Abstract
Human oncoproteins promote transformation of cells into tumours by dysregulating the signalling pathways that are involved in cell growth, proliferation and death. Although oncoproteins were discovered many years ago and have been widely studied in the context of cancer, the recent use of high-throughput sequencing techniques has led to the identification of cancer-associated mutations in other conditions, including many congenital disorders. These syndromes offer an opportunity to study oncoprotein signalling and its biology in the absence of additional driver or passenger mutations, as a result of their monogenic nature. Moreover, their expression in multiple tissue lineages provides insight into the biology of the proto-oncoprotein at the physiological level, in both transformed and unaffected tissues. Given the recent paradigm shift in regard to how oncoproteins promote transformation, we review the fundamentals of genetics, signalling and pathogenesis underlying oncoprotein duality.
One of the most fundamental challenges in cancer biology is to understand how the normal cell becomes a malignant cancer cell. Several decades ago, oncoproteins were discovered to hijack the signalling pathways that regulate cell proliferation, growth, death, differentiation and metabolism. We now understand that oncoproteins significantly contribute to many, if not all, steps of cancer formation and progression, and designing therapies that specifically block their activity has resulted in unprecedented clinical benefit. Next-generation sequencing has revolutionized the oncology field and helped identify genetic variants in human cancers. However, the impact of genome analysis has not been restricted to oncology, but rather genome analysis has impacted all fields of medicine1. For instance, an emerging number of congenital disorders caused by either somatic or germline pathogenic variants have been reported to be caused by mutations in genes that are well-known cancer drivers1–3. While most of these oncoprotein-driven germline syndromes and somatic mosaicisms are characterized by specific signs and symptoms, basic human architecture and organ structure remain histologically normal despite the presence of the mutation. These remarkable observations are in line with recent studies that demonstrate the presence of clonal oncogenic mutat ions in normal tissues, such as the oesophagus and skin4–6. The expression of oncoproteins in different cell lineages provides a unique opportunity to address two crucial questions: (1) the mechanism that leads to cell transformation or abnormal function in the clinically affected tissue and (2) the therapeutic potential behind the molecular mechanisms that avoid transformation in clinically unaffected tissues. Hence, understanding the reasons why certain tissues are phenotypically normal despite the presence of an oncoprotein is as important as understanding those instances in which tissues are transformed7. It now seems clear that certain tissues are extremely sensitive to transformation by specific oncoproteins, while others require many additional steps, or hits, to promote tumour growth. This observation is also true when individual alleles are taken into consideration and explains the mutation bias characteristic of many tumour types8,9. Orthogonal experimental approaches that take into consideration the molecular, cellular and organismal effect of oncoproteins will be required to better understand the role that specific oncoproteins play during transformation, as well as to predict epistatic interactions that lead to tumour initiation and growth.
In this Review, we provide a comprehensive catalogue of oncoproteins that are known to cause cancer and congenital disorders (a property that we have termed 'oncoprotein duality'). The genetic basis behind these conditions will be established and the signalling pathways that oncoproteins affect to promote tissue transformation will be discussed. Finally, we summarize some of the efforts made to model oncoproteins in the mouse and examine the therapeutic approaches being developed to target oncoproteins in the clinic.
Do oncoproteins cause transformation?
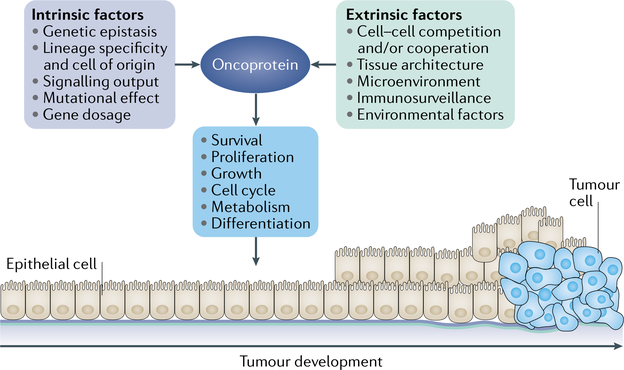
Historically, oncoproteins were identified on the basis of their ability to transform monolayer cell cultures, such as fibroblasts (FIG. 1). Expression of oncoproteins in these cultures can lead to insensitivity to contact inhibition, growth factor independence or tumorigenic potential when they are injected in animal hosts. While these properties are still used routinely in many laboratories as a proof of concept to define novel oncoproteins, the ability to transform non-malignant cell lines, or to what extent, is highly variable. Similarly, the expression of certain oncoproteins in animal models can transform specific tissues, but not all. The factors that underlie the ability of an oncoprotein to promote transformation are presumably tissue specific10. In an effort to categorize such factors, these have been defined as intrinsic, referring to the oncoprotein itself, and extrinsic, which refers to the environmental effects that modify indirectly the outcome of the oncoprotein (FIG. 2).
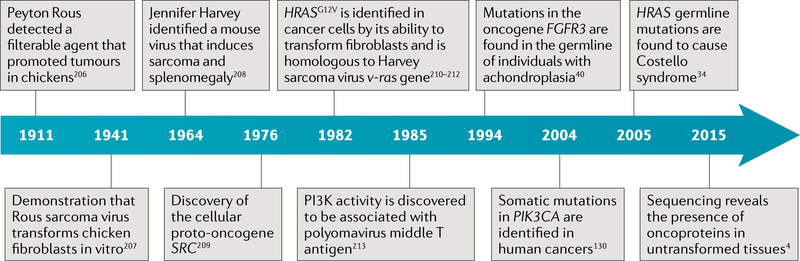
Fig. 1 |.
Timeline of the key events in the history of and research into oncoproteins4,34,40,130,206–213.
Fig. 2 |. Factors that influence the ability of oncoproteins to cause transformation.
The presence of oncogenic mutations in a tissue can lead to its transformation. However, this dogmatic view appears to be the exception rather than the rule. This figure depicts some of the factors that are likely to contribute to the process of oncoprotein-mediated transformation, which are dependent either on the characteristics of the oncoprotein itself (intrinsic factors) or the microenvironment and macroenvironment (extrinsic factors). Some of these factors are at the crossroads of this binary classification. The exact combination of factors required to facilitate tissue transformation is unknown, but it is reasonable to expect that many are needed and each tissue would require a different combination.
Intrinsic factors.
Intrinsic factors can include the role of gene dosage, such as amplifications of a cancer-associated gene that result in increased activity and downstream signalling11. Similarly, loss of the wild-type allele is seen in certain tumours, for example in patients with HRAS-mutant Costello syndrome who develop embryonal rhabdomyosarcoma12. The downstream effects of the mutation itself can also result in a different ability to transform tissues. For example, the different mutations in the phosphoinositide 3-kinase catalytic subunit-α gene (PIK3CA) use different molecular mechanisms to induce gain of function13,14, and lead to differential phenotypic effects that in some cases are probably linked to their distinct ability to activate certain signalling pathways in a tissue-specific manner. For instance, KRAS-G12R, an oncoprotein commonly found in pancreatic adenocarcinoma, is unable to bind and activate PI3K15. Furthermore, the signalling output of the KRAS-A146T mutant, the encoding allele of which is found only in gastrointestinal tumours, is strikingly different from that of the most common G12D variant, though still oncogenic16. The differences in downstream signalling that lead to or prevent tissue transformation are not necessarily post-translational, because tissues exhibit specific epigenetic landscapes that can result in different transcriptional responses17. All these factors contribute to cell-lineage specificity, a commonality which is essential to understand the ability of oncoproteins to transform certain tissues. Oncoprotein expression in particular lineages is also an important factor. For example, the oncoprotein anaplastic lymphoma kinase (ALK) is mostly expressed in embryonic and adult neuronal tissue — a pattern that can explain the predisposition to neuroblastoma in families carrying ALK activating mutations in the germline18,19. Because the expression of some of these oncoproteins leads to cell cycle arrest, terminal differentiation or senescence, it is not surprising that genetic epistatic interactions often occur with tumour suppressors involved in these cellular processes20. Many of these additional genetic hits are sequential and are required to promote complete transformation to overt cancers21.
Extrinsic factors.
Some of the factors that can be considered as extrinsic include the relationship between the mutant cell and the surrounding wild-type cells. Cell competition, for example, is a key mechanism that has been widely studied in flies, but is poorly understood in mammals22,23. Understanding how cells interact and compete with mutant populations is of great relevance to mosaicisms and tumours. In experimental epithelial monolayers, cells expressing the oncoprotein HRAS-G12V are apically protruded and eliminated by the surrounding wild-type neighbours24. This observation underlines the importance of tissue architecture in limiting oncogenesis25. For instance, in the skin, the presence of HRAS mutations (which are characteristic of squamous carcinomas), does not lead to abnormal growth because tissue architecture preserves homeostasis by correcting the mutant clones26 or because of compartmentalization in the hair follicle27. Similarly, the diversity of cell lineages present in the tissue can modulate the process of transformation, especially in certain conditions, such as tissue inflammation, that have been shown to cooperate with specific oncoproteins28. In a similar context, the ability of the immune cells to detect and remove populations of mutant cells could be compromised in cases in which the mutant population was present from birth, as seen in many congenital disorders and tumour predisposition syndromes29. Other environmental factors can promote the transformation potential of an oncoprotein, by causing secondary mutations or an inflammatory response, as seen with radiation and other carcinogens30,31. In summary, the ability of an oncoprotein to transform a specific tissue will be the result of a combination of factors that facilitate this process in a cell-autonomous and/or cell-non-autonomous manner.
Allele bias.
In the context of clinical genetics, this is when a specific mutation in a gene is far more frequent than expected.
Modifying alleles.
Single-nucleotide polymorphisms that can either decrease or exacerbate a clinical phenotype driven by a pathogenic mutation.
Mosaicism.
Characterized by the presence of cells with at least two distinct genetic make-ups.
Oncoprotein transmission
Oncogenic mutations can occur spontaneously and stochastically in any cell during development (FIG. 3a) or adulthood, as the result of either external mutagens or error-prone replication bypass and replication errors32. The origin of such mutations can be retrospectively investigated by the presence of mutational signatures in the tumour that are characteristic of specific genetic events, such as homology recombination deficiency, or genotoxins, such as UV exposure33. Depending on the timing and tissues in which such mutations occur, the phenotypes can range from a complete lack of phenotype (that is, silent mutations present in otherwise healthy tissues) to a severe and extensive transformation of the normal tissues. To add another layer of complexity, different variants of the same oncogene can lead to signi ficant variations in phenotypic outcomes as well; this phenomenon leads to the allele bias observed in tumours and congenital disorders. For example, HRASG12S is the most common mutation in Costello syndrome, while papillary thyroid cancer and pheochromocytoma are driven by HRASQ61L or HRASQ61R (REFS34–36).
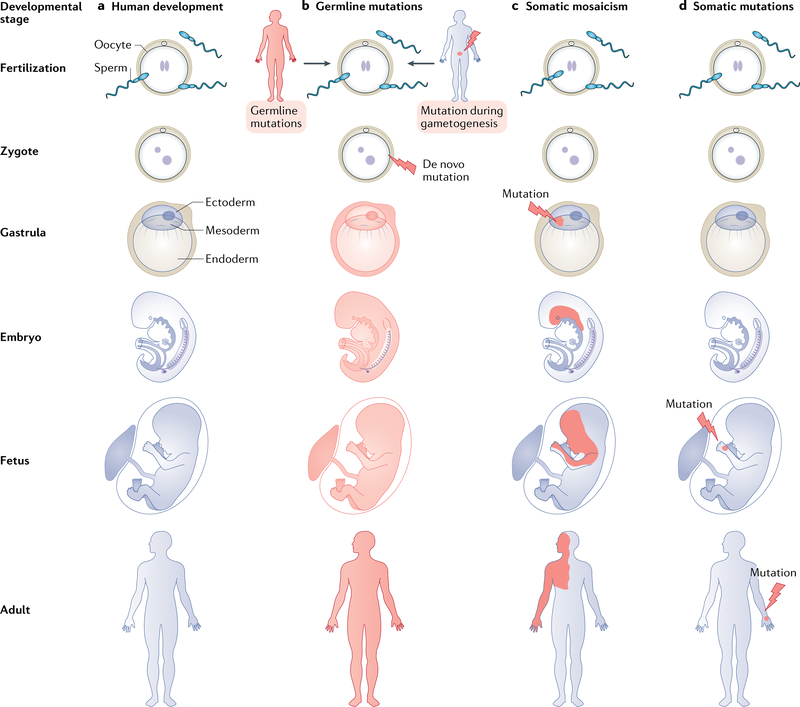
Fig. 3 |. Genetic transmission of oncoproteins.
Mutations that lead to oncoproteins can occur during human development (panel a). Depending on the time and location where such mutations occur, the affected tissue will exhibit different patterns in the adult. Germline inheritance affects all lineages of the body and, in the case of oncoproteins, with autosomal dominant segregation. Oncogenic mutations can be transmitted from an affected parent who acts as a carrier or can occur spontaneously (de novo) during gametogenesis or in the zygote. Examples include achondroplasia and Costello syndrome, respectively (panel b). Postzygotic somatic mutations that occur in the blastocyst, during gastrulation or during embryogenesis lead to different degrees of mosaicism. In this example, a mutation that occurs in the ectoderm and mesoderm will affect various lineages that arise from these germ layers, including skin, brain, muscle and endothelial cells. An example is Sturge-Weber syndrome (panel c). Somatic mutations occur late during embryogenesis, during development or in adulthood. These mutations typically affect a single lineage and display clonality, such as those seen in arteriovenous malformations and cancer (panel d).
Inheriting oncoproteins.
When oncogenic mutations occur de novo in germ cells, their inheritance pattern is termed 'germline transmission' and can be transmitted to the offspring. The age of the father can have a direct impact on the germline transmission of certain oncogenic variants, because these 'selfish' variants give a growth advantage to cells producing sperm in these individuals37–39. Although there are some examples of oncoproteins that are inherited through the germline, there is a clear restriction in this sense: the oncoprotein has to be compatible with embryonic development and, ultimately, with fertility (FIG. 3b). For instance, germline oncogenic mutations in the fibroblast growth factor receptor 3 gene (FGFR3) lead to a disorder termed 'achondroplasia', which results in dwarfism and macrocephaly, yet is compatible with life40. In contrast, oncogenic mutations in the PIK3CA gene have not been detected in the germline, consistent with the embryonic lethality observed in mouse models expressing the oncoprotein PI3Kα41. In this sense, it is important to introduce the concept of the division of genes encoding oncoproteins into weak versus strong alleles. This arbitrary separation is based on the ability of an oncoprotein, or a specific allele, to interfere with normal development, leading to a lethal or non-lethal embryonic phenotype. This is exemplified by the oncoprotein KRAS; mutations in the classical hotspots found in cancer (that is, G12, G13 and Q61) are highly transforming in cellbased assays, and the expression of such mutants in the germline results in embryonic lethality due to a severe phenotype that includes cardiomegaly and abnormal brain development42,43. However, KRAS mutations have been found in the germline of individuals with Noonan syndrome. These mutations never occur in the cancer hotspot alleles, but rather occur in secondary alleles such as KRASV14I, KRAST58I and KRASD153V (REF.44). Such mutations render KRAS active, but to an intermediate point where there is a balance between embryo survival and hyperactive signalling that results in a specific congenital disorder. Since these are weak activating mutations, their phenotype can often be attenuated or exacerbated by so-called modifying alleles. In some instances, affected individuals exhibit a mild, subclinical phenotype, which is not diagnosed. These individuals often transmit their oncogenic mutations to the offspring, who might be more severely affected and, hence, may receive a diagnosis. Examples of such pedigrees have been described in cardiofaciocutaneous syndrome, carrying oncogenic mutations in the MAPK/ERK kinase 1 gene (MEK1, also known as MAP2K1) or MEK2 (also known as MAP2K2)45.
Oncoproteins surviving embryonic development and human chimaeras.
Postzygotic mutations during embryonic development give rise to organisms with different genetic populations. These mutations can range from single variants to large chromosomal abnormalities in autosomes and/or sex chromosomes. The resulting clonal mosaicisms are relatively frequent in healthy individuals and could contribute to genetic conditions such as cancer or congenital disorders46–49. According to the Happle hypothesis (BOX 1), when an oncoprotein is incompatible with embryonic development (lethal gene), it can still survive through mosaicism50 (FIG. 3c). This explains how certain strong alleles can still be found in extensive parts of the body and highlights the importance of the balance between survival of the oncoprotein and survival of the embryo. Then, one can expect that there is a level of mosaicism in the individual that cannot be surpassed, otherwise the development of the embryo would be interrupted. In KRAS, where strong alleles in the germline are lethal, it could be expected that some of these strong alleles are found in the form of mosaicism. Consistently, Schimmelpenning–Feuerstein–Mims syndrome, a disorder characterized by the presence of sebaceous nevi and cataracts, is caused by KRASG12V and KRASG12D mutations in the neuroectodermal lineage as a result of mosaicism51. Many of the mosaicisms caused by oncoproteins can be easily visualized in affected individuals when there is a cutaneous involvement, as these follow a particular pattern termed the 'Blaschko lines'52. These lines represent the vestigial route of cell migration during skin development and become highly evident in mosaicism involving melanocytic lesions, such as the epidermal nevi driven by postzygotic FGFR3, HRAS or PIK3CA mutations53–55. The degree and lineage specificity of the mosaicism will be the result of the precise moment at which the oncogenic mutation occurs during embryonic development and whether the mutation might exhibit a positive or a negative advantage to the developing embryo. Mutations arising as early as the morula stage will give rise to highdegree mosaicisms affecting all tissue lineages, while mutations that occur during or after gastrulation will likely be restricted to a specific germ layer and/or cell fate56,57. By contrast, if the oncogenic mutation occurred at the very end of embryo development, then the effect will most likely be local unless such a mutation affects the migration patterns of certain cells (for example, melanocytes derived from the neural crest).
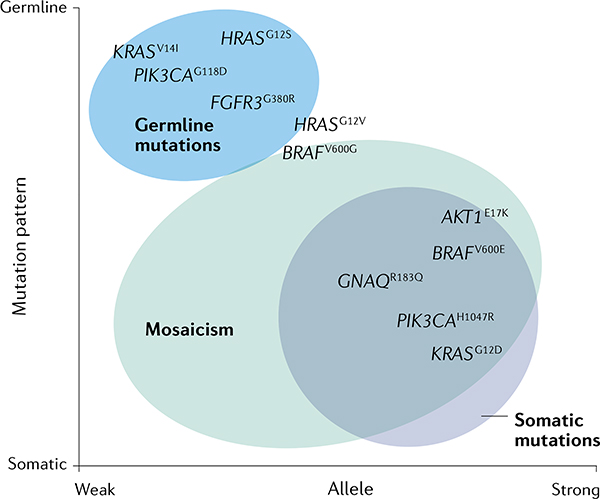
Box 1 |. The Happle hypothesis.
On the basis of the observation that McCune-Albright syndrome is not an inheritable disorder, Rudolf Happle, a German dermatologist, postulated what is now considered the Happle hypothesis50. McCune-Albright syndrome is characterized by the presence of fibrous dysplasia, endocrine dysfunction and cutaneous lesions118. Happle observed that the pigmented skin lesions follow the patterns of the Blaschko lines, described earlier by Alfred Blaschko52. Happle realized that skin lesions following such lines visualize the dorsoventral patterning of two cell populations during embryogenesis, suggesting that individuals with McCune-Albright syndrome have two genetic clones and are, therefore, chimaeras. Indeed, later genetic studies showed that these patients carry a mosaic GNAS mutation117. On the basis of the fact that all cases of McCune-Albright syndrome are sporadic, he speculated that this disorder was the result of a dominant lethal gene that survives only through mosaicism. The analysis of other cutaneous mosaicisms, such as in Schimmelpenning-Feuerstein-Mims syndrome, Proteus syndrome and Klippel-Trenaunay syndrome, among others, confirmed his observations; none of these disorders can be transmitted through the germline. With the recent development of mouse models, we can now confirm that these causative genes are incompatible with life. Following the Happle hypothesis, it is predicted that 'strong alleles' cannot be found in the germline, in contrast to 'weaker alleles'. Examples of these are summarized in the graph (see the figure).
Schimmelpenning–Feuerstein–Mims syndrome.
Neuro-oculocutaneous mosaicism characterized by the presence of skin lesions and pigmentation abnormalities, epilepsy, epibulbar dermoids, cloudy cornea, eyelid colobomas and arteriovascular defects, among other manifestations.
Blaschko lines.
Skin patterns found in adults that recapitulate the normal cell development during embryogenesis. These can be often appreciated in individuals with genetically driven skin stains.
Field cancerization.
The presence of large areas of tissue affected by carcinogenic mutations, which often contribute to malignant transformation. It is generally the result of a genotoxic exposure during a prolonged time and can lead to the presence of low-grade and high-grade tumours.
Somatic oncogenic variants and accumulating mutations.
When the mutation occurs in a single cell that will not give rise to other histological types, it can be considered a clonal somatic event. The current model for sporadic cancer initiation relies on this idea, where a single cell acquires an oncogenic mutation that drives tumour formation (FIG. 3d). This seems to be true in certain tumours (or overgrowths) that are monogenic in nature (such as sporadic venous malformations or epidermal nevi), but does not seem to explain how overt cancers are formed for several reasons: (1) tumours usually contain many other somatic mutations; (2) not all tumours remain addicted to the driver oncoprotein; and (3) expression of the oncoprotein is not sufficient to transform tissues in preclinical models. It was later proposed that, to promote cancer formation, cells require several additional mutations, or multiple hits, that would facilitate transformation. This concept, introduced by Nordling, and later known as the Knudson hypothesis, is exemplified by experiments in which the deletion of the tumour suppressor Trp53 highly accelerates the formation of tumours in mice expressing different oncoproteins58–60. Loss of heterozygosity at the RB1 locus was one of the first examples supporting the Knudson hypothesis in human cancers61. Additional genetic hits can also be boosted by a permissive microenvironment (that is, as a result of radiation or inflammation), as seen in the pancreatitis-induced models that cooperate with Kras mutations to induce pancreatic adenocarcinoma progression62. Therefore, a combination of a driver oncogenic mutation and secondary mutations is likely to result in tumour formation when present in a tissue susceptible to transformation.
While some tissues appear to be extremely sensitive to transformation by certain oncoproteins, others remain highly resistant. This is likely explained by the role that such proto-oncoprotein and its downstream effectors would play in the normal physiology of the tissue or cell of origin, resulting in constitutive growth and proliferation when its gene becomes mutated. For example, mutations in the G protein subunit αq gene (GNAQ) are drivers of uveal melanoma and congenital capillary malformations63,64 but are unlikely to transform other tissues. Hence, certain oncogenic mutations might be more common in normal tissues than we previously anticipated. Recent work has demonstrated that expansion of clones carrying an oncoprotein is frequent in histologically normal tissues, creating asymptomatic individuals with mosaic expression of common strong oncogenes (that is, KRASG12V and PIK3CAH1047R)4–6,65–67. This concept of clonal expansion can be termed 'silent oncogenic mosaicism' and needs to be recognized as an important, but understudied, discovery that potentially plays a role in cancer as an underlying factor. It is tempting to speculate that clonal expansion of a silent oncogenic mosaicism could give rise to a particular cancer after accumulating additional genetic mutations or hits; this idea is similar to the concept of field cancerization and has also recently been described in unaffected endometrial tissue surrounding endometrial tumours68,69 and in cases of early-onset bladder cancer, where a mosaicism for HRAS mutation was reported70. It is also important that, in the context of mosaicism, not all tissues expressing oncoproteins will develop a phenotype, or that tissues that exhibit a phenotype have to carry such mutations, because mutant cell clones can affect histologically normal tissues in a cell-non-autonomous manner. In an autopsy study on a patient with Proteus syndrome, a mosaicism driven by AKT1E17K, the correlation between tissues that exhibited histological changes and those with detectable mutational burden was rather poor. Certain tissues appeared highly affected microscopically, yet AKT1 mutations could not be found71. These observations clearly indicate that either small or distant populations of mutant cells have the ability to affect surrounding tissues.
Oncogenic pathways
Oncoproteins generally participate in mitogenic signalling, mostly the RAS-MAPK and PI3K-AKT pathways72. Despite the high frequency of mutations affecting these pathways, their effect is not universal. Oncoproteins exhibit both lineage specificity and allele specificity, underscoring the importance of specific gene products, or their downstream effectors, in the transformation process. To study this so-called oncoprotein dualism, a review of literature in PubMed, mutation data from tumour sequencing consortia using cBioPortal36 and congenital disorders using the Online Mendelian Inheritance in Man (OMIM) and NSEuronet databases and GeneReviews was conducted. Only oncoproteins that exhibit dualism and contribute to a significant fraction of the cases have been considered.
Receptor tyrosine kinase pathways, the upstream drivers of malignancy.
Growth factors are sensed through cell surface-anchored receptor tyrosine kinases (RTKs). Multiple protein ligands interact with RTKs to activate the signalling cascades that lead to specific cellular phenotypes73. While the activation of RTKs can trigger unique effectors, the RAS–MAPK and PI3K–AKT pathways appear to be downstream of most RTKs and are also involved in the pathogenesis of many cancers and congenital disorders. RTK genes are commonly mutated in cancer and the encoded proteins are considered bona fide oncoproteins, constitutively activated by point mutation, amplification, translocation or deletion of autoinhibitory regions74. In general, weakly activating mutations in these RTK genes are compatible with development and give rise to tumour predisposition syndromes rather than clinically unique syndromes (FIG. 4a). The best known RTKs contributing to human cancer are epidermal growth factor receptor (EGFR) and ERBB2. While EGFR mutations occur in a large number of lung adenocarcinomas (mostly L858R and E746–750del), ERBB2 variants are less frequent in the lung, but can be found in some patients with breast cancer. Gene amplification and/or protein overexpression of ERBB2 is a very common and subtype-defining event, and is found in breast and gastric adenocarcinomas. Most of these mutations are gain-of-function mutations because they promote ligand-independent activation of the receptor, but they have not been found in other congenital disorders. However, activating mutations in these genes have been detected in the germline of patients with familial predisposition to lung cancer75–77.
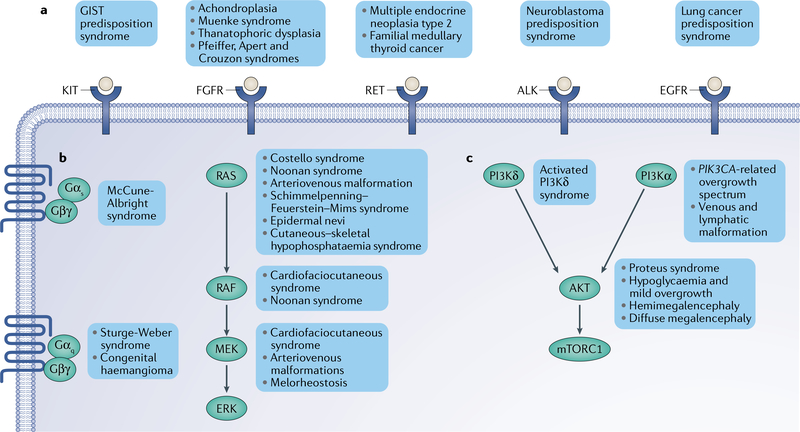
Fig. 4 |. The pathways of oncoprotein signalling in congenital disorders.
Most human oncoproteins are core components of the signalling pathways that regulate cell growth, division and proliferation. Among these, three major groups can be easily recognized: the receptor tyrosine kinase (RTK), PI3K-AKT and RAS-MAPK pathways. a | RTK pathway. Mutations in RTK genes often lead to ligand-independent activation of the receptor and signal through downstream pathways such as the PI3K, MAPK, signal transducer and activator of transcription (STAT) and SRC pathways. Most germline mutations in RTK genes lead to tumour predisposition syndromes. b | RAS-MAPK pathway. Mutations in genes in this pathway generally lead to cell cycle progression and proliferation. Activating mutations in the RAS gene isoforms (NRAS, HRAS and KRAS) are frequent in congenital disorders and cancer, but are mostly incompatible with life in the germline. Most germline syndromes include weak activating variants of the oncoproteins, such as the variants found cardiofaciocutaneous syndrome. Mutations in the genes encoding the trimeric G protein-associated GTPases GNAS, GNAQ and GNA11 cause syndromes characterized by their skin involvement, such as McCune-Albright syndome and Sturge-Weber syndrome. c | PI3K-AKT pathway. This is an important pathway that mainly regulates cell growth. Most mutations affecting PIK3CA, the PI3Ka isoform that generates phosphoinositide 3,4,5-trisphosphate, result in syndromes with severe overgrowth and vascular involvement. This is also true for AKT1 mutations in Proteus syndrome. Germline mutations in PIK3CD, the PI3Kδ isoform mostly expressed in lymphocytes, cause a syndrome characterized by immunodeficiency. ALK, anaplastic lymphoma kinase; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; GIST, gastrointestinal stromal tumour; mTORC1, mechanistic target of rapamycin complex 1.
Achondroplasia.
An autosomal dominant syndrome that is the most common form of skeletal dysplasia in humans and is caused by the FGFR3 mutation G380R. Patients exhibit macrocephaly and short limbs.
Acanthosis nigricans.
A hyperpigmentation and hyperkeratosis of the skin.
Another RTK gene, ALK, is mutated in both somatic tumours and families with cancer predisposition. Somatic events often result from translocation of the ALK kinase domain and other partners (for example, EML4 in lung cancer and TPM3 and/or TPM4 in anaplastic large B cell lymphoma) or as a result of gain-offunction alterations in the kinase domain that result in constitutive activation of the receptor, most frequently seen in neuroblastomas. In cases of family predisposition to neuroblastoma, ALK kinase domain alterations have been described in the germline, although these alleles do generally not overlap with those seen in sporadic cases78–80. RET, an RTK that plays a critical role in the biology of neural crest cells, displays similarities to the oncoprotein ALK; gene translocations are drivers in lung adenocarcinomas, activating point mutations are found in a specific tumours (medullary thyroid cancer) and there are germline mutations that predispose individuals to the same tumours. In the case of the oncoprotein RET, three highly overlapping conditions with autosomal dominant segregation have been described, namely multiple endocrine neoplasia type 2A and type 2B and familial medullary thyroid cancer81.
The platelet-derived growth factor receptor-α gene (PDGFRA) and KIT belong to the same subfamily of RTK genes and display similar patterns of oncogenesis. Strong activating variants in these genes are present in sporadic gastrointestinal stromal tumours, with PDGFRAD842V, KITD816N/V/Y/H and KITN822K as the main hotspots. Family members with gastrointestinal stromal tumour susceptibility have also been reported to be carriers of germline heterozygous mutations in these two oncogenes82–84. In addition to gastrointestinal stromal tumours, patients with germline PDGFRA and KIT mutations exhibit cutaneous mastocytosis. Other neoplasms driven by KIT oncogenic mutations include acute leukaemia and germ cell tumours.
The case of somatic and germline mutations in the fibroblast growth factor receptor (FGFR) gene family is of particular interest, given the gene-and allele-specific phenotypes resulting from gain-of-function mutations. Physiologically, FGFRs are activated by the FGF family of proteins, which differentially bind to FGFR1-FGFR4 to regulate key developmental processes such as mesodermal induction, limb formation, neural development and bone homeostasis73,85. The latter becomes apparent when one is studying the phenotypes of individuals carry ing germline gain-of-function mutations in FGFR1, FGFR2 or FGFR3, as these are generally characterized by dysmorphic features, such as craniosynostosis, and short limbs40. Activating mutations in FGFR3 give rise to clinically overlapping, but different, forms of bone affections, including achondroplasia (G380R), Muenke syndrome (P250R), thanatophoric dysplasia I and II (R248C and K650E, respectively), severe achondroplasia with developmental delay and acanthosis nigricans (K650M) and Crouzon syndrome with acanthosis nigricans (A391E)40,86–88. Some of these alterations occurat different functional domains of the protein (P250R at the extracellular immunoglobulin- like domain, G380R at the transmembrane domain and K650E at the intracellular kinase domain), which could potentially explain the differential traits of these patients. Somatic mutations in FGFR3 are very common in urothelial bladder carcinoma, with two main hotspots, R248C and S249C, that are also found in patients with thanatophoric dysplasia I. Such mutations also appear to be common in epidermal nevi and seborrheic keratosis, two frequent benign skin lesions characterized by, among other conditions, acanthosis54,89. Germline mutations in FGFR1 and FGFR2 are also a frequent cause of skeletal pathologies within this spectrum, for which Pfeiffer syndrome, Apert syndrome and Crouzon syndrome are widely recognized88,90,91. Although some uterine adenocarcinomas have FGFR2 hypermorphs similar to those in Apert syndrome, FGFR1 and FGFR2 mutations are relatively uncommon in cancer.
Although many other RTKs are known to be oncogenic, and often altered in tumours, they have not been discussed in this section because of limited evidence regarding gain-of-function mutations in congenital disorders and, hence, do not fall within the category of dual oncoproteins. These include ROS1, NTRK, MET and FLT3. This also applies in the opposite situation in cases such as TEK, INSR and DDR2.
Tumours and RASopathies driven by oncoproteins of the RAS-MAPK pathway.
The RAS-MAPK signalling network integrates multiple extracellular cues that result in activation of the canonical RAF-MEK-ERK axis, which promotes cellular proliferation and cell cycle progression. The RAS GTPases are central regulators of this signalling pathway and function as molecular switches that cycle between inactive and active conformers that promote the activation of the downsstream kinases RAF, MEK and ERK92. An increasing number of cancers and congenital disorders have been found to be driven by gain-of-function mutations in RAS, RAF or MEK gene isoforms (FIG. 4b). Mutations in the RAS oncogenes are very frequent across all human cancers72. KRAS mutations are found as a main driver in pancreatic adenocarcinoma, lung adenocarcinoma, colorectal adenocarcinoma, uterine carcinoma and carcinosarcoma, testicular germ tumours, multiple myeloma and gastric adeno carcinoma. HRAS mutations instead are mostly found in pheochromocytomas, paragangliomas, head and neck tumours, bladder and thyroid cancers, and melanoma. Mutations in NRAS are found in melanoma, acute myeloid leukaemias, and thyroid and colorectal cancers36.
Most mutations found in the RAS gene isoforms occur at three conserved hotspots (G12, G13 and Q61). Mechanistically, such mutations result in the loss of GTPase-activating protein-mediated hydrolysis, which results in constitutive RAS activation and signalling92. Most of the mutations that activate NRAS in melanoma are at codon 61, while in leukaemias they are at codon 12 (REF.93). The basis of this allelic imbal-ance is poorly understood as both alleles are considered strongly oncogenic. Such alleles have also been discovered in somatic monogenic disorders such as arteriovenous malformations94. Extracranial arteriovenous malformations can also carry mosaic KRAS mutations, although in a lower proportion95. KRAS mutations have also been identified in the form of mosaicism in nevus sebaceous and both oculoectodermal syndrome and Schimmelpenning-Feuerstein-Mims syndrome51,96. These syndromes are now considered part of the mosaic RASopathy spectrum given their molecular genetics and the overlapping clinical features, suggesting a common pathogenesis97. Within this group, other mosaic forms driven by oncogenic HRAS or NRAS mutations have also been described, including epidermal nevi, phacomatosis pigmentokeratotica, nevus spilus, woolly hair nevus, neurocutaneous melanosis/congenital giant melanocytic nevus and cutaneous-skeletal hypophosphataemia syndrome98–102. These disorders are characterized by their heavy cutaneous involvement, which, together with the fact that these mutations are also found in melanomas, highlights the importance of these proto-oncogenes in nnormal skin biology.
Arteriovenous malformations.
Abnormal blood vessels that tangle and allow direct connection between veins and arteries and can cause pain and severe haemorrhage if ruptured.
G protein-coupled receptor-associated GTPases.
Gα proteins are bound to Gβγ, forming an inactive trimeric complex that associates with G protein-coupled receptors (GPCRs). On GPCR stimulation, conformational changes in the receptor lead to Gβγ dissociation and Gα GTP loading and activation, resulting in the production of second messengers; for Gαs (encoded by GNAS) adenylate cyclase and production of cAMP, and for Gαq and Gαll (encoded by GNAQ and GNA11, respectively) phospholipase C, resulting in diacylglycerol and inositol trisphosphate.
Cutaneous–skeletal hypophosphataemia syndrome is of special interest because it exemplifies the result of both autonomous and non-autonomous effects caused by the oncoprotein RAS. This mosaicism is characterized by the presence of epidermal nevi and skeletal defects, mostly hypophosphataemic rickets and osteomalacia. HRAS and NRAS mutations have been identified in both skin and bone, suggesting a multilineage mosaicism arising from a multipotent cell progenitor103. Mutant bone exhibits dysplastic features and elevated secretion of FGF23, a hormone that regulates phosphorus homeostasis in the kidney (autonomous effect). These patients develop rickets in bones that do not contain the mutation as a result of paracrine and endocrine action of RAS mutant bone-derived FGF23 (non-autonomous effects), in a similar fashion as the paraneoplastic phenomenon observed in oncogenic osteomalacia104. Another mosaic RASopathy characterized by an extensive bone phenotype is melorheostosis, a rare disorder that causes excess bone growth with a classic 'dripping candle wax' pattern and is caused by activating mutations in MEK1 (REF.105). The effect of MAPK activation in the bone due to oncogenic mutations is not restricted to these syndromes, since skeletal abnormalities have been observed in other germline RASopathies, such as Noonan syndrome, cardiofaciocutaneous syndrome and Costello syndrome. These patients exhibit distinctive craniofacial dysmorphia and, in Costello syndrome, a dental pheno type that includes malocclusion and delayed tooth development106,107. Costello syndrome is the most seve of the germline RASopathies and is caused by de novo mutations in HRAS, mostly G12S. Although other mutations have been described, it is worth noting that strong alleles (that is, HRASG12V and HRASG12D) are rarely found in these patients, most likely due to embryonic lethality, but if found, they are associated with a severe phenotype12,34,108. Individuals affected by Costello syndrome exhibit classic RASopathy features, including difficulty to thrive, craniofacial dysmorphia, intellectual disabilities and specific phenotypes, such as hair and skin abnormalities and predisposition to tumours, mostly embryonal rhabdomyosarcoma and bladder cancer109. In the case of Noonan syndrome, several genes have been shown to contribute to the disorder due to gain-of-function mutations. Of these, many can be considered oncogenes, such as PTPN11, SOS1, RIT1 and RAF1 (REFS110–113). However, mutations in these genes are rather infrequent in human cancers. In the case of PTPN11 and SOS1, the alleles found in Noonan syndrome are not similar to those seen in cancers. In contrast, RIT1 and RAF1 hotspots are the same in Noonan syndrome and cancer. RIT1 mutations have been seen in a subset of patients with lung adenocarcinoma who do not harbour other typical driver mutations and these alleles are often the same as in patients with Noonan syndrome114.
Despite not being canonical components of the MAPK pathway, the heterotrimeric G protein-coupled receptor-associated GTPases are another emerging family of oncoproteins115. Three members of the Gα gene family have been recurrently found to be mutated in many cancers and disorders: GNAS, GNAQ and GNA11 (REF.116). GNAS gain-of-function germline mutations are not compatible with life, but can be found as postzygotic mosaicism in patients with McCune-Albright syndrome117. As with other mosaicisms, the clinical presentation depends on the affected tissues, but it is characterized by atypical café au lait macules, fibrous dysplasia and endocrine symptoms, including hyperthyroidism, Cushing disease and excessive growth hormone118. The common alleles GNASR201C and GNASR201H are also hotspots seen in certain epithelial cancers, such as gastric and pancreatic adenocarcinoma, as well as pituitary adenomas and pancreatic cysts119,120. These mutations were postulated to affect the GTP hydrolysis and promote effector activation due to constitutive GTP loading; however, recent structural studies suggest otherwise. Such mutations can subvert the GDP state, activating adenylate cyclase when it is bound to GDP121. Mutations in GNAQ and GNA11 are frequent in uveal melanomas and have also been described in other dermatological conditions63,64,122–124. For instance, in congenital haemangioma, patients exhibit the same variant found in uveal melanoma, Q209L/P, although the expression of the oncoprotein is restricted to the endothelial cells. It is likely that these mutations exhibit a similar mechanism as described for Gαs. The R183Q variant, which is never seen in uveal melanomas, is the driver of Sturge-Weber syndrome, a neuroectodermal mosaicism characterized by port-wine stains, glaucoma and leptomeningeal angiomatosis. The remarkable allele specificity seen in these conditions is, again, proof that not all mutations in oncogenes lead to similar clinical phenotypes.
Activating the PI3K-AKT pathway in cancer and PIK3Copathies.
The PI3K-AKT pathway is another major cellular sensor of growth factor stimuli, which can be regulated by upstream receptors and RAS proteins (FIG. 4c). A central component of the pathway is the lipid kinase PI3K, encoded by the PIK3CA, PIK3CB, PIK3CG and PIK3CD genes; while PI3Kα and PI3Kβ are widely expressed in most tissues, the other isoforms are restricted to immune cell lineages125,126. PI3K activation results in the activation of dowsntream AKT kinases, among others, to promote cell survival127,128. Gain-offunction mutations in PIK3CA were described in different tumour types and have been characterized at the cellular, biochemical and structural levels. These mostly occur at two hotspots found at the helical and kinase domains of the protein, resulting in increased cell growth, proliferation, survival and transformation when expressed in non-malignant cells13,129. While alterations in the helical domain have been shown to interfere with the inhibitory interaction of the regulatory subunit p85 (encoded by PIK3R1), alterations in the catalytic domain appear to promote kinase activity by increasing substrate availability14. In cancer, PIK3CA is the second most common oncogene and is particularly frequent in breast carcinomas, endometrial adenocarcinoma, head and neck tumours, and colorectal and bladder cancer, among other cancers130. An emerging number of congenital disorders characterized by overgrowth and vascular malformations have also been found to harbour monogenic mutations in PIK3CA. These disorders include congenital lipomatous overgrowth, vascular malformations, epidermal nevi, scoliosis syndrome (CLOVES), Klippel-Trenaunay syndrome, megalencephaly-capillary malformation syndrome, hemihypertrophy with multiple lipomatosis syndrome and many others with similar phenotypes131–134. All these syndromes are likely the result of different degrees of PIK3CA mutation mosaicism and, therefore, have been grouped under the umbrella term 'PIK3CA-related overgrowth spectrum' (PROS). While these somatic events occur in a postzygotic manner, the affected tissue lineage and the time of the mutation will determine the extent of the lesion, ranging from severe forms of CLOVES to localized overgrowth such as megadactyly, a disproportionate growth of one or multiple fingers135 Somatic mutations in PIK3CA have also been described in vascular malformations, including venous and lymphatic malformations. In these lesions, the mutation is found only in endothelial cells, which account for only a small fraction of the whole lesion. This observation highlights the effect of mutant cells on the surrounding cell populations, since these lesions often contain an increased number of non-mutant perivascular cells (that is, vascular smooth muscle cells)46,136–138. Germline PIK3CA mutations in the main hotspots have never been reported in humans, probably due to embryonic lethality. However, other less common mutations may be associated with a disorder resembling Cowden syndrome, which is classically caused by mutations in PTEN, which encodes the phosphatase that antagonizes PI3K enzymatic function139,140. Such PIK3CA mutations are weakly active, in contrast to the mutations found in patients with PROS. Strong germline gain-of-function mutations in PIK3CD (mostly E1021K) can be found in patients with a specific immunodeficiency, which has now been termed 'activated PI3Kd syndrome'. Given the restricted expression of PI3Kδ to lymphocytes, strong mutations appear to be compatible with embryonic development and have been seen inherited in many generations with an autosomal dominant segregation141. Somatic mutations in PIK3CD are also seen in a subset of patients with diffuse large B cell lymphoma, revealing the oncogenic potential of these alleles, and some individuals with activated PI3Kδ syndrome have developed different forms of B cell lymphoma142.
The most common AKT1 mutant is the E17K variant, which promotes constitutive membrane targeting as a result of the charge switch at the phosphoinositidebinding domain143. Analogous mutations have also been described in the AKT2 and AKT3 genes, although at a lower frequency. AKT1E17K mutations are mostly found in breast and uterine endometrioid carcinomas and have been shown to be transforming in cell culture assays. High-degree mosaicism of AKT1E17K leads to Proteus syndrome, a rare and severe progressive disorder characterized by asymmetric overgrowth of bone and adipose tissue, as well as vascular malformations and predisposition to benign and malignant tumours144. Germline heterozygous E17K mutations in the AKT2 gene have been shown in two individuals with hypoglycaemia and mild overgrowth, AKT3E17K somatic mutations are found in children with hemimegalencephaly and germline mutations are found in syndromic diffuse megalencephaly133,145. The phenotypic difference of these mutations suggests non-redundant roles between the AKT isoforms. Given the overlapping clinical phenotypes and the similarity to the RASopathies, we propose the term se 'PIK3Copathies' for all these disorders.
Modelling and new therapies
Genetically engineered mouse models have been used successfully to model the effects of germline and somatic mutations and/or mosaicism, using various approaches (BOX 2). One can predict that the use of these tools is highly convenient when one is addressing fundamental questions in the context of oncoprotein biology, including cell lineage tracing, cell competition between wild-type and mutant clones, and cell-autonomous versus cell-non-autonomous effects of the oncoprotein. With use of these approaches, many mouse models for either cancer or congenital disorders driven by oncoproteins have been described in the literature (TABLE 1). As hypothesized by Happle (BOX 1), many of the 'strong' oncogenes are incompatible with embryonic development, as demonstrated in mice carrying such alleles in line the germline. Examples of these include the KrasG12D, Pik3caH1047R, GnasR201H and Akt1E17K variants41,43,146,147. These same alleles have been successfully used to create models by means of somatic recombination in specific lineages. The use of animal models is not restricted to mice, since recent reports have described novel models using zebrafish that faithfully recapitulate some of the phenotypes of congenital disorders148. For instance, the cardiovascular defects and craniofacial dysmorphia observed in some RASopathies have been modelled in zebrafish149.
Box 2 |. Genetically engineered mouse models to study oncoproteins.
The Cre-loxP system allows us to conditionally express oncoproteins somatically, while CRISPR-Cas9 facilitates the generation of germline mutations by editing mouse zygotes214–216. In the Cre-loxP system, insertion of a loxP-STOP-loxP cassette in front of the oncogene prevents its expression until it is removed by Cre recombinase, for example in the KrasG12D conditional mouse model43. This strategy is not recommended when heterozygous compound mice exhibit a detrimental phenotype, since the conditional gene is a null allele. To overcome this limitation, many investigators have relied on the so-called safe locus (that is, Rosa26)217,218. However, the abnormal expression of the oncoprotein can lead to artefactual phenotypes. Alternatively, conditional knock-in mice allow expression of oncoproteins in their endogenous locus; the wild-type gene is normally expressed but, on Cre recombination, the mutant allele encoded in a downstream minigene replaces the endogenous gene. This is particularly useful in mutations that are found in the last coding exons, such as in the Pik3caH1047R mouse model219. To model somatic mosaicism, one approach is the use of latent alleles, which are based on the hit-and-run gene targeting technology and spontaneously recombine in vivo169. CreER mice express a fusion between Cre recombinase and a tamoxifen-dependent, but oestrogen-resistant, oestrogen receptor. Here, the degree of mosaicism can be dependent on the tamoxifen concentration achieved in the tissues of interest or the time at which recombination was induced220. In regard to modelling clinically relevant mosaicisms, it is important to highlight the importance of CreER strains that are specifically expressed during germ layer formation and can be used to express oncoproteins in the mesoderm, ectoderm or endoderm. Refining such strains will provide a powerful tool to replicate mosaicism in a more faithful manner. When one is phenocopying mosaicism, genetically engineered mouse models also provide a remarkable opportunity to track mutant cells by leveraging the use of reporter strains. This can be achieved either by use of a ubiquitous conditional reporter, such as fluorescent proteins, or, ideally, by insertion of the reporter downstream of the oncogene of interest42.
Table 1 |.
Genetically engineered mouse models to study cancer or congenital disorders
| Oncoprotein | Expression | Strain | Phenotype | Notes | Refs |
|---|---|---|---|---|---|
| KRAS | Pancreas | Pdx1–Cre, Ptf1–Cre, elastase–Cre; KrasG12D/G12V | PanIN | Develops PDAC on Tp53 deletion or pancreatitis; mice with KrasA146T fail to develop PanIN | 16,43,62,167,168 |
| Lung | Adeno–Cre, CCSP–Cre; KrasG12D/G12V | Lung adenoma | Progression to adenocarcinoma | 42,43,59,169 | |
| Muscle | Adeno–Cre; KrasG12D | RT | Develops rhabdomyosarcoma when Tp53 is inactivated | 60 | |
| Myeloid cells | Mx1–Cre; KrasG12D/A146T | Myeloproliferative disorder | NR | 16,170 | |
| Intestinal epithelia | Fabp1–Cre; KrasG12D/A146T/G13D | Hyperplasia | Develops colon adenomas and adenocarcinoma on Apc loss | 16,171,172 | |
| Germline | KrasG12D/G12V | Lethal | Mice surviving germline KrasG12V recombination were found to be mosaic | 42,43 | |
| Germline (weak) | KrasV14I | Noonan syndrome | NR | 150 | |
| HRAS | Bladder | Uroplakin II–HrasQ61L | Urothelial papillary tumours | NR | 173 |
| Germline | HrasG12V | Costello syndrome | HRASG12V variant is uncommon in patients with Costello syndrome (lethal); papillomas and angiosarcomas are frequent in adults | 174 | |
| Germline (weak) | HrasG12S | Costello syndrome | NR | 175 | |
| Endothelial cells | Cdh5–tTA; tetO–HrasG12V | Cerebrovascular malformation | HRASG12V variant is uncommon in arteriovenous malformations | 176 | |
| Melanocytes | Tyr–HrasG12V | Melanoma | NR | 177 | |
| NRAS | Melanocytes | Tyr–NrasQ61K | Melanoma | Cooperates with p16INK4A loss | 93,178 |
| Myeloid cells | Mx1–Cre; NrasG12D | Indolent myeloproliferative disorder | NR | 179 | |
| Intestinal epithelia | Fabp1–Cre; NrasG12D | RT | Develops tumours on occurrence of DSS-induced colitis | 171,180 | |
| RIT1 | Germline | Rit1M90I, Rit1A57G | Noonan syndrome | NR | 136,181 |
| GNAQ | Melanoblast | Mitf–Cre; R26–GnaqQ209L | Uveal melanoma | NR | 182 |
| GNA11 | Melanocyte | Tyr–CreER, R26–Gna11Q209L | Cutaneous, uveal and leptomeningeal melanoma | NR | 183 |
| GNAS | Transgenic | EF1-PGK–GnasR201H | Fibrous dysplasia | Human patients exhibit mosaicism | 184 |
| Germline | Sox2–Cre; GnasR201H | Lethal | NR | 146 | |
| Limb bud | Prrx1–Cre;GnasR201H | Fibrous dysplasia | NR | 146 | |
| RAF1 | Germline | Raf1L613V | Noonan syndrome | NR | 151 |
| BRAF | Lung | Adeno–Cre; BrafV600E | Lung adenoma | Progression to adenocarcinoma | 185 |
| Melanocytes | Tyr–CreER; BrafV600E | Melanocytic hyperplasia | NR | 186 | |
| Thyroid | Thyro–CreER; BrafV600E | Papillary thyroid carcinoma | NR | 187 | |
| Germline | BrafV600E | Lethal | NR | 185 | |
| Germline (weak) | BrafQ241R | Cardiofaciocutaneous syndrome | NR | 188 | |
| Pancreas | Pdx1–CreER; BrafV600E | PanIN | Develops PDAC on Tp53R270H mutation | 189 | |
| Intestinal epithelia | Villin–Cre; BrafV600E | Hyperplasia | NR | 190 | |
| MEK1 | Germline | Map2k1Y130C | Cardiofaciocutaneous syndrome | NR | 191 |
| PI3Kα | Mesoderm | T–CreER; Pik3caH1047R | Venous malformation | No overgrowth observed | 137 |
| Ubiquitous | CAG–CreER; R26–Pik3caH1047R | Venous malformation | NR | 136 | |
| Endothelial cells | Tie2–Cre; Pik3caH1047R | Lethal | NR | 41,136 | |
| Neural progenitor | GFAP–Cre; Pik3caH1047R | Megalencephaly–capillary malformation syndrome | NR | 192 | |
| Breast | MMTV–Cre, Wap1–Cre; Pik3caH1047R | Breast adenocarcinoma | NR | 58,193 | |
| Ubiquitous transgene | CAG–CreER; Pik3ca* | PIK3CA-related overgrowth spectrum | NR | 153 | |
| Pancreas | Ptf1–Cre; R26–Pik3caH1047R | PanIN | No PanIN when Pdx1–CreER is used | 189,194 | |
| PI3Kδ | Germline | Pik3cdE1020K | Primary immunodeficiency | Pik3cdE1020K is equivalent to human PIK3CDE1021K | 195 |
| AKT1 | Mosaic | R26–CreER; Akt1E17K | Proteus syndrome | NR | 147 |
| Breast | MMTV–tTA; tetO–Akt1E17K | Mammary gland hyperplasia | NR | 196 | |
| FGFR3 | Germline | Fgfr3G380R | Achondroplasia | NR | 197 |
| Epidermis | K5–Fgfr3S249C | Epidermal tumours | NR | 198 | |
| Bladder | Uroplakin II–Cre; Fgfr3K644E | RT | NR | 199 | |
| KIT | Germline | KitV658Δ | Familial gastrointestinal stromal tumours | NR | 200 |
| ALK | Germline | Actb–Cre; AlkF1174L | RT | NR | 201 |
| Neural crest | P0–Cre; AlkF1174L | Proliferation sympathetic ganglion progenitors | NR | 202 | |
| CD4+ cells | CD4–NPM/ALK | T cell lymphoma and plasma cell tumours | NR | 203 | |
| Lung | SPC–EML/ALK | Lung adenocarcinoma | NR | 204 | |
| EGFR | Lung | CCSP–rtTA; tetO–EgfrL858R | Lung adenocarcinoma | Similar phenotype in EgfrΔL747-S752 | 205 |
ALK; anaplastic lymphoma kinase; DSS, dextran sodium sulfate; EGFR, epidermal growth factor receptor; NR, not reported; PanIN, pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma; RT, resistant to transformation; tetO, tetracycline operator; tTA, tetracycline transactivator.
One of the most helpful uses of these models is the development of experimental therapies that are able to reverse or ameliorate the condition. In this context, some work has been done to determine the window of intervention, dosing and scheduling of targeted therapies in mouse models of congenital disorders such as Noonan syndrome, achondroplasia and vascular malformations136,150–153. Because most of the oncoproteins driving congenital disorders are considered drug targets for treating sporadic cancers, an extensive arsenal of drugs is clinically available or under development. The challenges in treating congenital disorders include determining the window during development in which intervention is expected to be effective, as well as dosing requirements and side effects in children, who may require long-term treatment. We describe some cases that exemplify how targeted therapies initially designed to treat cancer have the potential to become standard-of-care treatment for congenital disorders.
In achondroplasia, children need to be treated before the growth plate closes and is replaced by solid bone during adolescence154. Abnormal MAPK signalling driven by activated FGFR3 is suppressed by C-type natriuretic peptide155, and a phase III clinical trial is under way with vosoritide, an analogue of C-type natriuretic peptide that has caused increased growth in treated individuals156. Preliminary results in mice suggest that this treatment could be expanded to other congenital disorders with growth deficit, such as cardiofaciocutaneous syndrome157. Direct inhibition of FGFR3 by a small-molecule drug, infigratinib, will soon be investigated in phase II clinical trials. Infigratinib is also being tested in cancers driven by mutant FGFR and, while it is likely that treatment of these tumours will require complete inhibition of FGFR signalling to promote tumour regression158, reduced FGFR signalling might be sufficient to reverse the effects of hyperactive FGFR3 in achondroplasia, as proposed in a study that used a mouse model of dwarfism152. If so, lower doses of the drug are expected to be efficacious in this indication, with fewer side effects.
In the context of RASopathies, use of the MEK inhibitor selumetinib has recently led to beneficial responses in children with plexiform neurofibromas159,160. Individuals with Costello syndrome, cardiofaciocutaneous syndrome or Noonan syndrome might also benefit from treatment with MEK inhibitors, or other inhibitors of the MAPK pathway, as described in some preclinical trials using model organisms149,151,161. Recently, two infants with RIT1- driven Noonan syndrome who developed severe early-onset hypertrophic cardiomyopathy were treated with off-label trametinib, a potent allosteric MEK inhibitor. In both cases, the cardiac phenotype reversed, suggesting that MEK inhibitors could be efficacious in patients with RASopathy with extensive cardiovascular involvement162. Farnesyltransferase inhibitors block post-translational processing of HRAS at the plasma membrane. Tipifarnib, a farnesyltransferase inhibitor, is already in late-stage clinical trials for HRAS-mutant sporadic cancers and could, in principle, be tested in Costello syndrome. However, these syndromes, are characterized by multiple developmental and learning disorders, rather than focal tumours. Therefore, the clinical end points and the window of opportunity for these treatments are unclear163.
For PIK3Copathies, two therapeutic strategies are paving the way for targeted therapies in congenital disorders. First, the AKT inhibitor miransertib has proven preliminarily to be safe and active in a cohort of patients with Proteus syndrome, and patients are currently being enrolled for a registrational phase III study164,165. Second, a study involving 19 patients with PROS has recently reported dramatic therapeutic effects with low doses of the PI3K inhibitor alpelisib, which is approved by the FDA for treatment of metastatic breast cancer153. This study has led to a phase III clinical trial that will evaluate the efficacy of the compound in a larger cohort of patients with PROS. In oncology trials, AKT inhibitors and PI3K inhibitors have both shown adverse events, with the most significant being hyperglycaemia166 However, because the doses given to patients with PROS are expected to be lower, such secondary effects might not be an issue, even in long-term treatments. An alternative to overcome these problems, especially for patients with isolated vascular malformations, is topical treatment. As previously reported in preclinical models, this approach could help deliver high local doses without systemic toxicity136. Although many other efforts are being pursued in the field, these examples offer some insight into promising therapies for many congenital disorders.
Conclusions
The latest advances in next-generation sequencing and clinical genetics have challenged the dogma that defines oncoproteins as entities capable of transforming quiescent tissues. The expression of oncoproteins is far more common than we anticipated; they are present in many histologically normal tissues without exhibiting any phenotype but are also drivers of particular congenital disorders as well as cancer. Therefore, it is important to acknowledge the versatility of oncoproteins and begin to study them in a more comprehensive manner. At the organismal level, the study of oncoproteins will likely shed light on the key processes underlying embryonic development and tissue homeostasis. Moreover, identifying genes involved in phenotypically similar syndromes can lead to the discovery of novel components and regulators of signalling pathways. Among some outstanding questions, it is tempting to propose that elucidating the mechanisms that promote and restrict oncogenesis in certain tissues will be of great interest. This could be addressed through the use of -omic approaches that reveal the effect of oncoprotein expression in different tissues (that is, transforming versus non-transforming). One could also undertake genetic analysis in patients who have congenital disorders that are prone to neoplasia by comparing malignant and healthy tissues. In this context, genetically engineered mouse models can be of great interest. Finally, it is of vital importance to keep developing therapies that inhibit oncoprotein function, which could be used not only for patients with cancer but also for patients with congenital disorders.
Acknowledgements
The authors thank all the scientists who have contributed to this exciting field and apologize to those colleagues they were unable to cite. P.C. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This research was supported by the Thrasher Research Fund Early Career Award programme (to P.C.), the University of California, San Francisco Program for Breakthrough Biomedical Research Independent Postdoctoral Research Fellow (to P.C.) and the NIH/NCI grant R35CA197709-01 (to F.M.).
Footnotes
Competing interests
P.C. is a co-founder and advisory board member of Venthera. F.M. is a consultant for Aduro Biotech, Amgen, Daiichi, Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, PellePharm, Pfizer, PMV Pharma, Portola Pharmaceuticals and Quanta Therapeutics, has received research grants from Daiichi and Gilead Sciences and is a consultant for and cofounder of BridgeBio Pharma, DNAtrix, Olema Pharmaceuticals, and Quartz.
References
- 1.Adams DR & Eng CM Next-generation sequencing to diagnose suspected genetic disorders. N. Engl. J. Med 379, 1353–1362 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bamshad MJ et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet 12, 745–755 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Erickson RP Somatic gene mutation and human disease other than cancer: an update. Mutat. Res 705, 96–106 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Martincorena I et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).In this study, ultradeep sequencing of skin eyelids reveals the presence of an elevated number of somatic mutations, including many cancer-associated mutations.
- 5.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yizhak K et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haigis KM, Cichowski K & Elledge SJ Tissue-specificity in cancer: the rule, not the exception. Science 363, 1150–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Haigis KM KRAS alleles:the devil is in the detail. Trends Cancer 3, 686–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Balmain A & Counter CM A model for RAS mutation patterns in cancers: finding the sweet spot. Nat. Rev. Cancer 18, 767–777 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Schneider G, Schmidt-Supprian M, Rad R & Saur D Tissue-specific tumorigenesis: context matters. Nat Rev. Cancer 17, 239–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller S et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 554, 62–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estep AL, Tidyman WE, Teitell MA, Cotter PD & Rauen KA HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A 140, 8–16 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Zhao L & Vogt PK Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl Acad. Sci. USA 105, 2652–2657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke JE, Perisic O, Masson GR, Vadas O & Williams RL Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA). Proc. Natl Acad. Sci. USA 109, 15259–15264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs GA et al. Atypical KRASG12R mutant is impaired in PI3K signaling and macropinocytosis in pancreatic cancer. Cancer Discov 10, 104–123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulin EJ et al. Tissue-specific oncogenic activity of KRASA146T. Cancer Discov 9, 738–755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavahan WA, Gaskell E & Bernstein BE Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwahara T et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 14, 439–449 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Vernersson E et al. Characterization of the expression of the ALK receptor tyrosine kinase in mice. Gene Expr. Patterns 6, 448–461 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ Principles of tumor suppression. Cell 116, 235–246 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Schwartz R & Schaffer AA The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet 18, 213–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merino MM, Levayer R & Moreno E Survival of the fittest: essential roles of cell competition in development, aging, and cancer. Trends Cell Biol 26, 776–788 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Bowling S, Lawlor K & Rodriguez TA Cell competition: the winners and losers of fitness selection. Development 146, dev167486 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Hogan C et al. Characterization of the interface between normal and transformed epithelial cells. Nat. Cell Biol 11, 460–467 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Nelson CM & Bissell MJ Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol 22, 287–309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown S et al. Correction of aberrant growth preserves tissue homeostasis. Nature 548, 334–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineda CM et al. Hair follicle regeneration suppresses Ras-driven oncogenic growth. J. Cell Biol 218, 3212–3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greten FR & Grivennikov SI Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swann JB & Smyth MJ Immune surveillance of tumors. J. Clin. Invest 117, 1137–1146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Little JB Radiation carcinogenesis. Carcinogenesis 21, 397–404(2000). [DOI] [PubMed] [Google Scholar]
- 31.Loeb LA & Harris CC Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res 68, 6863–6872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steen HB The origin of oncogenic mutations: where is the primary damage? Carcinogenesis 21, 1773–1776(2000). [DOI] [PubMed] [Google Scholar]
- 33.Petljak M et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y et al. Germ line mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet 37, 1038–1040 (2005).This study identifies germ line HRAS mutations as the causative factor for Costello syndrome, a RASopathy. The authors show that mutations in codon 12 are frequent in these patients, similar to HRAS mutations found in certain human sporadic tumours.
- 35.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goriely A & Wilkie AOM Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am. J. Hum. Genet 90, 175–200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maher GJ et al. Visualizing the origins of selfish de novo mutations in individual seminiferous tubules of human testes. Proc. Natl Acad. Sci. USA 113, 2454–2459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maher GJ et al. Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes. Genome Res 28, 1779–1790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiang R et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell 78, 335–342 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Hare LM et al. Heterozygous expression of the oncogenic Pik3caH1047R mutation during murine development results in fatal embryonic and extraembryonic defects. Dev. Biol 404, 14–26 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Guerra C et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4, 111–120 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Tuveson DA et al. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5, 375–387 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Schubbert S et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet 38, 331–336 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Rauen KA et al. Molecular and functional analysis of a novel MEK2 mutation in cardio-facio-cutaneous syndrome: transmission through four generations. Am. J. Med. Genet. A 152A, 807–814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Laguna L et al. Somatic activating mutations in PIK3CA cause generalized lymphatic anomaly. J. Exp. Med 216, 407–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobs KB et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet 44, 651–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laurie CC et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet 44, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forsberg LA et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet 46, 624–628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Happle R Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J. Am. Acad. Dermatol 16, 899–906 (1987).In this article, the author hypothesizes the reason for which certain dermatological conditions are mosaicisms driven by mutations that do not follow a Mendelian inheritance pattern.
- 51.Groesser L et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat. Genet 44, 783–787 (2012).In this article, oncogenic HRAS and KRAS mutations are identified to be the cause of a syndromic mosaicism that is characterized by the presence of sebaceous nevi.
- 52.Bolognia JL, Orlow SJ & Glick SA Lines of Blaschko. J. Am. Acad. Dermatol 31, 157–190 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Hafner C et al. Oncogenic PIK3CA mutations occur in epidermal nevi and seborrheic keratoses with a characteristic mutation pattern. Proc. Natl Acad. Sci. USA 104, 13450–13454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hafner C et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J. Clin. Invest 116, 2201–2207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafner C et al. Keratinocytic epidermal nevi are associated with mosaic RAS mutations. J. Med. Genet 49, 249–253 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Biesecker LG & Spinner NB A genomic view of mosaicism and human disease. Nat. Rev. Genet 14, 307–320 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Fernández LC, Torres M & Real FX Somatic mosaicism: on the road to cancer. Nat. Rev. Cancer 16, 43–55 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Adams JR et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res 71, 2706–2717 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Jackson EL Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirsch DG et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat. Med 13, 992–997 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Cavenee WK et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305, 779–784 (1983).This study identifies the acquisition of secondary hits in the RB1 gene in patients with retinoblastoma, confirming the Knudson hypothesis.
- 62.Guerra C et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Ayturk UM et al. Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am. J. Hum. Genet 98, 789–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirley MD et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med 368, 1971–1979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai X et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep 8, 1280–1289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Genovese G et al. Clonal hematopoiesis and bloodcancer risk inferred from blood DNA sequence. N. Engl. J. Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yokoyama A et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Anglesio MS et al. Cancer- associated mutations in endometriosis without cancer. N. Engl. J. Med 376, 1835–1848 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suda K et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep 24, 1777–1789 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Hafner C, Toll A & Real FX HRAS mutation mosaicism causing urothelial cancer and epidermal nevus. N. Engl. J. Med 365, 1940–1942 (2011).In this case report, the authors describe the presence of FGFR3-mutant mosaicism giving rise to both urothelial cancer and epidermal nevi.
- 71.Doucet ME, Bloomhardt HM, Moroz K, Lindhurst MJ & Biesecker LG Lack of mutationhistopathology correlation in a patient with Proteus syndrome. Am. J. Med. Genet. A 170, 1422–1432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanchez-Vega F et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 173, 321–337.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lemmon MA & Schlessinger J Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson SC, Tynan JA & Donoghue DJ RTK mutations and human syndromes when good receptors turn bad. Trends Genet 16, 265–271 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Bell DW et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat. Genet 37, 1315–1316 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Oxnard GR, Nguyen K-SH & Costa DB Germline mutations in driver oncogenes and inherited lung cancer risk independent of smoking history. J. Natl Cancer Inst 106, djt361 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto H et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J. Natl Cancer Inst 106, djt338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Janoueix-Lerosey I et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Mossé YP et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morris S et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 263, 1281 (1994). [DOI] [PubMed] [Google Scholar]
- 81.Mulligan LM RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 14, 173–186 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Chompret A et al. PDGFRA germline mutation in a family with multiple cases of gastrointestinal stromal tumor. Gastroenterology 126, 318–321 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Hirota S et al. Gain- of- function mutations of c- kit in human gastrointestinal stromal tumors. Science 279, 577–580 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Nishida T et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat. Genet 19, 323–324 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Farrell B & Breeze AL Structure, activation and dysregulation of fibroblast growth factor receptor kinases: perspectives for clinical targeting. Biochem. Soc. Trans 46, 1753–1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muenke M et al. A unique point mutation in the fibroblast growth factor receptor 3 gene (FGFR3) defines a new craniosynostosis syndrome. Am. J. Hum. Genet 60, 555–564 (1997). [PMC free article] [PubMed] [Google Scholar]
- 87.Tavormina PL et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat. Genet 9, 321–328 (1995). [DOI] [PubMed] [Google Scholar]
- 88.Reardon W et al. Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat. Genet 8, 98–103 (1994). [DOI] [PubMed] [Google Scholar]
- 89.Hafner C et al. High frequency of FGFR3 mutations in adenoid seborrheic keratoses. J. Invest. Dermatol 126, 2404–2407 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Muenke M et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat. Genet 8, 269–274 (1994). [DOI] [PubMed] [Google Scholar]
- 91.Wilkie AO et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet 9, 165–172 (1995). [DOI] [PubMed] [Google Scholar]
- 92.Simanshu DK, Nissley DV & McCormick F RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burd CE et al. Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov 4, 1418–1429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nikolaev SI et al. Somatic activating KRAS mutations in arteriovenous malformations of the brain. N. Engl. J. Med 378, 250–261 (2018).In this work, the authors identify the presence of cancer-associated KRAS mutations in arteriovenous malformations of the brain.
- 95.Al-Olabi L et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J. Clin. Invest 128, 1496–1508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peacock JD et al. Oculoectodermal syndrome is a mosaic RASopathy associated with KRAS alterations. Am. J. Med. Genet. A 167, 1429–1435 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Hafner C & Groesser L Mosaic RASopathies. Cell Cycle 12, 43–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Groesser L et al. Phacomatosis pigmentokeratotica is caused by a postzygotic HRAS mutation in a multipotent progenitor cell. J. Invest. Dermatol 133, 1998–2003 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Levinsohn JL et al. Somatic HRAS p.G12S mutation causes woolly hair and epidermal Nevi. J. Invest. Dermatol 134, 1149–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kinsler VA et al. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J. Invest. Dermatol 133, 2229–2236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Charbel C et al. NRAS mutation is the sole recurrent somatic mutation in large congenital melanocytic nevi. J. Invest. Dermatol 134, 1067–1074 (2014). [DOI] [PubMed] [Google Scholar]
- 102.Lim YH et al. Multilineage somatic activating mutations in HRAS and NRAS cause mosaic cutaneous and skeletal lesions, elevated FGF23 and hypophosphatemia. Hum. Mol. Genet 23, 397–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lim YH et al. Cutaneous skeletal hypophosphatemia syndrome (CSHS) is a multilineage somatic mosaic RASopathy. J. Am. Acad. Dermatol 75, 420–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Florenzano P, Gafni RI & Collins MT Tumor-induced osteomalacia. Bone Rep 7, 90–97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang H et al. Somatic activating mutations in MAP2K1 cause melorheostosis. Nat. Commun 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tidyman WE & Rauen KA The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev 19, 230–236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goodwin AF et al. Craniofacial and dental development in Costello syndrome. Am. J. Med. Genet. A 164A, 1425–1430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gripp KW et al. Costello syndrome: clinical phenotype, genotype, and management guidelines. Am. J. Med. Genet. A 179, 1725–1744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gripp KW Tumor predisposition in Costello syndrome. Am. J. Med. Genet. C. Semin. Med Genet 137C, 72–77 (2005). [DOI] [PubMed] [Google Scholar]
- 110.Tartaglia M et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet 29, 465–468 (2001). [DOI] [PubMed] [Google Scholar]
- 111.Tartaglia M et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet 39, 75–79 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Aoki Y et al. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet 93, 173–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Razzaque MA et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet 39, 1013–1017 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Berger AH et al. Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene 33, 4418–4423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lyons J et al. Two G protein oncogenes in human endocrine tumors. Science 249, 655–659 (1990). [DOI] [PubMed] [Google Scholar]
- 116.O'Hayre M et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 13, 412–424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weinstein LS et al. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N. Engl. J. Med 325, 1688–1695 (1991).In this article, GNAS is identified as the causative gene for McCune–Albright syndrome. The mutations in this gene are similar to those in endocrine tumours.
- 118.Albright F, Butler AM, Hampton AO & Smith P Syndrome characterized by osteitis fibrosa disseminata, areas of pigmentation and endocrine dysfunction, with precocious puberty in females. N. Engl. J. Med 216, 727–746 (1937). [Google Scholar]
- 119.Song Z-J et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res 26, 1255–1259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu J et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Transl. Med 3, 92ra66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu Q & Shokat KM Disease-causing mutations in the G protein Gas subvert the roles of GDP and GTP. Cell 173, 1254–1264.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Raamsdonk CD et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue nevi. Nature 457, 599–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Van Raamsdonk CD et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med 363, 2191–2199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klebanov N et al. Use of targeted next-generation sequencing to identify activating hot spot mutations in cherry angiomas. JAMA Dermatol 155, 211–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bilanges B, Posor Y & Vanhaesebroeck B PI3K isoforms in cell signalling and vesicle trafficking. Nat. Rev. Mol. Cell Biol 20, 515–534 (2019). [DOI] [PubMed] [Google Scholar]
- 126.Fruman DA et al. The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Manning BD & Toker A AKT/PKB signaling: navigating the network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Saxton RA & Sabatini DM mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carson JD et al. Effects of oncogenic p110α subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem. J 409, 519–524 (2008). [DOI] [PubMed] [Google Scholar]
- 130.Samuels Y et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004). [DOI] [PubMed] [Google Scholar]
- 131.Madsen RR, Vanhaesebroeck B & Semple RK Cancer-associated PIK3CA mutations in overgrowth disorders. Trends Mol. Med 24, 856–870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kurek KC et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet 90, 1108–1115 (2012).In this work, the authors identify mosaic cancer-associated PIK3CA mutations in patients with CLOVES, a syndrome that is now part of PROS.
- 133.Rivière J-B et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet 44, 934–940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lindhurst MJ et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet 44, 928–933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Keppler-Noreuil KM et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am. J. Med. Genet. A 164A, 1713–1733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Castel P et al. Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med 8, 332ra42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Castillo SD et al. Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med 8, 332ra43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luks VL et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr 166, 1048–1054.e1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Orloff MS et al. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am. J. Hum. Genet 92, 76–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liaw D et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet 16, 64–67 (1997). [DOI] [PubMed] [Google Scholar]
- 141.Angulo I et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science 342, 866–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kracker S et al. Occurrence of B-cell lymphomas in patients with activated phosphoinositide 3-kinase d syndrome. J. Allergy Clin. Immunol 134, 233–236.e3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carpten JD et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007). [DOI] [PubMed] [Google Scholar]
- 144.Lindhurst MJ et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med 365, 611–619 (2011).Sequencing of samples from a patient with Proteus syndrome reveals the presence of mosaic cancer-associated hotspot mutation AKT1E17K, elucidating the causative gene in this syndrome.
- 145.Hussain K et al. An activating mutation of AKT2 and human hypoglycemia. Science 334, 474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khan SK et al. Induced Gnas R201H expression from the endogenous Gnas locus causes fibrous dysplasia by up-regulating Wnt/β-catenin signaling. Proc. Natl Acad. Sci. USA 115, E418–E427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lindhurst MJ et al. A mouse model of proteus syndrome. Hum. Mol. Genet 28, 2920–2936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lieschke GJ & Currie PD Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet 8, 353–367 (2007). [DOI] [PubMed] [Google Scholar]
- 149.Anastasaki C, Rauen KA & Patton EE Continual low-level MEK inhibition ameliorates cardio-facio-cutaneous phenotypes in zebrafish. Dis. Model. Mech 5, 546–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hernández-Porras I et al. K-RasV14I recapitulates Noonan syndrome in mice. Proc. Natl Acad. Sci. USA 111, 16395–16400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu X et al. MEK-ERK pathway modulation ameliorates disease phenotypes in a mouse model of Noonan syndrome associated with the Raf1L613V mutation. J. Clin. Invest 121, 1009–1025 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Komla-Ebri D et al. Tyrosine kinase inhibitor NVP-BGJ398 functionally improves FGFR3-related dwarfism in mouse model. J. Clin. Invest 126, 1871–1884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Venot Q et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 558, 540–546 (2018).In this work, the authors describe 19 patients with PROS treated under compassionate use with an inhibitor targeting PI3Kα, which was developed and approved to treat metastatic breast cancer.
- 154.Pauli RM Achondroplasia: a comprehensive clinical review. Orphanet J. Rare Dis 14, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wendt DJ et al. Neutral endopeptidase-resistant C-type natriuretic peptide variant represents a new therapeutic approach for treatment of fibroblast growth factor receptor 3-related dwarfism. J. Pharmacol. Exp. Ther 353, 132–149 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Savarirayan R et al. C-type natriuretic peptide analogue therapy in children with achondroplasia. N. Engl. J. Med 381, 25–35 (2019). [DOI] [PubMed] [Google Scholar]
- 157.Inoue S-I, Morozumi N, Yoshikiyo K, Maeda H & Aoki Y C-type natriuretic peptide improves growth retardation in a mouse model of cardio-facio-cutaneous syndrome. Hum. Mol. Genet 28, 74–83 (2019). [DOI] [PubMed] [Google Scholar]
- 158.Javle M et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J. Clin. Oncol 36, 276–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dombi E et al. Activity of selumetinib in neurofibromatosis type 1–related plexiform neurofibromas. N. Engl. J. Med 375, 2550–2560 (2016).In this study, and in reference 160, the authors report the safety and efficacy profiles of the MEK1/MEK2 inhibitor selumetinib for the treatment of plexiform neurofibromas in patients with neurofibromatosis type 1.
- 160.Gross AM et al. Selumetinib in children with inoperable plexiform neurofibromas. N. Engl. J. Med 382, 1430–1442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rauen KA et al. Proceedings of the fifth international RASopathies symposium: when development and cancer intersect. Am. J. Med. Genet. A 176, 2924–2929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Andelfinger G et al. Hypertrophic cardiomyopathy in Noonan syndrome treated by MEK-inhibition. J. Am. Coll. Cardiol 73, 2237–2239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rauen KA The RASopathies. Annu. Rev. Genomics Hum. Genet 14, 355–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Keppler-Noreuil KM et al. Pharmacodynamic study of miransertib in individuals with Proteus syndrome. Am. J. Hum. Genet 104, 484–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leoni C et al. First evidence of a therapeutic effect of miransertib in a teenager with Proteus syndrome and ovarian carcinoma. Am. J. Med. Genet. A 179, 1319–1324 (2019). [DOI] [PubMed] [Google Scholar]
- 166.Rodon J & Tabernero J Improving the armamentarium of PI3K inhibitors with isoformselective agents: a new light in the darkness. Cancer Discov 7, 666–669 (2017). [DOI] [PubMed] [Google Scholar]
- 167.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS & Sandgren EP Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res 63, 2016–2019 (2003). [PubMed] [Google Scholar]
- 168.Gidekel Friedlander SY et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell 16, 379–389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Johnson L et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001). [DOI] [PubMed] [Google Scholar]
- 170.Braun BS et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl Acad. Sci. USA 101, 597–602 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Haigis KM et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet 40, 600–608 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johnson CW et al. Isoform-specific destabilization of the active site reveals a molecular mechanism of intrinsic activation of KRas G13D. Cell Rep 28, 1538–1550.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang Z-T et al. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene 20, 1973–1980 (2001). [DOI] [PubMed] [Google Scholar]
- 174.Schuhmacher AJ et al. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J. Clin. Invest 118, 2169–2179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Oba D et al. Mice with an oncogenic HRAS mutation are resistant to high-fat diet-induced obesity and exhibit impaired hepatic energy homeostasis. EBioMedicine 27, 138–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li Q-F, Decker-Rockefeller B, Bajaj A & Pumiglia K Activation of Ras in the vascular endothelium induces brain vascular malformations and hemorrhagic stroke. Cell Rep 24, 2869–2882 (2018). [DOI] [PubMed] [Google Scholar]
- 177.Chin L et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev 11, 2822–2834 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ackermann J et al. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res 65, 4005–4011 (2005). [DOI] [PubMed] [Google Scholar]
- 179.Li Q et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117, 2022–2032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Y et al. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer Discov 3, 294–307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Takahara S et al. New Noonan syndrome model mice with RIT1 mutation exhibit cardiac hypertrophy and susceptibility to β-adrenergic stimulationinduced cardiac fibrosis. EBioMedicine 42, 43–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang JL-Y, Urtatiz O & Van Raamsdonk CD Oncogenic G protein GNAQ induces uveal melanoma and intravasation in mice. Cancer Res 75, 3384–3397 (2015). [DOI] [PubMed] [Google Scholar]
- 183.Moore AR et al. GNA11 Q209L mouse model reveals RasGRP3 as an essential signaling node in uveal melanoma. Cell Rep 22, 2455–2468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Saggio I et al. Constitutive expression of GsαR201C in mice produces a heritable, direct replica of human fibrous dysplasia bone pathology and demonstrates its natural history. J. Bone Miner. Res 29, 2357–2368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dankort D et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 21, 379–384 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dankort D et al. BrafV600E cooperates with Pten loss to induce metastatic melanoma. Nat. Genet 41, 544–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Charles R-P, Iezza G, Amendola E, Dankort D & McMahon M Mutationally activated BRAFV600E elicits papillary thyroid cancer in the adult mouse. Cancer Res 71, 3863–3871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Inoue S-I et al. Activated Braf induces esophageal dilation and gastric epithelial hyperplasia in mice. Hum. Mol. Genet 26, 4715–4727 (2017). [DOI] [PubMed] [Google Scholar]
- 189.Collisson EA et al. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2, 685–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rad R et al. A genetic progression model of BrafV600E-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell 24, 15–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Aoidi R et al. Mek1Y130C mice recapitulate aspects of human cardio-facio-cutaneous syndrome. Dis. Model. Mech 11, dmm031278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Roy A et al. Mouse models of human PIK3CA-related brain overgrowth have acutely treatable epilepsy. eLife 4, e12703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Stratikopoulos EE et al. Mouse ER+/PIK3CAH1047R breast cancers caused by exogenous estrogen are heterogeneously dependent on estrogen and undergo BIM-dependent apoptosis with BH3 and PI3K agents. Oncogene 38, 47–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Eser S et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 23, 406–420 (2013). [DOI] [PubMed] [Google Scholar]
- 195.Avery DT et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J. Exp. Med 215, 2073–2095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mancini ML, Lien EC & Toker A Oncogenic AKT1E17K mutation induces mammary hyperplasia but prevents HER2-driven tumorigenesis. Oncotarget 7, 17301–17313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang Y et al. A mouse model for achondroplasia produced by targeting fibroblast growth factor receptor 3. Proc. Natl Acad. Sci. USA 96, 4455–4460 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Logié A et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum. Mol. Genet 14, 1153–1160 (2005). [DOI] [PubMed] [Google Scholar]
- 199.Ahmad I et al. K-Ras and β-catenin mutations cooperate with Fgfr3 mutations in mice to promote tumorigenesis in the skin and lung, but not in the bladder. Dis. Model. Mech 4, 548–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sommer G et al. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the Kit receptor tyrosine kinase. Proc. Natl Acad. Sci. USA 100, 6706–6711 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ono S, Saito T, Terui K, Yoshida H & Enomoto H Generation of conditional ALK F1174L mutant mouse models for the study of neuroblastoma pathogenesis. Genesis 57, e23323 (2019). [DOI] [PubMed] [Google Scholar]
- 202.Berry T et al. The ALKF1174L mutation potentiates the oncogenic Activity of MYCN in Neuroblastoma. Cancer Cell 22, 117–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chiarle R et al. NPM- ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood 101, 1919–1927 (2003). [DOI] [PubMed] [Google Scholar]
- 204.Soda M et al. A mouse model for EML4-ALK-positive lung cancer. Proc. Natl Acad. Sci. USA 105, 19893–19897 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Politi K et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 20, 1496–1510 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rous P A transmissible avian neoplasm. (Sarcoma of the common fowl.). J. Exp. Med 12, 696–705 (1910). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Halberstaedter L, Doljanski L & Tenenbaum E Experiments on the cancerization of cells in vitro by means of Rous sarcoma agent. Br. J. Exp. Pathol 22, 179–187 (1941). [Google Scholar]
- 208.Harvey JJ An unidentified virus which causes the rapid production of tumours in mice. Nature 204, 1104–1105 (1964). [DOI] [PubMed] [Google Scholar]
- 209.Stehelin D, Varmus HE, Bishop JM & Vogt PK DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260, 170–173 (1976). [DOI] [PubMed] [Google Scholar]
- 210.Santos E, Tronick SR, Aaronson SA, Pulciani S & Barbacid M T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB-and HarveyMSV transforming genes. Nature 298, 343–347 (1982). [DOI] [PubMed] [Google Scholar]
- 211.Parada LF, Tabin CJ, Shih C & Weinberg RA Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature 297, 474–478 (1982). [DOI] [PubMed] [Google Scholar]
- 212.Der CJ, Krontiris TG & Cooper GM Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc. Natl Acad. Sci. USA 79, 3637–3640 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Whitman M, Kaplan DR, Schaffhausen B, Cantley L & Roberts TM Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature 315, 239–242 (1985). [DOI] [PubMed] [Google Scholar]
- 214.Gurumurthy CB & Lloyd KCK Generating mouse models for biomedical research: technological advances. Dis. Model. Mech 12, dmm029462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kersten K, de Visser KE, van Miltenburg MH & Jonkers J Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med 9, 137–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Heyer J, Kwong LN, Lowe SW & Chin L Non- germline genetically engineered mouse models for translational cancer research. Nat. Rev. Cancer 10, 470–480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zambrowicz BP et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA 94, 3789–3794 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet 21, 70–71 (1999). [DOI] [PubMed] [Google Scholar]
- 219.Kinross KM et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Invest 122, 553–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.McLellan MA, Rosenthal NA & Pinto AR Cre-loxP-mediated recombination: general principles and experimental considerations. Curr. Protoc. Mouse Biol 7, 1–12 (2017). [DOI] [PubMed] [Google Scholar]