xiao J., et al. Associations of short-term and long-term exposure to ambient air pollutants with hypertension: A systematic review and meta-analysis. Hypertension. 2016;68:62–70. doi: 10.1161/HYPERTENSIONAHA.116.07218. [DOI] [PubMed] [Google Scholar]
- Center for International Earth Science Information Network - CIESIN - Columbia University. 2018. Gridded population of the world, version 4 (gpwv4): Population count, revision 11. Palisades, NY:NASA Socioeconomic Data and Applications Center (SEDAC).
- Cole, M., Ozgen, C., Strobl, E., 2020. Air pollution exposure and covid-19. [DOI] [PMC free article] [PubMed]
- Conticini E., Frediani B., Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in northern italy? Environ. Pollut. 2020;261 doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Q., Amini H., Shi L., Kloog I., Silvern R., Kelly J., et al. Assessing NO2 concentration and model uncertainty with high spatiotemporal resolution across the contiguous United States using ensemble model averaging. Environ. Sci. Technol. 2019;54:1372–1384. doi: 10.1021/acs.est.9b03358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, Q., Amini, H., Shi, L., Kloog, I., Silvern, R., Kelly, J., et al., 2019b. An ensemble-based model of PM2.5 concentration across the contiguous united states with high spatiotemporal resolution. Environ. Int. 130, 104909. [DOI] [PMC free article] [PubMed]
- EpiCell. 2020. Techincal summary: Public Health England data series on deaths in people with COVID-19. Public Health England.
- Giorgini P., Di Giosia P., Grassi D., Rubenfire M.D., Brook R., Ferri C. Air pollution exposure and blood pressure: An updated review of the literature. Curr. Pharm. Des. 2016;22:28–51. doi: 10.2174/1381612822666151109111712. [DOI] [PubMed] [Google Scholar]
- Gómez-Rubio V., Bivand R.S., Rue H. Bayesian model averaging with the Integrated Nested Laplace Approximation. Econometrics. 2020;8:23. [Google Scholar]
- Hauser A., Counotte M.J., Margossian C.C., Konstantinoudis G., Low N., Althaus C.L., et al. Estimation of SARS-CoV-2 mortality during the early stages of an epidemic: A modeling study in Hubei, China, and six regions in Europe. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis V., Bennett J.E., Rashid T., Parks R.M., Pearson-Stuttard J., Guillot M., et al. Magnitude, demographics and dynamics of the effect of the first wave of the COVID-19 pandemic on all-cause mortality in 21 industrialized countries. Nat. Med. 2020 doi: 10.1038/s41591-020-1112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Sarran C. Controlling for unmeasured confounding and spatial misalignment in long-term air pollution and health studies. Environmetrics. 2015;26:477–487. doi: 10.1002/env.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Shi L., Zhao J., Liu P., Sarnat J.A., Gao S., et al. Urban air pollution may enhance COVID-19 case-fatality and mortality rates in the united states. The Innovation. 2020;1 doi: 10.1016/j.xinn.2020.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, A., 2019. Air quality in Europe-2019 report. European Environment Agency, Luxembourg.
- Rue H., Martino S., Chopin N. Approximate Bayesian inference for latent Gaussian models by using Integrated Nested Laplace Approximations. J. Roy. Statist. Soc.: Ser. B (Statist. Methodol.) 2009;71:319–392. [Google Scholar]
- Scheers H., Jacobs L., Casas L., Nemery B., Nawrot T.S. Long-term exposure to particulate matter air pollution is a risk factor for stroke: Meta-analytical evidence. Stroke. 2015;46:3058–3066. doi: 10.1161/STROKEAHA.115.009913. [DOI] [PubMed] [Google Scholar]
- Simpson D., Rue H., Riebler A., Martins T.G., Sørbye S.H. Penalising model component complexity: A principled, practical approach to constructing priors. Statist. Sci. 2017;32:1–28. [Google Scholar]
- Office for National Statistics. 2020. Coronavirus (COVID-19) related mortality rates and the effects of air pollution in England.
- Travaglio, M., Yu, Y., Popovic, R., Leal, N.S., Martins, L.M., 2020. Links between air pollution and COVID-19 in England. medRxiv. [DOI] [PMC free article] [PubMed]
- Van Donkelaar A., Martin R.V., Li C., Burnett R.T. Regional estimates of chemical composition of fine particulate matter using a combined geoscience-statistical method with information from satellites, models, and monitors. Environ. Sci. Technol. 2019;53:2595–2611. doi: 10.1021/acs.est.8b06392. [DOI] [PubMed] [Google Scholar]
- Villeneuve P.J., Goldberg M.S. Methodological considerations for epidemiological studies of air pollution and the sars and COVID-19 coronavirus outbreaks. Environ. Health Perspect. 2020;128:95001. doi: 10.1289/EHP7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J., Best N., Waller L. Bayesian approaches to disease mapping. Spatial Epidemiol.: Methods Appl. 2000:104–127. [Google Scholar]
- Wakefield J. Disease mapping and spatial regression with count data. Biostatistics. 2006;8:158–183. doi: 10.1093/biostatistics/kxl008. [DOI] [PubMed] [Google Scholar]
- Wakefield J. Ecologic studies revisited. Annu. Rev. Public Health. 2008;29:75–90. doi: 10.1146/annurev.publhealth.29.020907.090821. [DOI] [PubMed] [Google Scholar]
- Williamson E.J., Walker A.J., Bhaskaran K., Bacon S., Bates C., Morton C.E., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Nethery R.C., Sabath M.B., Braun D., Dominici F. Air pollution and COVID-19 mortality in the United States: Strengths and limitations of an ecological regression analysis. Sci. Adv. 2020;6(eabd4049) doi: 10.1126/sciadv.abd4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: A systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang J., Lu W. Exposure to nitrogen dioxide and chronic obstructive pulmonary disease (COPD) in adults: A systematic review and meta-analysis. Environ. Sci. Pollut. Res. 2018;25:15133–15145. doi: 10.1007/s11356-018-1629-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.