Abstract
The histone deacetylases (HDACs)-dependent mechanisms regulating gene transcription of the Na+/Ca+ exchanger isoform 3 (ncx3) after stroke are still unknown. Overexpression or knocking-down of HDAC4/HDAC5 down-regulates or increases, respectively, NCX3 mRNA and protein. Likewise, MC1568 (class IIa HDACs inhibitor), but not MS-275 (class I HDACs inhibitor) increased NCX3 promoter activity, gene and protein expression. Furthermore, HDAC4 and HDAC5 physically interacted with the transcription factor downstream regulatory element antagonist modulator (DREAM). As MC1568, DREAM knocking-down prevented HDAC4 and HDAC5 recruitment to the ncx3 promoter. Importantly, DREAM, HDAC4, and HDAC5 recruitment to the ncx3 gene was increased in the temporoparietal cortex of rats subjected to transient middle cerebral artery occlusion (tMCAO), with a consequent histone-deacetylation of ncx3 promoter. Conversely, the tMCAO-induced NCX3 reduction was prevented by intracerebroventricular injection of siDREAM, siHDAC4, and siHDAC5. Notably, MC1568 prevented oxygen glucose deprivation plus reoxygenation and tMCAO-induced neuronal damage, whereas its neuroprotective effect was abolished by ncx3 knockdown. Collectively, we found that: (1) DREAM/HDAC4/HDAC5 complex epigenetically down-regulates ncx3 gene transcription after stroke, and (2) pharmacological inhibition of class IIa HDACs reduces stroke-induced neurodetrimental effects.
Keywords: HDAC class II, MC1568, NCX3, neuroprotection
Introduction
Several evidence have established that the plasma membrane Na+/Ca2+ exchanger 3 (NCX3) has a neuroprotective role in stroke and in other neurological disorders.1–5 Indeed, over the years, it has been shown that NCX3 activity and expression are essential (1) to limit brain ischemic damage;5–7 (2) to mediate the preconditioning/postconditioning-induced neuroprotection; (3) to prevent an impairment in hippocampal long-term potentiation and in spatial learning and memory,8 and (4) to reduce the activation of caspase-12 in an in vitro model of Alzheimer Disease.9
In particular, in the brain, ncx3 gene expression is increased by the transcriptional activator adenosine 3ʹ,5ʹ cyclic monophosphate (cAMP) response element binding protein (CREB),10 and is decreased by the transcriptional repressor downstream regulatory element antagonistic modulator DREAM11 through a genetic mechanism. Previous studies have reported that the other central nervous system (CNS) expressed sodium–calcium exchanger 1 (ncx1) is activated by the transcription factor hypoxia-inducible factor 1 (HIF-1)12 and is inhibited by RE1-silencing transcription factor (REST), through a genetic and epigenetic mechanism.13,14 On the other hand, the epigenetic mechanisms regulating the gene transcription of ncx3 are presently unknown.
Among the various epigenetic mechanisms implicated in the cerebral ischemia pathophysiology, histone nucleosomal acetylation is one of the most widely investigated.15–17 Both acetylation and deacetylation reactions are reversible dynamic processes determined by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. In mammals, HDACs are divided into four classes: class I (HDACs 1–3 and 8), class II (HDACs 4–7 and 9,10), class III (Sirtuins 1–7) and class IV (HDAC11). Class II HDACs can be further divided into class IIa: HDAC 4, 5, 7, and 9, and IIb: HDAC6 and 10.18 Notably, the isoforms HDAC4 and HDAC5 are predominantly expressed in neurons, but not in astrocytes and microglia.19 Considering these premises and taking into account that: (1) NCX3 is a pro-survival factor4,5,20; (2) DREAM transcriptionally regulates NCX3 expression11;(3) HDACs modulate gene expression by binding transcription factors14 and are involved in stroke pathophysiology,16 the aim of the present work is to investigate the effect of the class I HDAC inhibitor MS275 and the class IIa HDAC inhibitor MC1568 on NCX3. Furthermore, we studied whether DREAM is involved in this epigenetic mechanism and which HDAC isoforms can be potentially involved in NCX3 down-regulation. Finally, by using in vitro and in vivo models of stroke, we examined the role of DREAM and HDACs in regulating ischemia-induced NCX3 reduction and the effect of MC1568 on stroke-induced brain damage.
Materials and methods
Animals and reagents
All restriction enzymes, DNA modifying enzymes, luciferase reporter kits and luciferase vectors were from Promega (Italy). Synthetic oligonucleotides were from Eurofins Genomics (Ebersberg, Germany). All siRNAs used were purchased from Qiagen (Italy) and raised against rat. Specifically, siRNAs for HDAC4 (siHDAC4), HDAC5 (siHDAC5), HDAC7 (siHDAC7), HDAC9 (siHDAC9), NCX3 (siNCX3), and negative control siCONTROL (siCTL) have used as previously published,21,22 whereas siRNAs for DREAM (siDREAM) (SI03061079) was used at 50 nM. Two sets of siRNAs were tested for DREAM; the set that more efficiently reduced NCX3 expression was used for the experiments (data not shown).
The constructs overexpressing HDAC4 (Addgene plasmid # 30485) were a gift from Tso-Pang Yao, the plasmids for HDAC5 (Addgene plasmid # 13821), HDAC7 (Addgene plasmid # 13824) were a gift from Eric Verdin.23 For HDAC9 overexpression, the construct was kindly provided by Prof. Edward Seto (Washington University, USA).24 The empty vector used was pcDNA3.1. The HDACs inhibitors TSA (T8552), MS-275 (EPS002), and MC1568 (M1824) were all purchased from Sigma (Italy). All drugs were resuspended in dimethyl sulfoxide (DMSO), which was diluted in culture medium to reach a final concentration of 0.2%. All animal procedures were performed in accordance with the Animal Care Committee of Federico II University of Naples by Italian Ministerial Authorization (DL 116/92 art.7). All protocols and procedures of the handling of animals were according to the international guiding principles for biomedical research proposed by the Council for International Organizations of Medical Sciences and ARRIVE (Animal Research: Reporting In Vivo Experiments) guideline.25,26
Cell cultures
The human glioma cells U87 were purchased from American Type Culture Collection and grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Italy) supplemented with 10% fetal bovine serum in a humidified incubator with 5% CO2 at 37°C and usually passaged every three days. Primary cortical neurons were prepared as previously reported22 and used after seven to nine days. In brief, neurons were treated with the antimitotic agent cytosine arabinoside (2.5 μM) on day 2 to reduce glial contamination. For LDH and luciferase experiments, neurons were seeded in 12-well plates at a density of 1 × 106 cells/well; for qRT-PCR, 5 × 106 cells were plated in 60 mm well dishes; for Western blot and chromatin immunoprecipitation (ChIP) analyses, cells were plated in 100 mm well plates at a density of 15 × 106 cells cells/well. For the luciferase assay in U87, the cells were seeded into a 12-well plate at a density of 1.5 × 105 cells/well.
The plates and the density of neurons plated for lactate dehydrogenase (LDH) assay, luciferase assay, Western blotting, qRT-PCR, chromatin immunoprecipitation (ChIP) and immunoprecipitation (IP) experiments were the same as previously published.14
Transfection with plasmids or small interfering RNA, and luciferase reporter assay in cortical neurons
Cortical neurons and U87 cells were transfected with 50 nM of siDREAM, siHDAC4, siHDAC5, siHDAC7, siHDAC9, or siNCX3. To overexpress HDAC4, HDAC5, HDAC7, and HDAC9, neurons were transfected with 15 μg of each construct in 100 mm plates. Each transfection was performed with lipofectamine LTX (15338-100, Invitrogen, Italy) in Opti-MEM, as recommended by the producer. After 2 h of transfection, the medium was replaced with a fresh one. Luciferase assay experiments were performed in 12-well plates. A 650-bp genomic fragment containing the human minimal ncx3 promoter region and exon 1 was amplified, by using primers as already published,11 and inserted in pGL3-basic vector, this new construct was named pGL3-ncx3. Cells were co-transfected with 1 μg of total DNA vectors, i.e., 200 ng of pRL-TK control vector expressing the Renilla luciferase gene, and 800 ng of one of the following reporter constructs: (1) pGL3-basic, (2) pGL3-ncx3, (3) pGL3-ncx3-DREmut. In the last construct, the DRE-site contained in the pGL3-ncx3 was mutated by a GT>CA substitution (GCGCGGCTTCACACACAGTG). Mutagenesis of the DRE-site on pGL3-ncx3 was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Italy). After 2 h of incubation, the medium was changed with a fresh one and the experiment was analyzed after 24 h with Dual-Luciferase Reporter Assay System kit (E1910 Promega, Italy). Transfection efficiency was ≅25% (data not shown).
Combined oxygen and glucose deprivation and reoxygenation
OGD was performed in cortical neurons incubated in a medium previously saturated with 95% N2 and 5% CO2 for 20 min and containing: NaCl 116 mM, KCl 5.4 mM, MgSO4 0.8 mM, NaHCO3 26.2 mM, NaH2PO4 1 mM, CaCl2 1.8 mM, glycine 0.01 mM, and 0.001% w/v phenol red. Afterwards, cells were placed in a hypoxic chamber for 3 h.14 To start, reoxygenation (Reoxy) neurons were removed from the hypoxic chamber and placed in a normal medium for 24 or 48 h. Transfections with siCTL, siDREAM, siHDAC4, siHDAC5 and siNCX3 were performed at 5 DIV; after 48 h, cells were subjected to OGD/Reoxy. MC1568 (5 μM) was added 1 h before and during the entire OGD period.
Quantitative real-time PCR analysis
First-strand cDNA and quantitative real-time PCR were carried out as previously described.13,15 In brief, quantitative real-time PCR was carried out in a 7500-fast real-time PCR system (Applied Biosystems, Monza, Italy) with Fast SYBR Green Master Mix (cod. 4385610; Applied Biosystems, Monza, Italy). Triplicate samples were amplified simultaneously in one assay as follows: heating 2 min at 50°C, denaturation 10 min at 95°C, amplification and quantification 40 cycles of 30 s at 60°C, with a single fluorescence measurement. PCR data were collected by using ABI Prism 7000 SDS software (Applied Biosystems). The data were normalized by using hypoxanthine phosphoribosyl transferase (HPRT) as internal control. The oligonucleotide sequences for NCX1, NCX3, and HPRT used were previously described.13,21
Western blotting
Cells and brain tissues were collected in ice-cold lysis buffer already reported13 and containing anti-protease cocktail (P8340 Sigma, Italy); 70 µg of total proteins were used to detect NCX1, HDAC4, HDAC5, HDAC7, HDAC9, and acetyl-histone H4 expression; 25 µg of total proteins were used to detect NCX3 and DREAM. NCX1, NCX3, HDAC4, HDAC5, HDAC7, and HDAC9 were separated on 8% SDS polyacrylamide gels, whereas DREAM and acetyl-histone H4 were separated on 12% and 15% SDS polyacrylamide gels, respectively. For cytosolic and nuclear fractions, tissues were lysed by homogenization in a buffer containing the following: 5 mM Hepes, 1 mM MgCl2, 2 mM EGTA, 1 mM DTT, and 300 mM sucrose, supplemented with the same protease inhibitor mixture. The obtained cell lysate was centrifuged at 3200 r/min (10 min at 4°C) to separate cytosol (supernatant) from the nuclear (pellet) fraction.27 The gels were transferred onto Hybond ECL nitrocellulose membranes (Amersham, Italy). Membranes were first blocked with 5% non-fat dry milk in 0.1% Tween 20 (Sigma, Italy) (2 mM Tris-HCl and 50 mM NaCl pH 7.5) for 2 h at room temperature. Then, they were incubated overnight at 4°C in the blocking buffer with the monoclonal antibodies against β-actin (A 4700; Sigma, Italy) and α-tubulin (T5168; Sigma, Italy) at the concentrations of 1:2000 and 1:5000, respectively, or with the polyclonal antibodies against NCX1 (R3F1; Swant, Bellinzona, Switzerland), NCX3 (95209; Swant, Bellinzona, Switzerland), HDAC4 (sc-11418; Santa Cruz Biotechnology, CA, USA), HDAC5 (sc-11419; Santa Cruz Biotechnology, CA, USA), HDAC7 (sc-11491; Santa Cruz Biotechnology, CA, USA), HDAC9 (sc-28732; Santa Cruz Biotechnology, CA, USA), DREAM (sc-9142; Santa Cruz Biotechnology, CA, USA), anti-histone H4 acetylated (06-866, Millipore) and anti-histone H3,22 all at the concentration of 1:1000. Finally, after incubation with primary antibodies, membranes were first washed with 0.1% Tween 20 and then incubated with secondary antibodies for 1 h at room temperature. The immunoreactive bands were detected with the ECL reagent (Amersham). The optical density of the bands, normalized to α-tubulin or β-actin, was determined by NIH's Image J software.
Chromatin immunoprecipitation
Brain tissues and cortical neurons were cross-linked with 1% formaldehyde, which was stopped by adding glycine at a final concentration of 0.125 M, both were washed three times in cold PBS containing proteinase inhibitors and then lysed in a buffer containing 50 mM Tris pH 8.1, 1% SDS, 10 mM EDTA, and anti-protease cocktail. DNA was sheared by sonication into 200–500 bp fragments by a Bandelin Sonopuls HD 2070 ultrasonic homogenizer (Bandelin, Berlin, Germany). Removal of cellular debris was achieved by centrifugation. The samples were then diluted with ChIP dilution buffer (16.7 mm Tris–HCl, pH 8.1, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl) to a final volume of 1 ml and incubated with protease inhibitors. Thirty microliters of the pre-immunoprecipitated lysate were saved as “input” for later normalization. Chromatin lysates were incubated overnight with 10 μl of antibody for HDAC4 (sc-11418; Santa Cruz Biotechnology, CA, USA), HDAC5 (sc-11419; Santa Cruz Biotechnology, CA, USA), DREAM (sc-9142; Santa Cruz Biotechnology, CA, USA), acetyl-Histone H4 (06-866, Millipore). Normal rabbit IgG was used as negative control. After immunoprecipitation, 40 μl of salmon sperm DNA/protein A-agarose beads (16-157, Millipore, Italy) was added to the lysates in order to precipitate the proteins bound to DNA. After rotating for 2 h at 4°C on a spinning wheel, the beads were washed sequentially with each of the following buffers: high-salt buffer (0.1% SDS, 1% Triton, 2mM EDTA, 20mM TrisHCl pH 8.1, 500mMNaCl), low-salt buffer (0.1% SDS, 1% Triton, 2mM EDTA, 20mM TrisHCl pH 8.1, 150mM NaCl), LiCl buffer (0.25M LiCl, 1% NP40, 1% deoxycholate, 1mM EDTA, 10mM TrisHCl pH 8.1), and twice with TE buffer (10mM Tris pH 8.1 and 1mM EDTA). The precipitates were eluted using immunoprecipitation elution buffer (1% SDS and 0.1M NaHCO3) and extracted via thermal cross-link reversion by adding NaCl (5 M) to the eluates. DNA fragments were first purified by phenol–chloroform extraction, ethanol precipitation and then dissolved in sterile water. DNA was analyzed by qRT-PCR using Fast SYBR Green Master Mix (cod 4385610; Applied Biosystems, Italy). The oligonucleotides used for the amplification of the immunoprecipitated DNA of rat NCX3 promoter were the following: forward 5′-CGTCACCCTATCAGCTGGGAG-3′ and reverse 5′-GGTCAGGACGAGTCGCCTCC-3′. In particular, DNA sequences/samples were heated for 2 min at 50°C and denatured for 10 min at 95°C. Amplification and quantification, achieved after 40 cycles at 95°C for 30 s, were followed by single fluorescence measurements for 1 min at 60°C. Binding activity was normalized for the total input of chromatin and graphically represented as percentage of control or sham. For each amplification, melting curves and gel electrophoresis of the PCR product were used to verify their identities. Samples were amplified simultaneously in duplicate in one assay run.
Immunoprecipitation
Cell lysates were performed by using immunoprecipitation lysis buffer (50 mMTris–HCL, pH 7.5, 1, Triton 1%, β-glicerol 10 mM, NaF 100 mM, Na3VO4 100 mM, 150 mMNaCl, and 1 mM EDTA), with the addition of protease inhibitor cocktail (1/100) P-8140 (Sigma). Aliquots of 1500 μg were immunoprecipitated overnight at 4°C using 3 μg of rabbit polyclonal anti-DREAM (sc-9142; Santa Cruz Biotechnology, CA, USA) or normal rabbit IgG. Next,20 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) was added to each sample in order to precipitate the proteins bound to the antibodies. After a brief centrifugation, the pellets were washed twice and resuspended in 20 μl of loading buffer, boiled for 10 min, and centrifuged. The supernatants were then subjected to Western blot analysis as previously described.28 Membranes were incubated overnight at 4°C with the following antibodies: anti-HDAC4 (sc-11418; Santa Cruz Biotechnology) and anti-HDAC5 (sc-11419; Santa Cruz Biotechnology).
Confocal double immunofluorescence
Confocal immunofluorescence procedures were performed as previously described.29,30 Briefly, the animals were euthanized 24 h after sham surgery or tMCAO. The rats were anesthetized intraperitoneally with chloral hydrate (300 mg/kg) and perfused transcardially with 4% w/v paraformaldehyde in phosphate buffer. The brains were sectioned coronally (50 µm), and, after blockade sections were incubated with the following primary antisera: rabbit polyclonal anti-DREAM (1:500, sc-9142; Santa Cruz Biotechnology, CA, USA), rabbit polyclonal anti-HDAC4 (1:100, sc-11418; Santa Cruz Biotechnology), rabbit polyclonal anti-HDAC5 (1.50, sc-11419; Santa Cruz Biotechnology), and mouse monoclonal anti-NeuN (1:2000, Millipore). Subsequently, sections were incubated in a mixture of the fluorescent-labeled secondary antibodies (Alexa 488/Alexa 594-conjugated antimouse/antirabbit IgG). Images were observed with a Zeiss LSM510 META/laser scanning confocal microscope. Single images were taken with an optical thickness of 0.7m and a resolution of 1024 × 1024.
Quantification of neuronal injury
Neuronal injury was assessed by measuring LDH efflux into the medium. After 24 h of transfection, with siNCX3 or with siCTL, neurons were treated with MC1568 an hour before and during the entire OGD protocol. Other neurons were subjected to OGD/Reoxy alone. Cytosolic levels of LDH in the extracellular medium were measured with the LDH cytotoxicity Kit (1000882 Cayman, DBA, Italy). After 48 h of reoxigenation, the medium was removed and sampled for LDH content by measuring absorbance at 490 nm using a spectrophotometer BioPhotometer (Eppendorf, Hamburg, Germany). Neurons treated with 1% Triton X-100 (Sigma-Aldrich) were used as a positive control and their value was considered 100%.
In vivo studies
Experimental groups
One hundred and five male Sprague-Dawley rats (Charles River) weighing 250–300 g and aged between six and eight weeks were housed under diurnal lighting conditions (12 h darkness/light). Experiments were performed according to the international guidelines for animal research. Animals not showing a reduction in cerebral blood flow of at least 70% and those dying after ischemia induction, that represent 20% of total rats used, were excluded from the study. Sample size for stroke experiments measurements was calculated a priori (n = 5/group) by predicting a conventional effect size of 0.9 and an alpha error of 0.1 and a statistical power of 95%.
Transient focal ischemia
Transient focal ischemia was induced under a steromicroscope by suture occlusion of the middle cerebral artery (MCA) in male rats anesthetized with 1.5% sevoflurane, 70% N2O, and 28.5% O2.14 Achievement of ischemia was confirmed by monitoring regional cerebral blood flow through laser Doppler (PF5001; Perimed). Animals not showing a reduction in cerebral blood flow of at least 70% (four rats) and those dying after ischemia induction (two rats) were excluded from the study. Rats were divided into two experimental groups: (1) sham-operated (Sham) and (2) ischemic rats subjected to transient MCA occlusion (tMCAO). The sham-operated animals underwent the same experimental conditions, as those in the ischemic group, except that the filament was not introduced into the internal carotid artery. In the ischemic group, the MCA was occluded for 100 min. siRNAs or MC1568 were intracerebroventricularly (icv) administered in rats three times, viz, at 18 and 6 h before ischemia and 6 h after ischemia as follows: 5μl of siRNA for HDAC4 (siHDAC4), HDAC5 (siHDAC5), and DREAM (siDREAM) at the concentration of 10 μM; 5μl siRNA for NCX3 (siNCX3) at the concentration of 5 μM, whereas MC1568 at the dose of 8mg/5µl. The effectiveness of siRNAs was evaluated in non-ischemic rats at the same concentration. In rats positioned on a stereotaxic frame, a 23-g stainless steel guide cannula (Small Parts) was implanted into the right lateral ventricle using the stereotaxic coordinates of 0.4 mm caudal to bregma, 2 mm lateral and 2 mm below the dura. The cannula was fixed to the skull using dental acrylic glue and small screws.31,32 MC1568 was used in an appropriate intracerebral dose to give the concentration of 5 micromolar, which resulted neuroprotective in our in vitro experiments. Rectal temperature was maintained at 37 ± 0.5°C with a thermostatically controlled heating pad, and a catheter was inserted into the femoral artery to measure arterial blood gases before and after ischemia (Rapid Laboratory 860, Chiron Diagnostic). All animals were euthanized 24 or 48 h after tMCAO. Protein quantification was carried out on the temporoparietal cortex, which corresponds to the perinfarct area, harvested from sham-operated and ischemic animals after 24 or 48 h of reperfusion.
Evaluation of infarct volume
Rats were decapitated 24 h after ischemia. Ischemic volume was evaluated by 2,3,5-triphenyltetrazolium chloride staining. In brief, the brains were cut into 500 μm coronal slices with a vibratome (Campden Instrument, 752 M). Sections were incubated in 2% 2,3,5-triphenyltetrazolium chloride for 20 min and in 10% formalin overnight. The infarct area was calculated by image analysis software (Image-Pro Plus). The total infarct volume was expressed as a percentage of the volume of the hemisphere ipsilateral to the lesion.33
Neurological deficit scores
Neurological scores were assessed 24 h after reperfusion agreeing to two scales: a general neurological scale and a focal neurological scale, as previously reported.25 All surgical procedures have been performed in a blinded manner. In the general score, these six general deficits were measured: (1) hair conditions (0–2), (2) position of ears (0–2), (3) eye conditions (0–4), (4) posture (0–4), (5) spontaneous activity (0–4), and (6) epileptic behavior (0–12). In the focal score, these seven areas were assessed: (1) body symmetry, (2) gait, (3) climbing, (4) circling behavior, (5) front limb symmetry, (6) compulsory circling, and (7) whisker response. For each of these items, animals were rated between 0 and 4 depending on severity. The values were summed to give a general and focal score.
Statistical analysis
Data were analyzed by GraphPad Prism 5 software (Graph Pad Software, Inc.). All bars in the figures represent the mean ± S.D. Statistical differences between two experimental groups were analyzed with the Student t-test. Statistically significant differences between more than two experimental groups were evaluated by one-way ANOVA, followed by Tukey’s multiple comparison test, whereas a two-way ANOVA, followed by Newman–Keuls test was used to analyze the significance of the infarct volume measurements. Neurological deficits data were analyzed using the non-parametric Kruskal–Wallis test, followed by Dunn’s multiple comparison test. Statistical significance was accepted at the 95% confidence level (p < 0.05).
Results
The HDAC class IIa inhibitor MC1568 increased ncx3 promoter activity and ncx3 gene products in primary cortical neurons
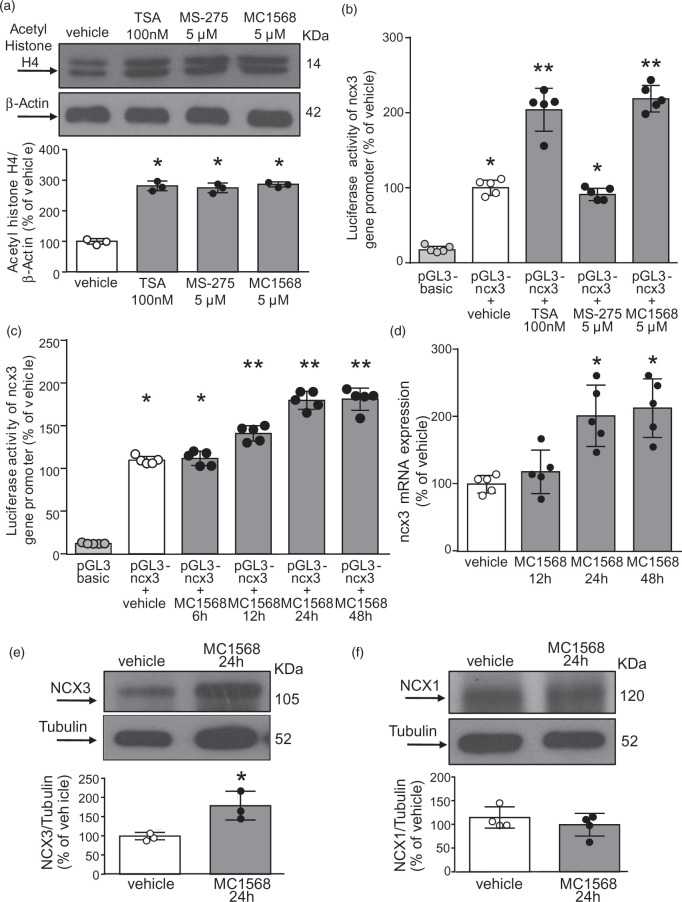
The pan-HDACs inhibitor TSA (100 nM),34 the class I HDAC inhibitor MS-275 (5 μM),14,35 and the class IIa HDACs inhibitor MC1568 (5 μM)36 substantially increased acetylaton of histone H4 in cortical neurons (Figure 1(a)). In particular, the selective class IIa HDAC inhibitor MC1568 and the pan-inhibitor TSA increased ncx3 gene promoter activity in cortical neurons transfected with a pGL3 construct containing human ncx3 minimal promoter region (pGL3-ncx3),10 whereas the selective class I HDAC inhibitor MS-275 was ineffective (Figure 1(b)). It is noteworthy that the stimulatory action triggered by MC1568 occurred at 12, 24 and 48 h on the ncx3 gene promoter and at 24 and 48 h on ncx3 mRNA, as shown in Figure 1(c) and (d). Furthermore, Western blotting analysis revealed that 24-h exposure to MC1568 increased NCX3 by up to 79% compared to control (Figure 1(e)). By contrast, class IIa HDACs inhibition did not affect NCX1 expression (Figure 1(f)). These results indicate that NCX3 is specifically modulated by HDACs belonging to the class IIA.
Figure 1.
Effect of class IIa HDAC inhibitor MC1568 on ncx3 promoter activity, gene, and protein expression in cortical neurons.
(a) Effect of 24 h of TSA (100 nM), MS-275 (5 µM), and MC1568 (5 µM) exposure on Acetylated-Histone H4 protein expression. Cells were treated with TSA, MS-275, and MC1568 for a 2 h pulse (n = 3). *p ≤ 0.05 versus vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (b) Effect of TSA (100 nM), MS-275 (5 µM), and MC1568 (5 µM) exposure on ncx3 promoter activity (n = 5). *p ≤ 0.05 versus pGL3-basic; **p ≤ 0.05 versus pGL3-basic and pGL3-ncx3 + vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (c) Effect of MC1568 (5 µM) on ncx3 promoter activity measured after 6, 12, 24 and 48 h treatment (n = 5). *p ≤ 0.05 versus pGL3-basic; **p ≤ 0.05 versus pGL3-basic and pGL3-ncx3 + vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (d) Effect of 12, 24, and 48 h of MC1568 (5μM) treatment on ncx3 gene expression (n = 5). *p ≤ 0.05 versus vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (e, f) Representative WB with quantification of NCX3 (e) and NCX1 (f) after 24 h treatment with MC1568 (5 µM) in cortical neurons (n = 3). *p < 0.05 versus vehicle, Student t test.
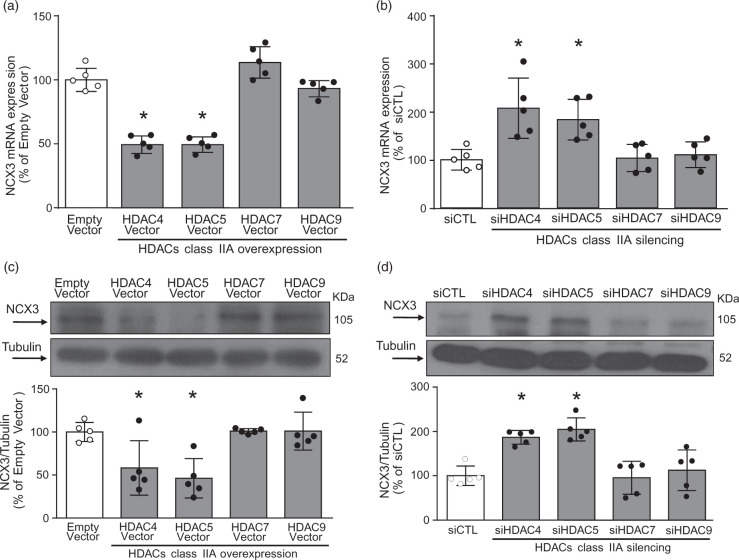
HDAC4 and HDAC5 overexpression reduced NCX3 mRNA and protein expression in primary cortical neurons, whereas siRNAs against HDAC4 and HDAC5 increased both ncx3 gene products
Cortical neurons transfected with cDNA for HDAC4, HDAC5, HDAC7, and HDAC9 substantially increased HDAC4, HDAC5, HDAC7 and HDAC9 protein expression by 83%, 64%, 40%, and 45%, respectively (Suppl. Figure 1(a) to (d)). Remarkably, only neurons overexpressing class IIa isoforms HDAC4 and HDAC5, but not HDAC7 and HDAC9, displayed a significant reduction in NCX3 mRNA (Figure 2(a)) and protein expression (Figure 2(b)). Transfection of specific siRNAs for HDAC4, HDAC5, HDAC7 and HDAC9 reduced HDAC4, HDAC5, HDAC7, and HDAC9 protein expression by 45%, 57%, 44%, and 58%, respectively (Suppl. Figure 1(e) to (h)). Intriguingly, siRNAs for HDAC4 and HDAC5, but not for HDAC7 and HDAC9, significantly increased NCX3 mRNA (Figure 2(c)) and protein expression (Figure 2(d)). Thus, only HDAC4 and HDAC5 regulate NCX3 gene transcription.
Figure 2.
HDAC4 and HDAC5 down-regulated ncx3 gene and protein expression. (a, b) ncx3 gene expression and representative WB with quantification in cortical neurons transfected with the following constructs: (1) Empty Vector, (2) HDAC4 Vector, (3) HDAC5 Vector, (4) HDAC7 Vector, and (5) HDAC9 Vector. (c, d) ncx3 gene expression and representative WB with quantification in cortical neurons transfected with the following siRNAs: (1) siCTL, (2) siHDAC4, (3) siHDAC5, (4) siHDAC7, and (5) siHDAC9. The cells were processed after 24 h of transfection for qRT-PCR and immunoblotting (n = 5). *p ≤ 0.05 versus empty vector or siCTL, one-way ANOVA, followed by Tukey’s multiple comparison test.
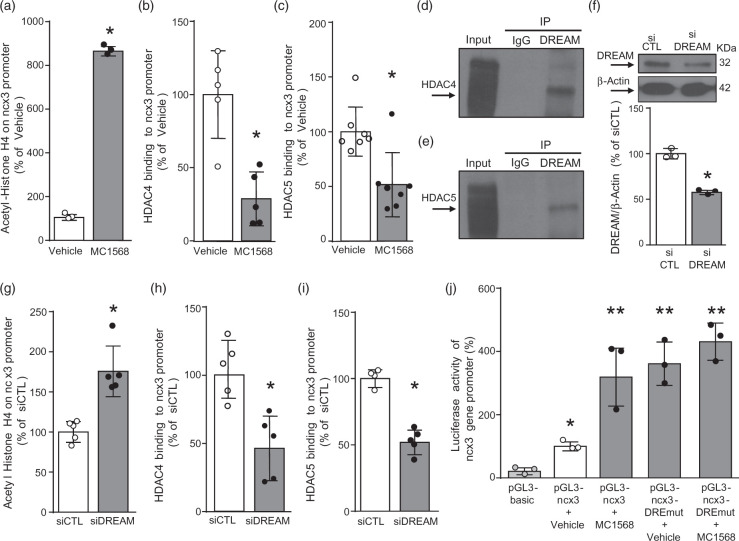
Silencing of downstream regulatory element antagonist modulator or the inhibition of HDAC4 and HDAC5 prevented HDAC4 and HDAC5 recruitment to the ncx3 promoter sequence
To investigate the hypothesis that HDAC4 and HDAC5 directly modulate ncx3 brain promoter, we performed ChIP assays on extracts from cortical neurons exposed to the class IIa HDAC inhibitor MC1568 (5 µM) for 24 h. As shown in Figure 3(a), when chromatin was precipitated with acetyl-histone H4 antibody, ncx3 brain promoter sequence resulted in hyper-acetylated neurons exposed to MC1568, compared to vehicle (Figure 3(a)). Importantly, HDAC4 (Figure 3(b)) and HDAC5 (Figure 3(c)) recruitment to the ncx3 promoter sequence was significantly reduced after MC1568 exposure. Immunoprecipitation experiments demonstrated that DREAM physically interacted both with HDAC4 (Figure 3(d)) and HDAC5 (Figure 3(e)), in primary cortical neurons. To further clarify the role of DREAM in modulating ncx3 gene transcription, we knocked-down DREAM with a specific siRNA (siDREAM). Silencing of DREAM reduced its protein expression by 41% (Figure 3(f)). Interestingly, ChIP analysis revealed that whereas siDREAM significantly increased the acetylation status of the ncx3 promoter sequence (Figure 3(g)), it significantly decreased HDAC4 (Figure 3(h)) and HDAC5 (Figure 3(i)) recruitment to the ncx3 promoter sequence. Notably, luciferase assay experiments, performed in cortical neurons transfected with a ncx3 promoter vector containing the DREAM binding mutated sequence (pGL3-ncx3-DREmut), showed that ncx3 promoter activity also increased in neurons exposed to the specific HDAC class IIa inhibitor MC1568 (Figure 3(j)). On the other hand, the treatment with MC1568 (5 µM), or singly transfection with siDREAM, siHDAC4 and siHDAC5, that increased ncx3 promoter activity in neurons, did not modify the transcriptional activity in U87 glial cells (Suppl. Figure 2), thus suggesting that this mechanism occurred only in neurons. Collectively, these data demonstrate that HDAC4 and HDAC5 bind DREAM on ncx3 gene promoter sequence in neurons.
Figure 3.
HDAC4 and HDAC5 bound to ncx3 brain promoter via downstream regulatory element antagonist modulator (DREAM).
(a–c) ChIP analysis of the ncx3 brain promoter performed with: (a) anti acetylated-histone H4, (b) anti-HDAC4, and (c) anti-HDAC5 in cortical neurons. Acetylation status of histone H4 (n = 3) and binding activity of HDAC4 (n = 5) and HDAC5 (n = 7) are graphically represented as the percentage of vehicle. *p ≤ 0.05 versus vehicle, Student t test. (d, e) Representative Western blot showing immunoprecipitation between DREAM and HDAC4 and between DREAM and HDAC5 in cortical neurons. IgG was used as a negative control. (f) Representative WB with quantification in cortical neurons after 24 h of transfection with siDREAM (n = 3). *p < 0.05 versus siCTL, Student t test. (g–i) ChIP analysis of the ncx3 brain promoter performed with: (g) anti-acetylated-histone H4, (h) anti-HDAC4, and (j) anti-HDAC5 in cortical neurons. Acetylation status of histone H4 (n = 5) and binding activity of HDAC4 (n = 5) and HDAC5 (n = 4/5) are graphically represented as the percentage of siCTL. *p ≤0.05 versus siCTL, Student t test. (j) Luciferase activity of ncx3 gene promoter wild type (pGL3-ncx3) or mutated in DRE-site (pGL3-ncx3-DREmut), with or without MC1568 (5 μM), in cortical neurons (n = 3). *p ≤ 0.05 versus pGL3basic; **p ≤ 0.05 versus pGL3basic and pGL3-ncx3 + vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test.
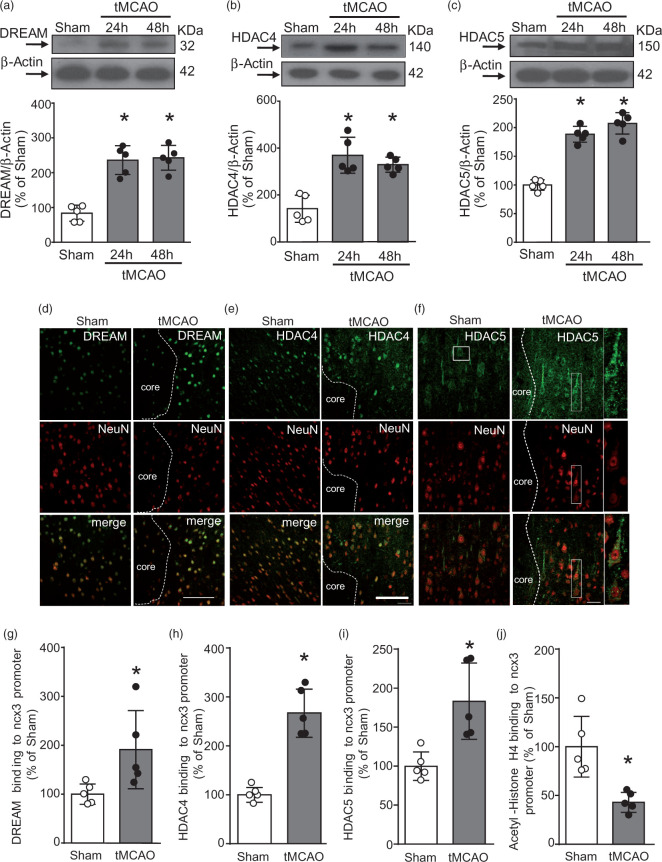
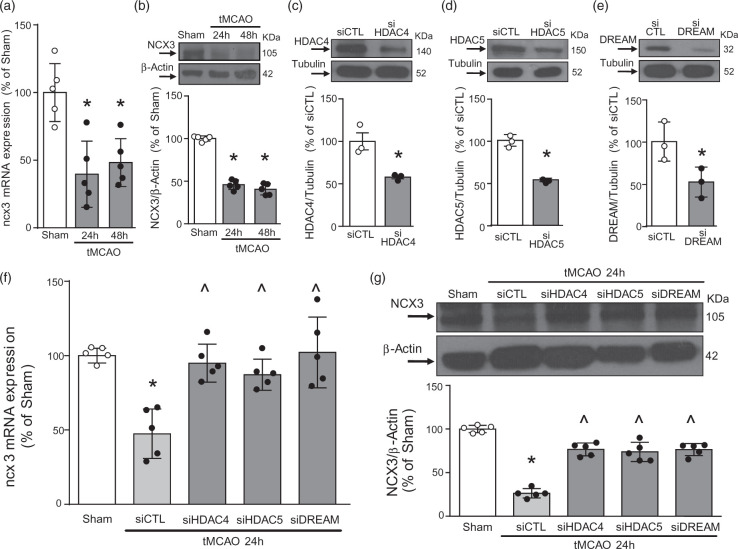
DREAM, HDAC4, and HDAC5 recruitment to the ncx3 promoter increased in periinfarct temporoparietal cortex after tMCAO
To investigate whether the transcription factor DREAM and the epigenetic erasers HDAC4 and HDAC5 were modified after stroke, DREAM, HDAC4, and HDAC5 protein expression was measured in the temporoparietal brain cortices of rats subjected to tMCAO. As shown in Figure 4(a) to (c), DREAM, HDAC4, and HDAC5 proteins significantly increased 24 and 48 h after stroke induction, as compared to sham-operated animals. Confocal double immunofluorescence experiments performed in the periischemic temporoparietal cortex region, where ncx3 mRNA and protein expression are reduced,20 revealed that DREAM (Figure 4(d)), HDAC4 (Figure 4(e)) and HDAC5 (Figure 4(f)) immunosignals were more intensely detected in the NeuN-positive neuronal nuclei, if compared to the corresponding ipsilateral cortical regions of sham-operated animals. Particularly, as shown in Supplemental Figure 3(a) and (b), after tMCAO HDAC4 had a predominant nuclear localization, whereas the increase of HDAC5 occurred both in the cytosolic and nuclear compartments. Notably, tMCAO treatment significantly increased DREAM (Figure 4(g)), HDAC4 (Figure 4(h)), and HDAC5 (Figure 4(i)) recruitment and reduced histone H4 acetylation on the ncx3 promoter sequence in the perinfarct area (Figure 4(j)). Therefore, tMCAO induced an increase of DREAM, HDAC4 and HDAC5 binding on ncx3 gene promoter sequence.
Figure 4.
DREAM, HDAC4, and HDAC5 bound to ncx3 brain promoter, determining its sequence deacetylation in the periischemic temporoparietal cortex of tMCAO rats. (a–c) Representative WB with quantification of DREAM, HDAC4, and HDAC5 in cortex of rats subjected to 24 h and 48 h of tMCAO (n = 5). *p < 0.05 versus Sham, one-way ANOVA, followed by Tukey’s multiple comparison test. Confocal double labeling experiments displaying the co-expression of DREAM (d), HDAC4 (e), and HDAC5 (f) with the neuronal marker NeuN in the ipsilateral temporoparietal cortex of sham-operated animals (left panels) and in the ipsilateral periischemic temporoparietal cortex 24 h after tMCAO surgery (right panels). Dashed lines delimit the cortical ischemic core and periischemic region. Scale bars 100 µm. ChIP analysis of the ncx3 brain promoter performed with: (g) anti-DREAM, (h) anti-HDAC4, (i) anti-HDAC5, and (j) anti-Acetylated-Histone H4 in cortex of rats subjected to 24 h of tMCAO. Binding activity of DREAM, HDAC4, HDAC5 and acetylation status of Histone H4 are graphically represented as the percentage of sham-operated animals (n = 5). *p ≤ 0.05 versus Sham, Student t test .
Silencing of HDAC4, HDAC5, and DREAM prevented ncx3 mRNA and protein reduction induced by tMCAO in the periinfarct temporoparietal cortex
To better clarify the role of the DREAM/HDAC4/HDAC5 complex in tMCAO-induced reduction in ncx3 mRNA (Figure 5(a)) and protein expression (Figure 5(b)), DREAM, HDAC4 and HDAC5 were knocked-down by siRNAs administration. As shown in Figure 5(c) to (e), icv injection of siDREAM, siHDAC4 and siHDAC5 in non-ischemic rats reduced their targets protein expression by 41%, 46%, and 57%, respectively. More importantly, these above-mentioned siRNAs even prevented ncx3 mRNA (Figure 5(f)) and protein (Figure 5(g)) reduction after stroke. Together, these findings indicate that single silencing of the each component of the DREAM/HDAC4/HDAC5 complex counteracts stroke-induced ncx3 reduction.
Figure 5.
HDAC4, HDAC5, and DREAM knockdown prevented ncx3 gene and protein expression reduction in the periischemic temporoparietal cortex oftMCAO rats. (a–b) Effect of 24 h and 48 h of tMCAO on: (a) ncx3 gene (n = 5) and (b) protein expression (n = 5). *p ≤ 0.05 versus Sham, one-way ANOVA, followed by Tukey’s multiple comparison test. (c–e) Representative WB with quantification in rats subjected to siRNAs intracerebroventricular (icv) injection against HDAC4, HDAC5 or DREAM and euthanized after 24 h (n = 3). *p < 0.05 versus siCTL, Student t test. Effect of 24 h of siHDAC4, siHDAC5, and siDREAM icv administration in rats subjected to 24 h of tMCAO on ncx3 (f) gene (n = 5) and (g) protein expression (n = 5). *p ≤ 0.05 versus Sham and ^p ≤ 0.05 versus Sham and tMCAO + siCTL, one-way ANOVA, followed by Tukey’s multiple comparison test.
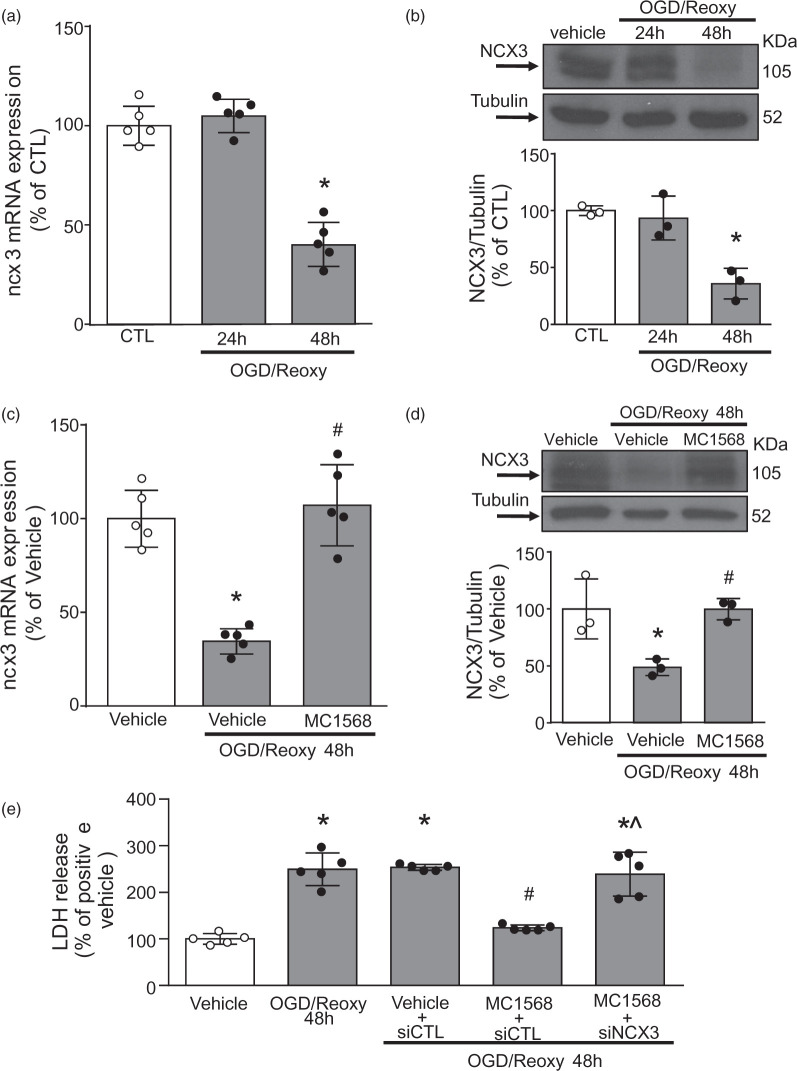
By preventing NCX3 reduction, siDREAM, siHDAC4, siHDAC5 and MC1568 attenuated OGD-induced damage of cortical neurons
To further confirm our hypothesis that HDAC4 and HDAC5 inhibition prevents ischemia-induced reduction in NCX3, we evaluated the effect of the class IIa HDAC inhibitor MC1568 (5 µM) in primary cortical neurons exposed to 3 h of oxygen and glucose deprivation (OGD) and followed by 24 and 48 h of reoxygenation (Reoxy). As shown in Figure 6(a) and (b), ncx3 mRNA and protein expression significantly decreased after 48 h of Reoxy. Notably, MC1568 completely prevented OGD/Reoxy-induced reduction of NCX3 mRNA (Figure 6(c)) and protein expression (Figure 6(d)). More important, MC1568 (5 µM) promoted a significant neuroprotective effect by preventing cell death after OGD/Reoxy, as revealed by the reduction in LDH release. Furthermore, to validate the role of NCX3 in MC1568-induced neuroprotection after OGD/Reoxy, we knocked-down ncx3 by siRNA transfection. Indeed, the MC1568-induced cell death reduction in neurons subjected to OGD/Reoxy was reverted by siNCX3 (Figure 6(e)). Additionally, protein expression of DREAM, HDAC4 and HDAC5 significantly increased after 24 and 48 h of OGD/Reoxy (Suppl. Figure 4(a) to (c)). Furthermore, the single silencing of DREAM, HDAC4 and HDAC5 prevented OGD/Reoxy-induced ncx3 mRNA reduction (Suppl. Figure 4(d)) and neuronal cell death (Suppl. Figure 4(f)). However, the double silencing of HDAC4 and HDAC5, although showed a positive effect on ncx3 expression (Suppl. Figure 4(e)) and a protective effect on neuronal cell death (Suppl. Figure 4(f)), did not show an additive effect, compared to the single siRNAs (Suppl. Figure 4(e) and (f)). These results demonstrate that MC1568 affords neuroprotection via ncx3.
Figure 6.
MC1568 reduced neuronal death in cortical neurons exposed to OGD/Reoxy.
(a, b) Effect of 24 h and 48 h of reoxygenation (Reoxy) after 3 h of OGD on: (a) ncx3 gene (n = 5) and (b) protein expression (n = 3) in cortical neurons. *p ≤ 0.05 vs CTL, one-way ANOVA, followed by Tukey’s multiple comparison test. (c, d) Effect of MC1568 (5μM) in cortical neurons subjected to OGD followed by 48 h of reoxygenation on: (c) ncx3 gene (n = 5) and (d) protein expression (n = 3). *p ≤ 0.05 versus Vehicle; #p ≤ 0.05 versus OGD/Reoxy 48 h + Vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (e) Effects of MC1568 (5μM) exposure or siNCX3 transfection on cell death of cortical neurons subjected to OGD/Reoxy for 48 h, as evaluated by LDH assay (n = 5). *p ≤ 0.05 versus vehicle; #p ≤ 0.05 versus OGD/Reoxy 48h; ^p ≤ 0.05 versus MC1568 + siCTL + OGD/Reoxy 48h, one-way ANOVA, followed by Tukey’s multiple comparison test.
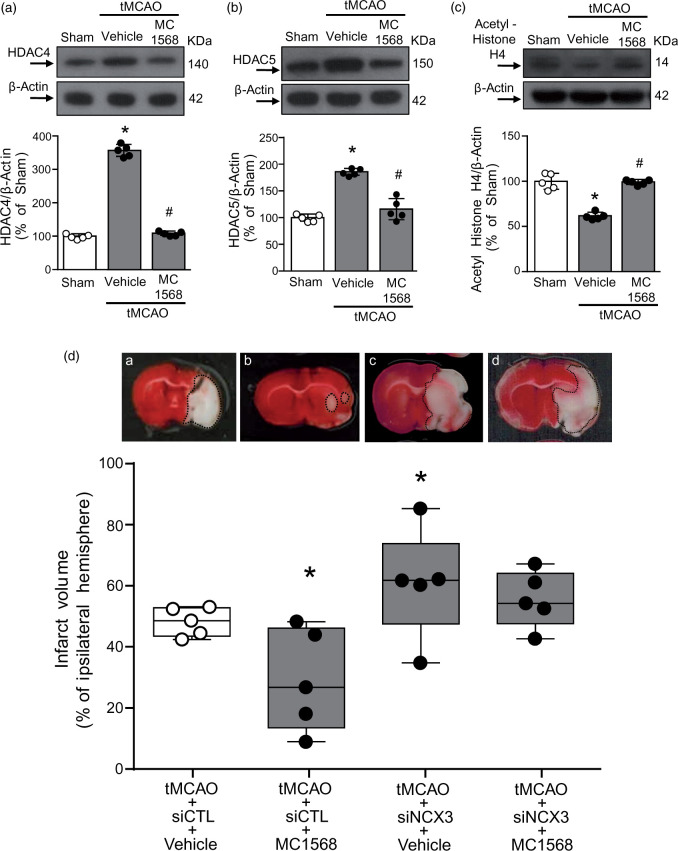
MC1568, by reducing tMCAO-induced HDAC4 and HDAC5 up-regulation, ameliorated ischemic damage via NCX3
The neuroprotective role played by MC1568 through NCX3 was also validated in vivo by using tMCAO. As shown in Figure 7(a) to (c), icv administration of MC1568 significantly prevented tMCAO-induced increase of HDAC4 and HDAC5 and reduction in the acetylated form of histone H4. Notably, MC1568-induced HDAC4 and HDAC5 protein reduction could be determined according to their sumoylation and consequent protein degradation, as reported by other studies.22,37 Significantly, MC1568 reduced the infarct volume in rats subjected to brain ischemia by preventing NCX3 down-regulation. Indeed, MC1568-induced neuroprotection was almost completely abolished by siNCX3 injection (Figure 7(d)). The efficiency of siNCX3 was already published.20 Importantly, the reduction in the ischemic volume induced by MC1568 was associated with an improvement in general (Suppl. Figure 5(a)) and focal scores (Suppl. Figure 5(b)). In parallel, it has been evaluated the effect of siDREAM, siHDAC4 and siHDAC5 icv administered on the infarct volume. As shown in Suppl. Figure 6, only siHADC5 significantly reduced the infarct size, whereas siDREAM and siHDAC4 did not show any significant effect on ischemic damage.
Figure 7.
MC1568 ameliorated ischemic damage in rats subjected to tMCAO. (a–c) Representative WB with quantification of HDAC4 (n = 5), HDAC5 (n = 5) and Acetylated-Histone H4 protein (n = 5) expression after 24 h of tMCAO and effect of icv administered MC1568. *p ≤ 0.05 versus Sham; #p ≤ 0.05 versus tMCAO + vehicle, one-way ANOVA, followed by Tukey’s multiple comparison test. (d) % of brain infarct volume in male rats subjected to tMCAO after icv administration of: (a) siCTL + Vehicle, (b) siCTL + MC1568, (c) siNCX3 + Vehicle, and (d) siNCX3 + MC1568. Representative brain slices from each experimental group are shown at the top of each column (n = 5). Ischemic rats were euthanized 24 h after tMCAO of the % of the infarct volume compared to the ipsilateral hemisphere. *p ≤0.05 vs tMCAO + siCTL + Vehicle, two-way ANOVA, followed by Newman–Keuls test.
Discussion
This work demonstrates for the first time that the transcriptional repressor DREAM, by recruiting HDAC4 and HDAC5 to the brain ncx3 promoter sequence, forms a complex that epigenetically down-regulates ncx3. Equally important, we also found that the formation of the DREAM/HDAC4/HDAC5 complex after stroke reduced the levels of ncx3 mRNA and protein expression in the periinfarct temporoparietal cortex. Interestingly, HDAC class IIA pharmacological inhibition prevented the decrease in ncx3 gene products, thereby ameliorating brain damage.
Our findings expand previous studies demonstrating that among the class IIa HDAC isoforms, HDAC4 and HDAC5 are expressed primarily in neurons.19 Indeed, we found that they both reduce ncx3 mRNA and protein levels by binding to the transcriptional repressor DREAM. Moreover, our immunoprecipitation assays revealed that the binding of these two deacetylases to the transcriptional repressor DREAM caused a reduction in NCX3 mRNA and protein levels. Instead, siDREAM hampered the recruitment of HDAC4 and HDAC5 to the ncx3 promoter sequence. This event, by disrupting DREAM/HDAC4/HDAC5 complex formation, led the hyperacetylation of histone H4, which in turn, determined NCX3 up-regulation. Furthermore, evidence that the site-directed mutation of DREAM consensus binding site (DRE) on the ncx3 promoter reduced HDAC4 and HDAC5 recruitment to this sequence demonstrates that the down-regulation of NCX3 by the two class IIA histone deacetylases occurs only via the formation of a complex with DREAM. This evidence was further confirmed by the fact that the increase in the ncx3 promoter activity exerted by the HDAC class IIa inhibitor MC1568 is abolished when DRE site is mutated. In addition, the epigenetic regulation of ncx3 gene seemed to occur exclusively in neurons, but not in glial cells, since luciferase assay experiment in U87 cells demonstrated that DREAM, HDAC4, HDAC5 and MC1568 did not modify ncx3 promoter activity. The mechanism by which DREAM represses its target gene has also been substantiated in previous studies showing that DREAM reduced the expression of the thyroglobulin gene, by binding to the thyroglobulin promoter and by blocking the thyroid transcription factor-1 (TTF-1)-mediated trans-activation.38 Interestingly, our experiments support the theory that the mechanism by which the transcription factor DREAM represses its target gene ncx3 involves histone deacetylation via HDACs. Notably, the notion that there is a tight interplay between the DREAM/HDAC4/HDAC5 complex and NCX3 expression was further substantiated by our experiments performed in in vivo model of brain ischemia. Indeed, our confocal microscopy images demonstrated that stroke induced a neuronal increase in DREAM, HDAC4, and HDAC5 specifically in the neurons of the periinfarct temporoparietal cortex, where NCX3 is reduced.21 It is noteworthy that experiments of cell fractionation (Suppl. Figure 3) demonstrated that after stroke, HDAC4 translocated mainly in the nucleus, suggesting that its effect predominantly occurred by modulating gene expression through the deacetylation of the histone proteins, whereas HDAC5 increased both at nuclear and cytoplasmic level, suggesting that it acts either at transcriptional level and through deacetylation of non-histone proteins. On the contrary, another research indicates that HDAC4 and HDAC5 were down-regulated by brain ischemia induced by tMCAO in adult rats.19 However, it should be underlined that differently from our study, in the work by He et al. the expression of HDAC4 and HDAC5 was evaluated not only in the penumbral vital tissue, but also in the ischemic core and the occlusion of the MCA was maintained longer,19 thus generating a bigger infarct and a more extended ischemic core. Importantly, we evidenced that stroke-induced NCX3 reduction was completely counteracted by icv injection of siRNAs against DREAM, HDAC4, and HDAC5. It should be noted that double silencing for HDAC4 and HDAC5 did not exert an additive effect on the increase of ncx3 gene expression and prevention of neuronal death induced by OGD/Reoxy, as compared to single HDACs silencing, thus suggesting that the simultaneously presence of HDAC4 and HDAC5 on ncx3 promoter is a required condition for the activation of the DREAM/HDAC4/HDAC5 complex. Other studies have shown that each of the components of the DREAM/HDAC4/HDAC5 complex contributes to neuronal cell death. One study, for instance, shows that DREAM, in cooperation with pro-apoptotic proteins, promotes cell death of specified sensory organ precursors in the peripheral nervous system.39 Other studies report that HDAC4, by increasing its protein expression in the nucleus: (1) determines neuronal cell death in in vivo and in vitro models of stroke40; (2) decreases both the length and complexity of dendrites of hippocampal neurons.41 Our in vitro results are in accordance with all previous mentioned works, since when DREAM, HDAC4, and HDAC5 are singly silenced in cortical neurons the neurodetrimental effect of OGD/Reoxy is significantly counteracted. On the contrary, in in vivo stroke experiments, only the knocking-down of HDAC5, but not of HDAC4 and DREAM, was able to significantly reduce the infarct volume size of ischemic rats. A possible explanation is that in the brain are present other cell populations beyond neurons, such as microglia, oligodendrocytes and astrocytes, that react in a different manner to the down-regulation of the different HDACs induced by siRNA. Furthermore, it is well established that in vitro models differ from in vivo stroke models in some features, such as: (1) the absence of blood vessels and blood circulation, that are relevant structural and functional components of the injury process; (2) a relatively longer resistance to anoxic or hypoxic stimulus; (3) a less severe ATP depletion; (4) a delayed glutamate’s release compared to stroke.42
A relevant result of this study is that HDAC class IIA pharmacological inhibition through MC1568, by blocking the epigenetic regulation of the ncx3 gene, counteracted its stroke-induced down-regulation, thus ameliorating ischemic neuronal damage in our in vitro and in vivo models of stroke. Meaningfully, this effect is accompanied by a notable amelioration in general and focal neurological scores of rats subjected to stroke. These obtained results are consistent with other studies demonstrating the neuroprotective action of MC1568 that: (1) protects neurite arbours against neurotoxic insult,43 (2) reduces diethylmercury-induced apoptotic cell death in rat prefrontal cortex,22 and (3) prompts a transient improvement of motor function in amyotrophic lateral sclerosis (ALS) mice.44 Importantly, it has been found that MC1568 administered via oral gavage 2 days after stroke induction, negatively affects neuronal remodeling and functional outcome 28 days after the initial insult.45 This difference could be due to several aspects: (1) the different administration route, oral versus intracerebroventricular; and (2) the different model of stroke, permanent versus transient middle cerebral artery occlusion.
Collectively, our findings have revealed one of the epigenetic mechanism underlying stroke-induced neuronal damage, and that the transcriptional repressor DREAM, by recruiting HDAC4 and HDAC5, down-regulated NCX3 in the brain, a major neuroprotective player in stroke1,2,4,5,20,21,46,47 Furthermore, we demonstrated that MC1568-induced neuroprotection occurred by preventing NCX3 reduction. Since no agent able to activate NCX3 is available at the moment, the modulation of HDAC class IIa may represent an innovative therapeutic strategy in stroke.
Supplemental Material
Supplemental material, JCB884742 Supplemetal Material for HDAC4 and HDAC5 form a complex with DREAM that epigenetically down-regulates NCX3 gene and its pharmacological inhibition reduces neuronal stroke damage by Luigi Formisano, Giusy Laudati, Natascia Guida, Luigi Mascolo, Angelo Serani, Ornella Cuomo, Maria Cantile, Francesca Boscia, Pasquale Molinaro, Serenella Anzilotti, Vincenzo Pizzorusso, Gianfranco Di Renzo, Giuseppe Pignataro and Lucio Annunziato in Journal of Cerebral Blood Flow & Metabolism
Acknowledgments
We thank Dr Paola Merolla for editorial revision and all the members of the Annunziato laboratory for their constructive comments on the data and the manuscript.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the following grants: Programma Operativo Nazionale (PON_01602, PON03PE_00146_1, PON PERMEDNET ArSol-1226) from Italian Ministry of Research, MIUR to L.A.; PRIN 2015, # 2015783N45 - CUP: E62F1600136001 to Research, MIUR to G.P., from Italian Ministry of Research, MIUR; Nanomirnictus, F/050139/01,02/X32 to L.A. from Italian Ministry of Economic Development, MISE; SIR 2014 Prot. N° RBSI14QECG to A.V., from Italian Ministry of Research, PRIN 2015 (2015 BEX2BR) by MIUR to L.F. and P.M., from Italian Ministry of Research, MIUR.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
Conceived and designed the experiments: LF, NG, GDR and LA. Performed the experiments: GL, NG, LM, AS, OC, FB, MC, PM, SA and VP. Analyzed the data: LF, GL, NG, AS, OC and FB. Contributed reagents/materials/analysis tools: LF, GDR, GP and LA. Wrote the paper: LF, NG, GDR, GP and LA.
Supplementary material
Supplemental material for this article is available online.
References
- 1.Annunziato L, Pignataro G, Di Renzo GF.Pharmacology of brain Na+/Ca2+ exchanger: from molecular biology to therapeutic perspectives. Pharmacol Rev 2004; 56: 633–654. [DOI] [PubMed] [Google Scholar]
- 2.Anzilotti S, Brancaccio P, Simeone G, et al. Preconditioning, induced by sub-toxic dose of the neurotoxin L-BMAA, delays ALS progression in mice and prevents Na(+)/Ca(2+) exchanger 3 downregulation. Cell Death Dis 2018; 9: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casamassa A, La Rocca C, Sokolow S, et al. Ncx3 gene ablation impairs oligodendrocyte precursor response and increases susceptibility to experimental autoimmune encephalomyelitis. Glia 2016; 64: 1124–1137. [DOI] [PubMed] [Google Scholar]
- 4.Formisano L, Saggese M, Secondo A, et al. The two isoforms of the Na+/Ca2+ exchanger, NCX1 and NCX3, constitute novel additional targets for the prosurvival action of Akt/protein kinase B pathway. Mol Pharmacol 2008; 73: 727–737. [DOI] [PubMed] [Google Scholar]
- 5.Molinaro P, Cuomo O, Pignataro G, et al. Targeted disruption of Na+/Ca2+ exchanger 3 (NCX3) gene leads to a worsening of ischemic brain damage. J Neurosci 2008; 28: 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pignataro G, Gala R, Cuomo O, et al. Two sodium/calcium exchanger gene products, NCX1 and NCX3, play a major role in the development of permanent focal cerebral ischemia. Stroke 2004; 35: 2566–2570. [DOI] [PubMed] [Google Scholar]
- 7.Boscia F, Gala R, Pignataro G, et al. Permanent focal brain ischemia induces isoform-dependent changes in the pattern of Na+/Ca2+ exchanger gene expression in the ischemic core, periinfarct area, and intact brain regions. J Cereb Blood Flow Metab 2006; 26: 502–517. [DOI] [PubMed] [Google Scholar]
- 8.Molinaro P, Viggiano D, Nistico R, et al. Na+ -Ca2+ exchanger (NCX3) knock-out mice display an impairment in hippocampal long-term potentiation and spatial learning and memory. J Neurosci 2011; 31: 7312–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pannaccione A, Secondo A, Molinaro P, et al. A new concept: Aβ1-42 generates a hyperfunctional proteolytic NCX3 fragment that delays caspase-12 activation and neuronal death. J Neurosci 2012; 32: 10609–10617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabellini N, Bortoluzzi S, Danieli GA, et al. Control of the Na+/Ca2+ exchanger 3 promoter by cyclic adenosine monophosphate and Ca2+ in differentiating neurons. J Neurochem 2003; 84: 282–293. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Villafuertes R, Torres B, Barrio J, et al. Downstream regulatory element antagonist modulator regulates Ca2+ homeostasis and viability in cerebellar neurons. J Neurosci 2005; 25: 10822–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valsecchi V, Pignataro G, Del Prete A, et al. NCX1 is a novel target gene for hypoxia-inducible factor-1 in ischemic brain preconditioning. Stroke 2011; 42: 754–763. [DOI] [PubMed] [Google Scholar]
- 13.Formisano L, Guida N, Valsecchi V, et al. NCX1 is a new rest target gene: role in cerebral ischemia. Neurobiol Dis 2013; 50: 76–85. [DOI] [PubMed] [Google Scholar]
- 14.Formisano L, Guida N, Valsecchi V, et al. Sp3/REST/HDAC1/HDAC2 complex represses and Sp1/HIF-1/p300 complex activates ncx1 gene transcription, in brain ischemia and in ischemic brain preconditioning, by epigenetic mechanism. J Neurosci 2015; 35: 7332–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Formisano L, Noh K, Miyawaki T, et al. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci U S A 2007; 104: 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langley B, Brochier C, Rivieccio M.Targeting histone deacetylases as a multifaceted approach to treat the diverse outcomes of stroke. Stroke 2009; 40: 2899–2905. [DOI] [PubMed] [Google Scholar]
- 17.Schweizer S, Meisel A, Märschenz S.Epigenetic mechanisms in cerebral ischemia. J Cereb Blood Flow Metab 2013; 33: 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazantsev AG, Thompson LM.Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 2008; 7: 854–868. [DOI] [PubMed] [Google Scholar]
- 19.He M, Zhang B, Wei X, et al. HDAC4/5-HMGB1 signalling mediated by NADPH oxidase activity contributes to cerebral ischaemia/reperfusion injury. J Cell Mol Med 2013; 17: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pignataro G, Boscia F, Esposito E, et al. NCX1 and NCX3: two new effectors of delayed preconditioning in brain ischemia. Neurobiol Dis 2012; 45: 616–623. [DOI] [PubMed] [Google Scholar]
- 21.Pignataro G, Esposito E, Cuomo O, et al. The NCX3 isoform of the Na+/Ca2+ exchanger contributes to neuroprotection elicited by ischemic postconditioning. J Cereb Blood Flow Metab 2011; 31: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guida N, Laudati G, Mascolo L, et al. MC1568 inhibits thimerosal-induced apoptotic cell death by preventing HDAC4 up-regulation in neuronal cells and in rat prefrontal cortex. Toxicol Sci 2016; 154: 227–240. [DOI] [PubMed] [Google Scholar]
- 23.Fischle W, Emiliani S, Hendzel MJ, et al. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem 1999; 274: 11713–11720. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z, Peng L, Radhakrishnan R, et al. Histone deacetylase 9 (HDAC9) regulates the functions of the ATDC (TRIM29) protein. J Biol Chem 2010; 285: 39329–39338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molinaro P, Sirabella R, Pignataro G, et al. Neuronal NCX1 overexpression induces stroke resistance while knockout induces vulnerability via Akt. J Cereb Blood Flow Metab 2016; 36: 1790–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilkenny C, Browne WJ, Cuthill IC, et al. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010; 8: e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyawaki T, Mashiko T, Ofengeim D, et al. Ischemic preconditioning blocks BAD translocation, Bcl-xL cleavage, and large channel activity in mitochondria of postischemic hippocampal neurons. Proc Natl Acad Sci U S A 2008; 105: 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guida N, Laudati G, Anzilotti S, et al. Methylmercury upregulates RE-1 silencing transcription factor (REST) in SH-SY5Y cells and mouse cerebellum. Neurotoxicology 2015; 52: 89–97. [DOI] [PubMed] [Google Scholar]
- 29.Boscia F, Casamassa A, Secondo A, et al. NCX1 exchanger cooperates with calretinin to confer preconditioning-induced tolerance against cerebral ischemia in the striatum. Mol Neurobiol 2016; 53: 1365–1376. [DOI] [PubMed] [Google Scholar]
- 30.Boscia F, Ferraguti F, Moroni F, et al. mGlu1alpha receptors are co-expressed with CB1 receptors in a subset of interneurons in the CA1 region of organotypic hippocampal slice cultures and adult rat brain. Neuropharmacology 2008; 55: 428–439. [DOI] [PubMed] [Google Scholar]
- 31.Pignataro G, Studer FE, Wilz A, et al. Neuroprotection in ischemic mouse brain induced by stem cell-derived brain implants. J Cereb Blood Flow Metab 2007; 27: 919–927. [DOI] [PubMed] [Google Scholar]
- 32.Pignataro G, Esposito E, Sirabella R, et al. nNOS and p-ERK involvement in the neuroprotection exerted by remote postconditioning in rats subjected to transient middle cerebral artery occlusion. Neurobiol Dis 2013; 54: 105–114. [DOI] [PubMed] [Google Scholar]
- 33.Cuomo O, Pignataro G, Gala R, et al. Antithrombin reduces ischemic volume, ameliorates neurologic deficits, and prolongs animal survival in both transient and permanent focal ischemia. Stroke 2007; 38: 3272–3279. [DOI] [PubMed] [Google Scholar]
- 34.Yildirim F, Gertz K, Kronenberg G, et al. Inhibition of histone deacetylation protects wildtype but not gelsolin-deficient mice from ischemic brain injury. Exp Neurol 2008; 210: 531–542. [DOI] [PubMed] [Google Scholar]
- 35.Guida N, Laudati G, Serani A, et al. The neurotoxicant PCB-95 by increasing the neuronal transcriptional repressor REST down-regulates caspase-8 and increases Ripk1, Ripk3 and MLKL expression determining necroptotic neuronal death. Biochem Pharmacol 2017; 142: 227–241. [DOI] [PubMed] [Google Scholar]
- 36.Koppel I, Timmusk T.Differential regulation of Bdnf expression in cortical neurons by class-selective histone deacetylase inhibitors. Neuropharmacology 2013; 75: 106–115. [DOI] [PubMed] [Google Scholar]
- 37.Scognamiglio A, Nebbioso A, Manzo F, et al. HDAC-class II specific inhibition involves HDAC proteasome-dependent degradation mediated by RANBP2. Biochim Biophys Acta 2008; 1783: 2030–2038. [DOI] [PubMed] [Google Scholar]
- 38.Rivas M, Mellström B, Naranjo JR, et al. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J Biol Chem 2004; 279: 33114–33122. [DOI] [PubMed] [Google Scholar]
- 39.Rovani MK, Brachmann CB, Ramsay G, Katzen AL.The dREAM/Myb-MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin. Dev Biol 2012; 372: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan J, Najafov A, Py BF.Roles of caspases in necrotic cell death. Cell 2016; 167: 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litke C, Bading H, Mauceri D.Histone deacetylase 4 shapes neuronal morphology via a mechanism involving regulation of expression of vascular endothelial growth factor D. J Biol Chem 2018; 293: 8196–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodruff TM, Thundyil J, Tang SC, et al. Pathophysiology, treatment, and animal and cellular models of human ischemic stroke. Mol Neurodegener 2011; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins LM, Adriaanse LJ, Theratile SD, et al. Class-IIa histone deacetylase inhibition promotes the growth of neural processes and protects them against neurotoxic insult. Mol Neurobiol 2015; 51: 1432–1442. [DOI] [PubMed] [Google Scholar]
- 44.Buonvicino D, Felici R, Ranieri G, et al. Effects of class II-selective histone deacetylase inhibitor on neuromuscular function and disease progression in SOD1-ALS mice. Neuroscience 2018; 379: 228–238. [DOI] [PubMed] [Google Scholar]
- 45.Kassis H, Shehadah A, Li C, et al. Class IIa histone deacetylases affect neuronal remodeling and functional outcome after stroke. Neurochem Int 2016; 96: 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scorziello A, Savoia C, Sisalli MJ, et al. NCX3 regulates mitochondrial Ca(2+) handling through the AKAP121-anchored signaling complex and prevents hypoxia-induced neuronal death. J Cell Sci 2013; 126(Pt 24): 5566–5577. [DOI] [PubMed] [Google Scholar]
- 47.Secondo A, Staiano RI, Scorziello A, et al. BHK cells transfected with NCX3 are more resistant to hypoxia followed by reoxygenation than those transfected with NCX1 and NCX2: possible relationship with mitochondrial membrane potential. Cell Calcium 2007; 42: 521–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, JCB884742 Supplemetal Material for HDAC4 and HDAC5 form a complex with DREAM that epigenetically down-regulates NCX3 gene and its pharmacological inhibition reduces neuronal stroke damage by Luigi Formisano, Giusy Laudati, Natascia Guida, Luigi Mascolo, Angelo Serani, Ornella Cuomo, Maria Cantile, Francesca Boscia, Pasquale Molinaro, Serenella Anzilotti, Vincenzo Pizzorusso, Gianfranco Di Renzo, Giuseppe Pignataro and Lucio Annunziato in Journal of Cerebral Blood Flow & Metabolism