Abstract
SARS-CoV-2 pandemic is having a devastating effect on human lives. Recent reports have shown that majority of the individuals recovered from COVID-19 have serious health complications, which is going to be a huge economic burden globally. Given the wide-spread transmission of SARS-CoV-2 it is almost impossible to test every individual in densely populated countries. Recent reports have shown that sewage-based surveillance can be used as holistic approach to understand the spread of the pandemic within a population or area. Here we have estimated the spread of SARS-CoV-2 in the city of Hyderabad, India, which is a home for nearly 10 million people. The sewage samples were collected from all the major sewage treatment plants (STPs) and were processed for detecting the viral genome using the standard Reverse Transcription Polymerase Chain Reaction (RT-PCR) method. Interestingly, inlet samples of STPs were positive for SARS-CoV-2, while the outlets were negative, which indicates that the standard sewage treatment methods are efficient in eliminating the SARS-CoV-2 viral particles. Based on the detected viral gene copies per litre and viral particle shedding per individual, the total number of individuals exposed to SARS-CoV-2 was estimated. Through this study we suggest that sewage-based surveillance is an effective approach to study the infection dynamics, which helps in efficient management of the SARS-CoV-2 spread.
Keywords: Sewage, COVID-19, Inactivation, RNA, RT-PCR, Infectivity
Graphical abstract
1. Introduction
The surveillance of disease prevalence during pandemic like Coronavirus Disease-19 (COVID-19) is a crucial task considering the spreading rate and high population in different parts of the world. The massive testing of the population to contain the spread of the virus is a challenge. Moreover, the problem is further compounded because a majority of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infected individuals are asymptomatic. Emerging studies have shown the after effects of COVID-19 is going to be a huge economic burden globally and therefore pressing the importance of not only managing the infected individuals but also to keep a check on the spread (McKibbin and Fernando, 2020). Asymptomatic and symptomatic infections result in significant uncertainty in the estimated extent of SARS-CoV-2 infection (Li et al., 2020). Considering the present testing capacity and cost incurred, it is impractical to test all the individuals. Thus, there is a need for alternative strategies to assess the disease spread and therefore efficiently allocate resources for disease management.
Even though SARS-CoV-2 is majorly a respiratory pathogen, the persistence and replication of virus in the gastrointestinal (GI) tract and shedding through faeces is established (Wang et al., 2020a, Wang et al., 2020b; Xiao et al., 2020a, Xiao et al., 2020b; Zhang et al., 2020; Young et al., 2020; Woelfel et al., 2020; Venkata Mohan et al., 2021). Different independent studies highlighted the presence and replication of SARS-CoV-2 in GI tract and the prolonged shedding of SARS-CoV-2 viral material through faeces during and after active infectious stage (Woelfel et al., 2020; Holshue et al., 2020; Kitajima et al., 2020; Cai et al., 2020; Ling et al., 2020; Wu et al., 2020; La Rosa et al., 2020a; Xiao et al., 2020a, Xiao et al., 2020b; Ahmed et al., 2020a; Wurtzer et al., 2020; La Rosa et al., 2020b).
In this scenario, wastewater-based epidemiology (WBE) studies are suitable to understand and estimate the virus spread in a given population for effective disease surveillance. WBE was earlier used to detect and manage viral diseases such as polio, rotavirus, noroviruses etc. (Ahmed et al., 2020a; Usman et al., 2020; Murakami et al., 2020; Lodder and de RodaHusman, 2020; Hata and Honda, 2020; Venkata Mohan et al., 2021). Recent reports employed WBE-based approaches to detect SARS-CoV-2 in sewage water and estimated the percentage of infected individuals in a given population (Wu et al., 2020; Ahmed et al., 2020a; Wurtzer et al., 2020; La Rosa et al., 2020b; Medema et al., 2020; Usman et al., 2020). Wastewater offers an aggregate sample representing an entire community and is more easily accessible than pooled clinical samples (Murakami et al., 2020). The monitoring of SARS-CoV-2 in wastewater could quantify the scale of infection prevailing among the community with a benefit of detecting virus from symptomatic, asymptomatic, or pre-symptomatic cases which manifest as an early-warning signal (Medema et al., 2020; Lodder and de RodaHusman, 2020; Hata and Honda, 2020; Mallapaty, 2020; Naddeo and Liu, 2020; Qu et al., 2020). WBE approach has the potential to minimize the outbreak spread and also serve as an alarm for future outbreaks (Daughton, 2018; Mao et al., 2020; Zhou et al., 2020).
Here we have studied the spread of SARS-CoV-2 infection in Hyderabad, one of the major and densely populated metropolitan cities of India. The wastewater infrastructure of the city was used as an effective resource to access and estimate the spread of SARS-CoV-2 across the city of Hyderabad. We have simplified the sample processing method for viral detection by RT-PCR. A minimum estimate of the number of infected individuals was calculated based on the concentration of SARS-CoV-2 RNA in wastewater.
2. Materials and methods
2.1. Sampling sites
The detection of SARS-CoV-2 genetic material in domestic sewage was performed by collecting samples from different sewage treatment plants (STPs) in Hyderabad Metropolitan City, India. Hyderabad (17.37°N 78.48°E) is fifth-largest urban economy in India and is the capital of Telangana state that is spread over ~625 km2. It is the fourth-most populous city in India with 10 million residents in the metropolitan region. The raw sewage samples were collected from inlet and outlet points from the STPs with a total coverage of 603.5 million litres per day (MLD) out of 735 MLD, that receive wastewater from all parts of the city (80% coverage of the existing STPs).
2.2. Sampling procedure
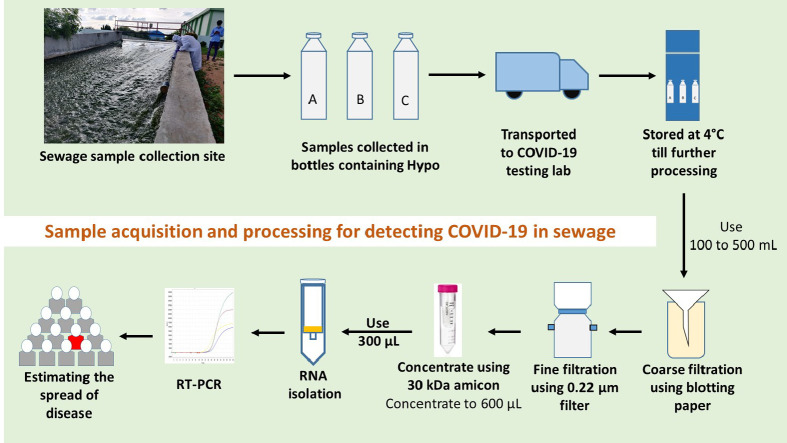
The sewage samples were collected from 8th July 2020 to 6th August 2020 (Table 1 ) taking all the safety measures as per the standard operating procedure (SOP) designed for this purpose. A total of 30 samples were collected from 14 inlets (equalization tanks outlet (ET)) and 14 treated wastewaters (outlets of secondary clarifiers (SC)) of 10 STPs and 2 samples from a gated community (outlet of collection tank prior to disposing to drains). A 10 MLD STP was selected for a time course study to understand the weekly variation in the viral load, where weekly samples were collected and analysed. The basis for sample collection from ET/SC of STP is that it would provide a composite sample accounting for a period of retention time (1–5 h). Grab sampling protocol (Rimoldi et al., 2020) was employed for sampling 1 L of sewage in a disposable bottle (plastic) of 1 L capacity with 20 mL of 0.1% of sodium hypochlorite (NaOCl)/L to inactivate the pathogens. After sampling the surface of the sample container was disinfected with 90% ethanol and sealed in multi-layered plastic covers, labelled and transported (2–4 °C) immediately to lab prior to storing at 4 °C until further processing. All the samples were processed within 12 h of the sampling event unless mentioned otherwise. During sampling, care was taken to follow all biosafety protocols. All the sampling activities were performed during the daytime when peak load was available (8 am to 4 pm), on the days with no report of rainfall events during last 24 h.
Table 1.
Sampling information: Sewage samples collected from various STP of Hyderabad Metropolitan City. ET=equalization tank; SC=secondary clarifier.
| Date of Collection | Capacity of STP | Sample Collection Point | Sample ID |
|---|---|---|---|
| 08-07-2020 | 10 MLD (W1) | ET | ET-1 |
| SC | SC-1 | ||
| 14-07-2020 | 20 MLD | ET | ET-2 |
| SC | SC-2 | ||
| 30 MLD | ET | ET-3 | |
| SC | SC-3 | ||
| 10 MLD (W2) | ET | ET-4 | |
| SC | SC-4 | ||
| 29-07-2020 | 10 MLD (W3) | ET | ET-5 |
| SC | SC-5 | ||
| 06-08-2020 | 339 MLD | ET | ET-6 |
| SC | SC-6 | ||
| 2.5 MLD | ET | ET-7 | |
| SC | SC-7 | ||
| 172 MLD | ET | ET-8 | |
| SC | SC-8 | ||
| 30 MLD | ET | ET-9 | |
| SC | SC-9 |
2.3. Optimization of disinfectant concentration
Optimum concentration of sodium hypochlorite (NaOCl) (Qualigens) addition during the sampling was optimized by various initial concentrations of sodium hypochlorite (0.1%, 0.5%, 1%, 2%, 3% and 4%) using samples collected from 10 MLD STP. Grab samples were collected from the ET outlet point of 10 MLD STP in disposable plastic bottles containing 20 ml of the above-mentioned concentrations of sodium hypochlorite. Collected samples were sealed and wrapped in plastic covers in two layers and transferred to the laboratory immediately and stored at 4 °C. Samples were processed within 12 h of sampling for the detection of SARS-CoV-2 RNA.
2.4. Processing of samples
Collected samples were subjected to gravity filtration with 1 mm thick blotting sheets to remove the debris or larger particles followed by filtration using 0.2 μm filtration units (Nalgene® vacuum filtration system; Thermofisher Scientific) to remove bacteria and other particles/debris. The filtrate was collected in 1000 mL sterile wide-mouth bottles (Borosil). 100 mL of the total filtrate was concentrated to ~600 μL using 15 mL 30 kDa Amicon® Ultra-15 (Merck Millipore) by centrifugation at 4000 rpm (4 °C; 10 min). The concentrated samples were further processed for RNA isolation. Sample filtration, concentration and processing till detection were performed in a Biosafety level 2 (BSL-2) facility. All the materials after use were discarded in biosafety bags followed by decontamination.
2.5. RNA extraction and RT-PCR
To quantify SARS-CoV-2 RNA in sewage samples, a total of 300 μl concentrate was used for RNA extraction using QIAamp Viral RNA isolation kit (Qiagen, Germany) by following manufacturer's protocol. The isolated RNA samples were tested for presence of SARS-CoV-2 RNA using FDA (Food and Drug Administration, USA Government) approved Fosun COVID-19 RT-PCR Detection Kit (Shanghai Fosun Long March Medical Science Co., Ltd., China) (https://www.fda.gov/media/137120/download, n.d). It contains primers and probes which targets the envelope protein coding gene (E-gene; ROX labelled), nucleocapsid gene (N-gene; JOE labelled) and open reading frame1ab (ORF1ab; FAM labelled) of SARS-CoV-2 and the RT-PCR was performed as per manufacturer recommendation on QuantStudio™5. Reaction conditions include two initial cycles, one at 50 °C for 15 min (Reverse transcription) and the other at 95 °C for 3 min (Initial denaturation) followed by 45 cycles at 95 °C for 5 s and 60 °C for 40 s (Initial 5 cycles without data acquisition and 40 with data collection). The signals of FAM (ORF1ab), JOE (N gene), ROX (E gene), and CY5 (Internal reference) fluorescence channels were collected at 60 °C. Positive and negative controls provided in the Fosun RT-PCR kit were also included in the amplification plates, and the CT values were in accordance with the manufacturer protocol proving to be efficient and devoid of contamination. All the samples were tested in triplicates.
2.6. Estimation of RT-PCR kit efficiency
To assess the performance and efficiency of the qRT-PCR kit used in this work, 2.14*107 pfu/mL viral culture was inactivated at 55 °C for 30 min and provided to us by Dr. H H Krishnan, CSIR-CCMB. RNA was isolated from the heat-inactivated SARS-CoV-2 which was followed by the preparation of log10 dilutions of the RNA. RT-PCR was performed in triplicates for each dilution. The R2 values obtained from linear regression and efficiency was calculated as described (Ginzinger, 2002).
2.7. Standard curve for copy number calculation
To calculate the number of RNA copies, present in the wastewater samples, the E gene amplified from the SARS-CoV-2 RNA was cloned into the vector pcDNA3.1 between KpnI and HindIII restriction sites. The cloned plasmid was then quantified using Qubit™ dsDNA HS Assay Kit (Invitrogen, USA) and Qubit™ 4 Fluorometer (Invitrogen, USA). The copy number per nanogram was calculated using the E gene and vector sequences retrieved from https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2?report=fasta&from=26245&to=26472 and https://www.addgene.org/browse/sequence_vdb/2093/, respectively. The plasmid was serially diluted from 9.01 log10 copies to 0.01 log10 copies and the RT-PCR reaction was performed as mentioned in Section 2.5 in triplicates. The CT values were plotted against the log copy number and a linear fit equation was obtained (Supplementary Table 3; Supplementary Fig. 1).
2.8. Virus recovery from sewage
To find out the recovery of SARS-CoV-2 from sewage samples, 1 mL of 2.14 × 107 pfu/mL SARS-CoV-2 virus (heat-inactivated) was added to 100 mL of sewage water and four log10 dilutions were prepared (with sewage water). As a control, similar dilutions were made using milliQ water (MQ). The RNA was isolated from all the samples and RT-PCR was performed in triplicates.
2.9. Calculation of number of infected people in a population
To identify the viral copy number present in the wastewater samples, the linear fit equation obtained from the standard curve for E-gene was used (Supplementary Fig. 1). We followed two different methods to calculate the number of infected individuals in a given population based on the average number of RNA copies present in the sewage water.
Method 1 (Ahmed et al., 2020a):
Faeces excreted/person/day = 128 g. (Rose et al., 2015).
One positive person sheds 107 RNA copies/g of faeces (maximum estimate, Foladori et al., 2020, Bivins et al., 2020).
Method 2 (Hellmér et al., 2014).
Number of RNA copies excreted per mL of faeces = 107 .
Volume of faeces excreted = 120 mL (calculated by considering the density of human faeces is 1.07 g/mL (Foladori et al., 2020).
3. Results and discussion
3.1. Determining the RT-PCR kit efficiency
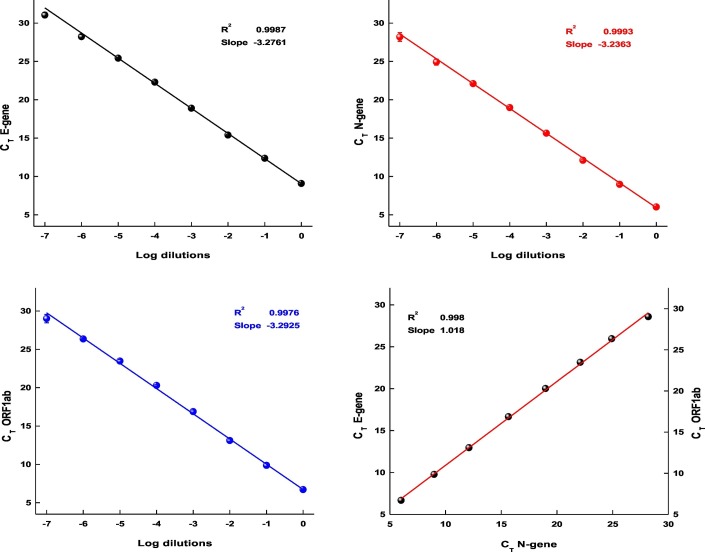
Earlier studies have shown that the regular SARS-CoV-2 kits which are used for testing individuals can be employed for the detection and quantification of the viral RNA from sewage (Wu et al., 2020; Rimoldi et al., 2020; Or et al., 2020). To independently examine the efficiency of the RT-PCR kit used in this study, we isolated RNA from 150 μl of 2.14 × 107 pfu/mL virus culture and subjected the RNA to log10 serial dilutions. The CT values and standard deviations (from triplicates) obtained from dilutions up to 10−7 are presented in Fig. 1 . The observed average CT difference of 3.2, between log10 dilutions for all three genes provides further proof for the performance of the kit and its usefulness to estimate the number of viral RNA molecules (Table 2 ). Based on the CT values a linear curve was plotted and subjected to regression analysis, where R2 0.9987, 0.9993, and 0.9976 and the calculated efficiency of 106.55%, 105.59%, and 103.01% for E-gene, N-gene and ORF1ab were noted respectively. Overall, the average slope of −3.2070 and R2 of 0.9985 obtained from linear regression with the calculated efficiency of 105.05% suggests the standard performance (nearly 100%) of the kit.
Fig. 1.
The standard curves of a) E- gene, b) N-gene and c) ORF1ab gene, plotted with CT values from log10 dilutions of SARS-CoV-2 RNA. d) Plot representing the linear fit of all three genes CT values.
Table 2.
The CT values of viral-specific genes from log10 dilution samples.
| Dilution | E Gene |
N gene |
ORF1ab |
|||
|---|---|---|---|---|---|---|
| Average Ct | SDa | Average Ct | SDa | Average Ct | SDa | |
| Undiluted | 9.08 | 0.10 | 6.01 | 0.10 | 6.71 | 0.03 |
| 10−1 | 12.36 | 0.10 | 8.97 | 0.12 | 9.86 | 0.31 |
| 10−2 | 15.40 | 0.08 | 12.10 | 0.16 | 13.10 | 0.20 |
| 10−3 | 18.89 | 0.03 | 15.64 | 0.03 | 16.87 | 0.22 |
| 10−4 | 22.30 | 0.14 | 18.97 | 0.16 | 20.29 | 0.11 |
| 10−5 | 25.43 | 0.01 | 22.10 | 0.03 | 23.48 | 0.21 |
| 10−6 | 28.23 | 0.28 | 24.92 | 0.38 | 26.35 | 0.07 |
| 10−7 | 31.06 | 0.33 | 28.20 | 0.57 | 29.01 | 0.54 |
| Efficiency (%) | 106.55 | 105.59 | 103.01 | |||
| Slope | −3.1744 | −3.1947 | −3.2519 | |||
| R2 | 0.9987 | 0.9993 | 0.9976 |
Standard deviation.
Fig. 1d, with CT values (log10 dilutions) of all three genes plotted together indicates the acceptable efficiency of all primer sets used in the RT-PCR. The R2 value 0.998 and slope 1.018 represents (Fig. 1d) a good linear regression fit and concordance among the primers performance in the given experimental conditions.
3.2. Recovery of SARS-CoV-2 RNA from sewage
One of the major bottlenecks in analysing sewage samples is to estimate the recovery of the samples compared to the actual presence. Earlier studies have shown that processing sewage samples by ultra-filtration followed by concentration lead to 70% loss of samples and therefore making the recovery to be only around 30%. (Ahmed et al., 2020b). To check the virus recovery and efficiency of the method implemented in this work, we performed the log10 dilutions from 100 ml of sewage water which was spiked with 1 ml of 2.14 × 107 pfu/mL SARS-CoV-2 virus. For comparison, similar dilutions were performed with MQ water to check the effect of sewage on virus recovery. The RNA was isolated from independently processed samples and RT-PCR was performed. The CT values from different dilutions and samples are presented (Table 3 ). The average CT value differences (all three genes) of 3.72 and 3.61 for sewage and MQ water dilutions, respectively, suggests efficient recovery from log dilution samples. A CT value difference of 3.74 between identical log10 dilutions of spiked sewage and MQ samples were observed, which is possible due to inhibitors in sewage and this difference largely depends on heterogeneity among different samples. To eliminate the unwanted large particles from sewage samples, which could damage the filter membrane we implemented prior filtration of samples using a blotting sheet. To rule out the effect of this, we processed two spiked samples with and without the extra filtration step. The average CT difference of 0.02 indicates the absence of unwanted effect due to blotting sheet filtration.
Table 3.
The CT values obtained from log10 dilutions of spiked sewage and MQ water samples.
| Sample | E gene |
N gene |
ORF1ab |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sewage |
MQ |
Sewage |
MQ |
Sewage |
MQ |
|||||||
| Average CT | SD⁎ | Average CT | SD⁎ | Average CT | SD⁎ | Average CT | SD⁎ | Average CT | SD⁎ | Average CT | SD⁎ | |
| UD+ | 11.29 | 0.41 | 8.50 | 0.04 | 8.13 | 0.44 | 5.44 | 0.03 | 9.74 | 0.49 | 6.32 | 0.08 |
| 1/10 | 17.02 | 0.35 | 12.96 | 0.04 | 14.02 | 0.34 | 9.61 | 0.18 | 16.25 | 0.37 | 11.48 | 0.08 |
| 1/100 | 19.73 | 0.30 | 15.06 | 0.08 | 16.33 | 0.45 | 11.85 | 0.20 | 18.73 | 0.39 | 13.55 | 0.09 |
| 1/1000 | 22.87 | 0.11 | 19.99 | 0.15 | 19.65 | 0.20 | 16.43 | 0.25 | 21.75 | 0.28 | 18.48 | 0.17 |
| 1/10000 | 25.88 | 0.47 | 22.79 | 0.13 | 22.95 | 0.48 | 19.48 | 0.04 | 24.97 | 0.50 | 21.35 | 0.13 |
|
Filtered (0.22 μm) |
11.15 | 0.14 | 8.36 | 0.18 | 9.58 | 0.51 | ||||||
Standard deviation; + Undiluted
To further understand the efficiency of viral recovery from both sewage and MQ dilutions, we performed a standard curve from CT values obtained. The percentage efficiency (94.85), slope (−3.45), and R2 values (0.96) achieved (from sewage) indicates a good recovery efficiency of the viral particles (Table 4 ).
Table 4.
Calculated values obtained from CT values of log10 dilutions of spiked sewage and MQ water samples.
| Gene | Efficiency (%) |
Slope |
R2 value |
|||
|---|---|---|---|---|---|---|
| Sewage | MQ | Sewage | MQ | Sewage | MQ | |
| E gene | 94.54 | 89.47 | −3.4601 | −3.6030 | 0.9750 | 0.9685 |
| N gene | 95.87 | 92.71 | −3.4251 | −3.5099 | 0.9657 | 0.9995 |
| ORF1ab | 94.13 | 87.11 | −3.4710 | −3.6752 | 0.9465 | 0.9711 |
| Average | 94.85 | 89.76 | −3.45 | −3.60 | 0.96 | 0.98 |
3.3. Optimization of disinfection concentration and storage
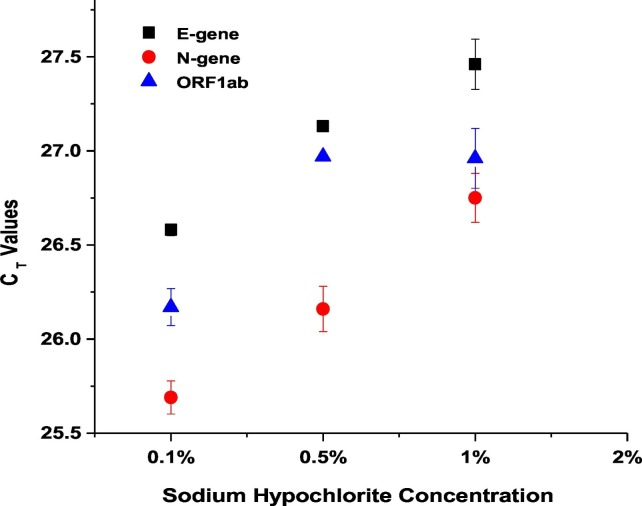
Sodium hypochlorite was used to disinfect the sewage samples collected from the STPs in order to reduce the possible pathogenicity of virus/bacteria during transport and processing as the scope of the study is only to evaluate the SARS-CoV-2 RNA. In order to find the least concentration of sodium hypochlorite that result in identifying maximum number of RNA copies using RT-PCR, we performed an optimisation step. We collected 1 L of real-field wastewater (from the 10 MLD STP) in the presence of 20 mL of six different initial concentrations of sodium hypochlorite (Supplementary Table 1). From the results it was observed that three concentrations of hypochlorite (0.1%, 0.5% and 1%) were positive for all the three genes E-gene, N-gene and ORF1ab of SARS-CoV-2 with a minimum difference of ~1 CT. Among the tested concentrations, samples collected with 20 ml of 0.1% hypochlorite resulted in better detection of SARS-CoV-2 RNA. It was also observed that the final concentration of ≥0.04% (initial concentration of ≥2%) did not yield any result for the same set of samples, indicating a possible complete inactivation of viral genetic material by sodium hypochlorite (Supplementary Table 1; Fig. 2 ). The results suggest that addition of 0.1% sodium hypochlorite did not affected the presence of viral RNA. Hence, we used 20 mL of 0.1% sodium hypochlorite/L of wastewater for all further samples collected due to the safety during transport. The earlier reports on SARS reported the complete inactivation of virus with ≥0.5 mg/L of free chlorine (FC) within 30 min of contact time at 22 ± 3 °C (Wang et al., 2005). Hospital based wastewater was detected to be positive to SARS-CoV-2 RNA even after the addition of sodium hypochlorite (Wang et al., 2020a, Wang et al., 2020b; Kataki et al., 2020).
Fig. 2.
Concentration of sodium hypochlorite affects the detection ability of SARS-CoV-2 RNA. Scatter plot showing the effect of different concentrations of sodium hypochlorite on the CT values of viral targets. Each dot represents average CT values obtained from two replicates and the bar represents the standard error of mean.
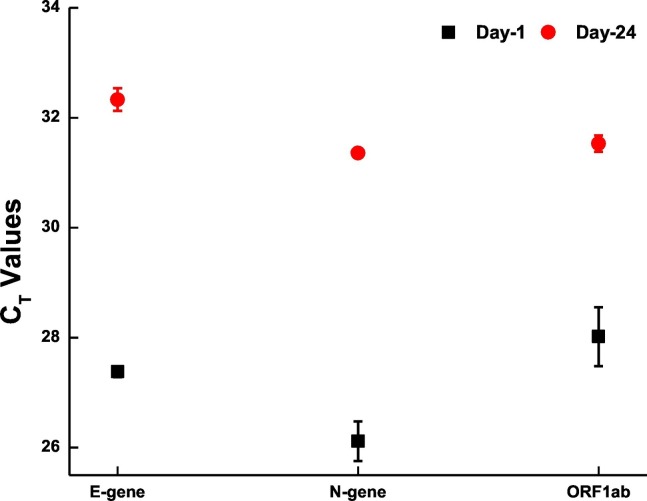
We also looked for the effect of sample storage on the detection of SARS-CoV-2 RNA from wastewater samples. For this, part of the sample collected on 7th July 2020 was filtered and processed within 24 h of collection and the rest (filtered) was stored at 4 °C. The stored sample was processed on 31st July 2020 (after 24 days of initial collection). RT-PCR showed a difference of approximately 4 CT values between the samples with the stored sample showing higher CT (Fig. 3 ). This indicates the presence of viral genome even after 3 weeks when the samples are stored at 4 °C, however, it is best to analysed the samples before 24 h for all practical purpose.
Fig. 3.
Degradation of Viral genome with time. Scatter plot showing the degradation of viral genome with the span of 24 days. Each dot represents average CT values obtained from two replicates and the bar represents the standard error of mean.
3.4. Detection of SARS-CoV-2 in various STPs and residential community
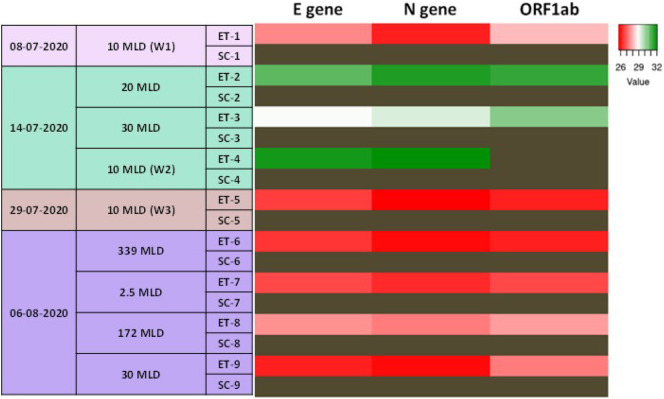
RT-PCR based detection of SARS-CoV-2 RNA was used for screening the inlet water in the STPs that cover about 80% of Hyderabad's STP capacity i.e. 603.5 MLD. SARS-CoV-2 RNA was detected in the inlets of all the tested STPs (Supplementary Table 2), indicating that the infection is widespread. We observed that the level of viral RNA in the STPs was dynamic, as implied by the changes in CT values of the samples collected on different days. As a testimony of efficient wastewater treatment, no viral RNA copies were detected in the outlet of the STPs that we sampled (Fig. 4 ; Supplementary Table 2). We also surveyed samples collected from a gated residential community where confirmed positive cases were reported during the sample collection period and observed the presence of SARS-CoV-2 RNA in the samples (Table 5 ).
Fig. 4.
SARS-CoV-2 RNA is present in Hyderabad's sewage water: Heat map showing the CT values of E gene, N gene, and ORF1ab in the wastewater samples collected from various STPs in the city of Hyderabad on different days during the pandemic. The experiments were performed in duplicates or triplicates. Dark brown cells correspond to samples with no amplification. MLD-Million Litres per Day; ET-Equalization Tank; SC-Secondary Clarifier. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 5.
Community surveillance: SARS-CoV-2 RNA CT values detected by real-time RT-PCR from raw sewage samples of selected gated community. n.d.=not detected; PC=positive control; NTC=no template control.
| Date of Collection | Capacity of STP | Sample ID | CT Values |
||
|---|---|---|---|---|---|
| E gene | N gene | ORF1ab | |||
| 14-07-2020 | 72 Houses | C1 | 31.87±0.317 | 31.93±0.453 | 31.65±0.570 |
| C2 | 31.06±1.047 | 32.56 | 33.32 | ||
| PC | 23.98 | 25.20 | 23.80 | ||
| NTC | n.d. | n.d. | n.d. | ||
3.5. Long term viral load monitoring
One of the STPs (10 MLD), was sampled at different time to assess the dynamics of disease spread with time. We observed a highly fluctuating pattern of viral RNA presence with time, from as low as 661 copies/L wastewater (on 14-07-2020) to as high as 24,469 copies/L wastewater (on 29-07-2020) (Table 6 ; Supplementary Table 3; Supplementary Fig. 1). The reason for variations could be sampling time, number of actually infected people and the amount of viral shedding by infected individuals, and temporal presence of other compounds (such as surfactants) that could affect the viral material stability in the domestic sewage.
Table 6.
Temporal SARS-CoV-2 monitoring: SARS-CoV-2 RNA CT values detected by real-time RT-PCR of raw sewage samples of selected STP (10MLD) for weekly monitoring. ET=equalization tank; SC=secondary clarifier; PC=positive control; NTC=no template control; n.d.=not detected; RNA copies were calculated based on the equation obtained from the standard curve (Supplementary Figure 1).
| Date of Collection | Week | Sample ID | CT Values – E gene | Average number of RNA copies/L water |
|---|---|---|---|---|
| 08-07-2020 | W1 | ET-1 | 27.38±0.098 | 13,964 |
| SC-1 | n.d. | |||
| PC | 23.00 | |||
| NTC | n.d. | |||
| 14-07-2020 | W2 | ET-4 | 31.73±0.388 | 661 |
| SC-4 | n.d. | |||
| PC | 23.98 | |||
| NTC | n.d. | |||
| 29-07-2020 | W3 | ET-5 | 26.58 | 24,469 |
| SC-5 | n.d. | |||
| PC | 25.94 | |||
| NTC | n.d. |
3.6. Calculation of number of infected people
We used two previously published methods for calculating the number of infected people from the number of RNA copies in the wastewater samples (Ahmed et al., 2020a; Hellmér et al., 2014). These methods take into account the number of RNA copies present in the wastewater and the number of RNA copies present in the faecal matter of infected individuals (Section 2.8). Previous studies have established these numbers and we used them for calculating the number of infected individuals. Existing reports suggest that an infected individual shed viral material in faeces for up to 47 days since the symptom onset and remains infectious till 14 days since symptom onset (Wu et al., 2020; Foladori et al., 2020). This suggests that for approximately 35 days, a person sheds viral material while not being infectious. This indicates that 2 in 5 infected people are infectious at any given point of time during the 30 days window. We used this fact to calculate the number of infected people in the active phase of infection (Table 7, Table 8 ). Owing to the uncertainty and difference in the number of viral particles excreted by infected individuals, we calculated the possible number of infected people for three different shedding rates within the reported range (105, 106, and 107 copies/mL faeces) (Table 8). Results indicate that the number of infected people might be anywhere between thirty thousand and three million during the study period (Table 8). Studies have reported the loss of 0.02 to 3000 viral RNA copies/mL during the transit of faeces from the point of excretion to the STP (Foladori et al., 2020). This could further influence the correct estimation of the number of infected individuals. Resampling from these sites periodically would give a better estimate to understand where the disease spread rate is decreasing or increasing with time. In addition, this study puts forth the necessity for large-scale studies on the excretion dynamics of viral particles by infected individuals which could help in estimating the near-precise number of infected individuals in a given locality.
Table 7.
Estimated number of RNA copies per litre of wastewater processed in each of the STPs; RNA copies were calculated based on the equation obtained from the standard curve (Supplementary Figure 1).
| Date of Collection | Capacity of the STP (MLD) | E Gene |
|
|---|---|---|---|
| Average CT | RNA copies/1 L | ||
| 08-07-2020 | 10 | 27.38 | 13,964 |
| 14-07-2020 | 20 | 30.83 | 1243 |
| 30 | 28.94 | 4677 | |
| 10 | 31.73 | 661 | |
| 29-07-2020 | 10 | 26.58 | 24,469 |
| 06-08-2020 | 339 | 26.4 | 27,760 |
| 2.5 | 26.64 | 23,461 | |
| 172 | 27.52 | 12,658 | |
| 30 | 26.12 | 33,782 | |
Table 8.
Disease dynamics: Estimate of the number of people infected during the sampling window, which includes individuals who are symptomatic, asymptomatic, and recovered.
| Capacity of the STP (MLD) | Per person contribution to STP (107 copies/mL faeces) | Method 1 | Method 2 | Per person contribution to STP (106 copies/mL faeces) | Method 1 | Method 2 | Per person contribution to STP (105 copies/mL faeces) | Method 1 | Method 2 |
|---|---|---|---|---|---|---|---|---|---|
| 10 | 120 | 109 | 116 | 12 | 1091 | 1164 | 1.20 | 10,909 | 11,636 |
| 20 | 60 | 19 | 21 | 6 | 194 | 207 | 0.60 | 1942 | 2071 |
| 30 | 40 | 110 | 117 | 4 | 1096 | 1169 | 0.40 | 10,961 | 11,692 |
| 10 | 120 | 5 | 6 | 12 | 52 | 55 | 1.20 | 517 | 551 |
| 10 | 120 | 191 | 204 | 12 | 1912 | 2039 | 1.20 | 19,116 | 20,391 |
| 339 | 4 | 7352 | 7842 | 0.35 | 73,521 | 78,423 | 0.04 | 735,213 | 784,227 |
| 2.5 | 480 | 46 | 49 | 48 | 458 | 489 | 4.80 | 4582 | 4888 |
| 172 | 7 | 1701 | 1814 | 0.7 | 17,009 | 18,143 | 0.07 | 170,092 | 181,432 |
| 30 | 40 | 792 | 845 | 4 | 7918 | 8446 | 0.40 | 79,177 | 84,456 |
| Total (603.5 MLD) | 10,325 | 11,013 | Total (603.5 MLD) | 103,251 | 110,134 | Total (603.5 MLD) | 1,032,510 | 1,101,344 | |
| Total (1800 MLD) | 30,796 | 32,849 | Total (1800 MLD) | 307,956 | 328,487 | Total (1800 MLD) | 3,079,565 | 3,284,869 | |
| Average estimate of infected individuals | 31,822 | 318,222 | 3,182,217 | ||||||
| Estimate of the population in active phase of the infection | 12,729 | 127,289 | 1,272,887 | ||||||
Based on the recent learning from SARS-CoV-2, it is evident that screening large population to contain the spread is an inconceivable task and it is more complex in urban areas with high population density. As the SARS-CoV-2 colonise the GI tract and is released through faeces the WBE studies provide an effective edge for mass surveillance to prevent the spread of virus. The work presented here covered nearly 80% of STPs capacity (603.5 MLD) in the metropolitan city Hyderabad, India, for the detection and estimation of SARS-CoV-2 infected individuals in a window of 30 days. Based on number of viral RNA copies present in the sewage samples collected from different locations, here, we clearly estimated a range of the number of infected individuals and the actively spreading population during the given time window. The estimations were done based on published independent studies and WHO guidelines (https://www.who.int/news-room/commentaries/detail/criteria-for-releasing-covid-19-patients-from-isolation, n.d). pertaining to SARS-CoV-2 infected individuals. The wastewater infrastructure has been previously shown to function as a surveillance system for poliovirus (Lodder et al., 2012). and Aichi virus (Lodder et al., 2013). WBE approach, apart from helping to minimize the existing outbreak spread, can also serve as a tool for future epidemics surveillance (Lodder and de RodaHusman, 2020; Mallapaty, 2020; Daughton, 2018). Considering the present and previous reports on SARS-CoV-2 WBE studies, we recommend collection of sewage samples with a window of 15 days from same localities to get a better estimation of cases.
This study provides a concrete evidence for the application of WBE as a potential method for disease as well as environmental surveillance. These results will be an immense resource for the healthcare and associated departments to vigilantly allocate the necessary resources to manage existing cases as well as to carefully contain the disease spread. Along with clinical data, WBE could provide critical monitoring of SARS-CoV-2 transmission within a community including the beginning, tapering, or reemergence of an epidemic (Bivins et al., 2020. Hence, sewage-based surveillance provides a holistic approach to manage the pandemic and also to monitor for future outbreaks, if any. The current study also offers a framework to monitor other pathogens to avoid future epidemic. Overall, this study provides a simplistic framework for WBE studies with basic resources available in majority of the labs towards sustainable environmental Surveillance. We strongly recommend the scientific community and the healthcare agencies to pursue similar studies periodically, for allocating resources appropriately to fight the pandemic.
CRediT authorship contribution statement
Manupati Hemalatha: Methodology, Investigation, Data curation, Writing – original draft. Uday Kiran: Methodology, Investigation, Data curation, Writing – original draft. Santosh Kumar Kuncha: Conceptualization, Investigation, Writing – original draft. Harishankar Kopperi: Methodology, Investigation, Writing – original draft. C.G. Gokulan: Methodology, Investigation, Formal analysis, Writing – original draft. S. Venkata Mohan: Conceptualization, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. Rakesh K. Mishra: Conceptualization, Supervision, Funding acquisition, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge the Officials of Hyderabad Metropolitan Water Supply and Sewerage Board (HMWSSB) and Hyderabad Metropolitan Development Authority (HMDA), Government of Telangana for providing the samples for this study. The work was supported by Council of Scientific and Industrial Research (CSIR), New Delhi, India in the form of project entitled ‘Testing for COVID-19 in wastewater as a community surveillance measure (6/1/COVID-19/2020/IMD)’. UK thanks UGC, CGG and MH thank CSIR for the financial support received. MH, KH, SVM acknowledge the Director, CSIR-IICT for the support. The authors thank Dr. H. H. Krishnan, Divya Gupta, and Dixit Kumar Tandel from CSIR-CCMB for providing the inactivated SARS-CoV-2 viral samples.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.144704.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J.W., Choi, P.M., Kitajima, M., Simpson, S.L., Li, J., Tscharke, B. 2020a. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 138764. [DOI] [PMC free article] [PubMed]
- Ahmed, W., Bertsch, P., Bivins, A., Bibby, K., Farkas, K., Gathercole, A., Haramoto, E., Gyawali, P., Korajkic, A., McMinn, B.R. and Mueller, J., 2020b. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 139960. [DOI] [PMC free article] [PubMed]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Xu L., Qu Z., Zhang Y., Zhang H., Jia R., Wang X., Ge Y., Xia A. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Monitoring wastewater for assessing community health: sewage chemical-information mining (SCIM) Sci. Total Environ. 2018;619-620:748–764. doi: 10.1016/j.scitotenv.2017.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;140444 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger D.G. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 2002;30(6):503–512. doi: 10.1016/s0301-472x(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Hata A., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with norovirus cases. Environ. Sci. Technol. 2020;54:6451–6452. doi: 10.1021/acs.est.0c02271. [DOI] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80(21):6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.fda.gov/media/137120/download
- https://www.who.int/news-room/commentaries/detail/criteria-for-releasing-covid-19-patients-from-isolation
- Kataki S., Chatterjee S., Vairale M.G., Sharma S., Dwivedi S.K. Concerns and strategies for wastewater treatment during COVID-19 pandemic to stop plausible transmission. Resour. Conserv. Recycl. 2020;105156 doi: 10.1016/j.resconrec.2020.105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;139076 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods-a scoping review. Water Res. 2020;115899 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, G., Iaconelli, M., Mancini, P., Ferraro, G.B., Veneri, C., Bonadonna, L., Lucentini, L., Suffredini, E. 2020b. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 139652. [DOI] [PMC free article] [PubMed]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Sci. 2020;3221 doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de RodaHusman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Buisman A.M., Rutjes S.A., Heijne J.C., Teunis P.F., de RodaHusman A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012;78(11):3800–3805. doi: 10.1128/AEM.07972-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W.J., Rutjes S.A., Takumi K., de RodaHusman A.M. Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 2013;19(8):1222. doi: 10.3201/eid1908.130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580(7802):176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- McKibbin, W. J., Fernando, R. 2020. The global macroeconomic impacts of COVID-19. Brookings Institute, no. March. 1-43.
- Medema G., Heijnen L., Elsinga G., Italiaander R.A., Brouwer Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Murakami M., Hata A., Honda R., Watanabe T. Letter to the editor: wastewater-based epidemiology can overcome representativeness and stigma issues related to COVID-19. Environ. Sci. Technol. 2020;20045880 doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6(5):1213–1216. [Google Scholar]
- Or I.B., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shi-razi R., Kramarsky-Winter E., Nir O., Abu-Ali O., Ronen Z., Rinott E., Lewis Y.E., Friedler E.F., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in thepopulation: a proof-of-concept for quantitative environmental surveillance. MedRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G., Li X., Hu L., Jiang G. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) Environ. Sci. Technol. 2020;3730-3732 doi: 10.1021/acs.est.0c01102. [DOI] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Critical reviews in environ. Sci. Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman M., Farooq M., Hanna K. Existence of SARS-CoV-2 in wastewater: implications for its environmental transmission in developing communities. Environ. Sci. Technol. 2020;54:7758–7759. doi: 10.1021/acs.est.0c02777. [DOI] [PubMed] [Google Scholar]
- Venkata Mohan S., Hemalatha M., Kopperi H., Ranjith I., Kumar A.K. SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021;405:126893. doi: 10.1016/j.cej.2020.126893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shen J., Ye D., Yan X., Zhang Y., Yang W., Li X., Wang J., Zhang L., Pan L. Disinfection technology of hospital wastes and wastewater: suggestions for disinfection strategy during coronavirus disease 2019 (COVID-19) pandemic in China. Environ. Pollut. 2020;114665 doi: 10.1016/j.envpol.2020.114665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126(1–2):171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. MedRxiv. 2020 doi: 10.1101/2020.03.05.20030502. [DOI] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer, S., Marechal, V., Mouchel, J.M., Maday, Y., Teyssou, R., Richard, E., Almayrac, J.L., Moulin, L. 2020. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. doi: doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroentero. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. Jama. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Cui X., Xiao J., Meng T., Zhou W., Liu J. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. BioRxiv. 2020 doi: 10.1101/2020.01.30.927806. [DOI] [Google Scholar]
- Zhou N.A., Fagnant-Sperati C.S., Komen E., Mwangi B., Mukubi J., Nyangao J., Hassan J., Chepkurui A., Maina C., Van Zyl W.B., et al. Feasibility of the bag-mediated filtration system for environmental surveillance of poliovirus in Kenya. Food Environ. Virol. 2020;12:35–47. doi: 10.1007/s12560-019-09412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material